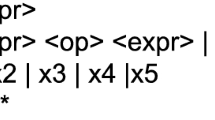Abstract
Sepsis-induced glucose fluctuations present major challenges in critical care, underscoring the importance of accurate glucose monitoring and forecasting to improve patient outcomes. This study introduces a suite of forecasting models trained using continuous glucose monitoring data from a diabetic patient with sepsis (19,621 data points). The models include four transformer-based ones (iTransformer, Crossformer, PatchTST, FEDformer), a dynamic linear model (DLinear), and an ensemble zero-shot inference method leveraging ChatGPT-4. Model performance was evaluated for 15-, 30-, and 60-minute prediction horizons with an optimized 30-minute lookback window. PatchTST achieved the lowest mean maximum percentage error (MMPE) for short-term forecasts (3.0% at 15 minutes), while DLinear excelled at longer horizons (7.46% and 14.41% MMPE at 30 and 60 minutes, respectively). The ensemble ChatGPT-4 approach also showed competitive results. Overall, this work offers a toolbox of advanced forecasting models for ICU glucose prediction and management. The comprehensive comparison among the models highlights the promise of machine learning models–particularly DLinear and PatchTST–in supporting glucose monitoring and ultimately digital twin implementations, paving the way toward personalized and adaptive glycemic control in septic patients.
Similar content being viewed by others
Introduction
Sepsis, a life-threatening condition characterized by a dysregulated host response to infection, remains a major challenge in critical care. As defined by the Third International Consensus Definitions for Sepsis (“Sepsis-3”)49, sepsis involves acute organ dysfunction resulting from systemic inflammation. Its pathophysiology includes immune hyperactivation, widespread inflammation, thrombosis, and microcirculatory disturbances, often culminating in multiple organ dysfunction syndrome (MODS). Despite medical advances, sepsis continues to be a leading cause of mortality among ICU patients worldwide29.
Clinical presentations of sepsis are highly heterogeneous, shaped by factors such as infection site, comorbidities, and timeliness of intervention4. This variability, alongside incomplete understanding of underlying mechanisms, complicates the development of universally effective treatments15,44,45. As a result, efforts have increasingly shifted to individualized interventions; however, no consensus has yet been reached on the optimal treatment strategy12.
One critical aspect of sepsis pathophysiology is its impact on glucose metabolism. In early stages, the stress response triggers increased glycogenolysis and gluconeogenesis, leading to elevated blood glucose levels–a condition known as stress hyperglycemia33,38. This response is mediated by elevated levels of stress hormones such as epinephrine, cortisol, and glucagon, which promote insulin resistance and hepatic glucose output9,10,27,32,58.
Several studies2,53,59 have demonstrated that hyperglycemia in sepsis is associated with worsened outcomes due to heightened inflammation, immune suppression, and prolonged recovery. Furthermore, elevated glucose levels are correlated with increased mortality, reinforcing a vicious cycle of inflammation and metabolic stress42. While glycemic control is a central therapeutic target, the optimal glucose range remains debated. Some advocate for tight regulation26,52, while others caution against the risks of hypoglycemia and endorse more moderate approaches19.
Large-scale clinical trials6,51,52 underscore the importance of precise glucose management in improving sepsis outcomes, emphasizing the need for individualized treatment strategies guided by clinical judgment. Recent developments in artificial intelligence (AI), machine learning (ML), and digital health technologies–including digital twins (DTs) and non-invasive bedside monitoring–offer new opportunities to advance personalized sepsis care18,30.
AI can uncover complex disease patterns and enable predictive modeling based on historical and real-time patient data, thereby supporting clinical decision-making13,54. DT technologies in healthcare construct virtual representations of patients or disease states, integrating domain knowledge, real-time monitoring, and AI/ML-driven models. This enables continuous tracking of disease progression and personalized prediction of treatment responses, laying the foundation for dynamic, adaptive care strategies30.
The integration of AI and DTs holds particular promise for sepsis management, especially in ICU or emergency settings, where time-sensitive interventions are critical. By harnessing data-driven modeling and virtual simulation, clinicians can tailor treatment strategies to individual patients, potentially improving patient outcomes in real time.
Modern AI/ML pipelines have demonstrated significant predictive power in demanding scientific domains, such as civil and hydraulic engineering. For example, CatBoost models accurately predict the seepage parameters for hydraulic structures 17 and substantially improve construction cost estimation 16. Explainable XGBoost and Random Forest models have been applied to forecast saltwater intrusion in aquifers 5 and predict the structural capacity of hybrid columns 62. Furthermore, ensemble classifiers such as LightGBM effectively categorize concrete compressive strength 3. SHapley Additive exPlanations (SHAP) has been adopted in many studies involving neural networks when identifying dominant physical factors and ranking their importance.
In sepsis research, interpretable AI/ML pipelines have also been successfully applied to stratify the risk for sepsis-related outcomes by leveraging large ICU electronic health records (EHRs). These models have successfully predicted sepsis-associated delirium by identifying key clinical predictors 48 and incident sepsis in high-risk cohorts such as ischemic stroke patients 37. Research also highlights the prognostic value of novel biomarkers and laboratory ratios, including the glucose-to-albumin ratio for mortality 21 and metabolism-inflammation indices in pancreatitis 60. Moreover, models for complications such as hypoalbuminemia have demonstrated strong generalizability across cohorts 56. Overall, these findings establish a paradigm of combining disease-specific features with interpretable ML and multi-cohort validation, which guides the present work.
This study extends the ML application to the more demanding challenge of real-time forecasting. To achieve this, a suite of machine learning models is applied to longitudinal data on glucose levels and insulin administration, collected continuously at the patient’s bedside. The main objective is to construct a personalized, AI-driven, predictive toolbox for individual septic patients with hyperglycemia. This toolbox is designed to predict patient-specific glucose trajectories, providing a foundation for real-time feedback and therapeutic adjustment. Once trained, the models in the toolbox can be efficiently adapted to new patients via transfer learning or hyperparameter tuning, supporting scalable and generalizable deployment across broader clinical populations.
Traditionally, longitudinal physiological data have been modeled using mechanistic approaches, primarily systems of ordinary differential equations (ODEs). One of the most well-known models in this context is the Bergman minimal model, which describes glucose-insulin dynamics using ODEs grounded in medically established mechanisms7. Since then, several models have been developed and calibrated against coarse-grained clinical data to simulate glucose-insulin interactions8,14,24,25,34,41,50. While these models can qualitatively capture short-term glucose dynamics, their long-term predictive power is limited. This limitation stems largely from the sensitivity of dynamical systems to initial and boundary conditions and their over-simplified mechanisms and structures. Errors in parameter estimation or inaccurate initial states can lead to significant deviations in long-term predictions.
This challenge is exacerbated by the inherent complexity of the human body, which exhibits high dimensionality and variability. Modeling such a complex system using low-dimensional ODEs with simplified structures inherently restricts the model’s ability to capture the full physiological dynamics. Consequently, individualized modeling that accounts for uncertainties and patient-specific variability is more suitable, especially for predicting outcomes in critically ill patients.
Recent advances in ML have opened new venues for data-driven modeling in healthcare13,54. ML-based diagnostic systems have been developed for various diseases. For example, Khanna et al. proposed a decision-support system for osteoporosis risk prediction using feature selection and explainable AI to enhance interpretability31; Nayak et al. developed a deep learning framework for Monkeypox detection using transfer learning and explainable methods39; and Chadaga et al. introduced a stacked ML model for distinguishing mild to moderate COVID-19 from other respiratory conditions based on hematological biomarkers11. While these tools support early diagnosis, their applications in managing ongoing critical conditions like sepsis are still limited.
In contrast, this work focuses on real-time management by leveraging continuous glucose monitoring (CGM) data and ML models to forecast glucose levels over clinically relevant time horizons in ICU settings. The goal is to provide timely, actionable insights for improving patient care.
Some studies have begun to integrate ML into glucose monitoring systems. For instance, Fitzgerald et al. demonstrated the use of Continuous Time Recurrent Neural Networks (CTRNNs) to address challenges of predicting blood glucose in complex medical environments with irregular sampling intervals20. CTRNNs maintain hidden state dynamics between observations, overcoming constraints of traditional time series models. This capability positions CTRNNs as promising tools for ICU glucose forecasting and broader irregular time-series applications in healthcare.
Despite advancements in CGM technology, practical challenges such as data sparsity, noise, and sensor errors persist. Selecting appropriate models that are well-matched to data characteristics is essential for achieving accurate and robust predictions. This work leverages high-resolution bedside CGM data to develop and benchmark several ML models for individualized glucose forecasting in septic patients.
Figure 1 presents a complete workflow of the data-driven approach, including patient data acquisition, preprocessing, model training, and predictive analysis. A central challenge in this pipeline is identifying models capable of robust forecasting using a fixed-length lookback window–a sequence of recent observations used to predict future values. To address this challenge, the study systematically evaluates five advanced machine learning models, training and validating them using continuous glucose data from a single patient, and evaluating their performance on a holdout dataset comprising approximately 20% of the collected data.
Since transformer-based models have recently shown remarkable success in time series forecasting due to their ability to model long-range dependencies, four such models are evaluated in this study: PatchTST40, iTransformer35, Crossformer64, and FEDformer65. Each model employs unique architectural innovations to balance prediction accuracy and computational efficiency. In addition, DLinear63 is assessed as well, a simple yet powerful linear forecasting model that has outperformed many Transformer variants in recent benchmarks. Finally, this study explores an ensemble zero-shot inference approach using ChatGPT-4 for forecasting23. This zero-shot inference method treats the time series as a textual input, offering predictions without task-specific retraining. To reduce uncertainties and promote robustness of the prediction, we adopt an ensemble approach to produce the desired result. I.e., after making k repeated inquiries to ChatGPT-4 using a well-defined prompt, the median of the responses is taken as the final predicted value. Its potential for rapid, low-overhead deployment makes it an appealing candidate for real-world clinical settings.
While these models have demonstrated impressive performance on generic time series benchmarks, their applications to clinical glucose prediction remains underexplored. This study makes up the gap by conducting a comprehensive, head-to-head comparison of advanced ML models applied to continuous glucose data collected from a patient in the ICU. These findings facilitate model selection and shape deployment strategies for enhancing glucose management in critically ill patients in ICU.
Research novelty
This research addresses a critical gap in the application of data-driven machine learning for the real-time management of sepsis by focusing on glucose forecasting. The main contributions and novel aspects of this work are summarized as follows:
-
This study introduces a suite of glucose forecasting models and presents a comprehensive evaluation of their performance for glucose prediction in septic patients. The machine learning models include Transformer-based architectures (iTransformer, Crossformer, PatchTST, and FEDformer), DLinear, and an ensemble zero-shot inference approach using ChatGPT-4.
-
The evaluation of models on the extensive longitudinal patient dataset highlights their distinct strengths across different forecasting horizons: PatchTST delivers superior accuracy for short-term predictions, while DLinear proves more effective for medium- to long-term forecasting, striking a balance between simplicity and performance.
-
A 30-minute lookback window is identified as optimal for glucose forecasting, effectively simplifying the data requirements for clinical deployment with continuously collected bedside data.
-
ChatGPT-4, when applied in the ensemble zero-shot inference, achieved performance comparable to supervised models in selected scenarios, highlighting its potential utility for rapid deployment in resource-constrained environments where task-specific training may not be feasible.
-
Together, these models provide a versatile decision-support toolbox, enabling physicians to make timely and informed choices in the ICU setting.
These findings showcase the promise of integrating predictive models into future digital twin frameworks for personalized patient management and decision making. By leveraging machine learning and continuous physiological monitoring, this work contributes to advancing glycemic control and improving clinical outcomes for critically ill septic patients.
The rest of this paper is structured as follows: Section 3 details the process of data acquisition. Section 4 outlines the dataset characteristics and describes the methodological framework, the machine learning models, and the metrics used for performance assessment. Section 5 presents the comprehensive results of the assessment in glucose forecasting experiments using the continuous glucose monitoring dataset. Section 6 discusses the broader implications and limitations. Finally, Section 7 concludes the study.
Data acquisition
The data used in this work is from a patient who suffered from diabetes and sepsis. A 39-year-old female suddenly experienced dysarthria, convulsions, and loss of consciousness at home without any apparent cause. Emergency services brought the patient to Sichuan Provincial People’s Hospital. Diazepam was administered en route, which slightly relieved her symptoms. Upon arrival at the emergency department, the patient was in a state of confusion. Blood gas analysis showed pH 7.1, PCO2 38mmHg, PO2 44mmHg, BE -17.2mmol/L, Glu> 27.8mmol/L. Venous blood biochemical tests indicated glucose levels of 33.52mmol/L. An urgent cranial Computed Tomography (CT) scan revealed patchy and striped low-density shadows in the left fronto temporo parietal lobe and the right temporo-parietal lobe. The emergency physician considered the patient was suffering from diabetic ketoacidosis (DKA) complicated by seizures, and immediately administered sodium valproate to control seizures, and insulin to lower blood glucose levels. After the intubation and ventilator support, the patient was transferred to the emergency intensive care unit (EICU) for further treatment.
Upon admission to the EICU, physicians inquired about the patient’s medical history from her family. The patient had a history of diabetes, with unknown blood glucose control and no recent monitoring. Additionally, she had a history of encephalitis and epilepsy over 10 years ago, for which she had been taking medication regularly and had a relatively good control of the symptom. Two days prior, the patient had experienced a fall, besides that there were no other notable events. Her Acute Physiology and Chronic Health Evaluation II (APACHE II) score was 19. The initial diagnoses upon admission were: 1. Diabetic ketoacidosis; 2. Seizures. In the EICU, sodium valproate, levetiracetam, and lamotrigine were used to control seizures. Further head CT, Magnetic Resonance Angiography (MRA), and Diffusion-Weighted Imaging (DWI) examinations revealed generalized brain atrophy and a softening lesion in the left parietal lobe, not ruling out the possibility of a viral infection or inflammatory changes. A lumbar puncture was performed to obtain cerebrospinal fluid, but no abnormalities were found, ruling out an infection. On the third day of hospitalization, the patient developed bilateral pleural effusions, and her blood tests showed a white blood cell count of \(10.02\,\times \, 10^{9}\)/L and a high-sensitivity C-reactive protein level of 52.62mg/L. Considering pulmonary infection caused sepsis, piperacillin/tazobactam was administered for an antimicrobial treatment. On the 7th day of hospitalization, after the evaluation by a neurology specialist, the patient’s condition was deemed stable enough to receive the hyperbaric oxygen therapy. After a series of symptomatic and supportive treatments, the patient’s consciousness improved on the 11th day of hospitalization and she could open her eyes and follow simple directions. On the 15th day, she was transferred to a regular ward for the continued treatment. Additional seven days later, she was discharged from the hospital.
Data from continuous glucose monitoring device
After the patient was enrolled in the study, she was put on a CGM device. In this study, the FreeStyle Libre hospital CGM system (Abbott Diabetes Care Ltd., UK) was used. The real-time glucose data from the previous day is downloaded daily at 8 AM. When the patient was discharged from the EICU, the attending nurse removed the device and stored it properly for unified collection and filing.
Clinical data
The patient’s basic information and admission status were collected from the emergency medical record system (EMRS), including the gender, age, admission date, ICU admission date, ICU discharge date, hospital discharge date, department outcome, hospital discharge outcome, admission diagnosis, consciousness status, mechanical ventilation, height, weight, Body Mass Index (BMI), APACHE II score, Sequential Organ Failure Assessment (SOFA) score, and Nutrition Risk Screening (NRS-2002) score.
Biochemical indicators were collected from the laboratory information system (LIS). The patient’s nutritional support status was extracted from the hospital information system (HIS), including enteral and parenteral nutrition regimens and dosages, as well as the patient’s insulin dosage. In addition, continuous data from ventilators, infusion workstations, and electrocardiogram monitors were gathered using the EICU clinical research unit’s real-time automatic data collection device.
Datasets
This study focuses on models for glucose and insulin of the patient alone, ignoring any possible influence from other biomarkers/bioindicators. With this simplification, other factors are expected to have a marginal impact within the predictive time scales of interest. The dataset comprises blood glucose measurements for a patient from December 4th to 18th, acquired minute-by-minute, yielding a total of 19,621 data points. These are divided into training, validation, and testing sets in a 7:1:2 ratio. Table 1 summarizes commonly used statistics of glucose and insulin levels of the dataset. Figure 2 shows the time series of glucose (blue line) and insulin (orange line), with specific regions highlighted for data segmentation. The green-shaded area denotes the validation set, while the purple-shaded area represents the test set.
Time series of glucose (blue) and insulin (orange) levels marked with dataset divisions: the training set is uncolored (70% of the dataset), the validation set is highlighted in green (10% of the dataset), while the test set (20% of the dataset) is highlighted in purple. The model training is carried out using data from the training set+the validation set. The model performance is evaluated using the testing set. The insulin intervention is administered on a need basis decided by attending physicians when glucose levels exceeding certain threshold values. It is pumped in during a 2 to 4 hour interval at a constant rate.
Figure 3 illustrates the statistical distribution of the patient’s glucose and insulin data. The boxplots (left) show that insulin levels are low and right-skewed, while glucose levels are higher and more widely dispersed. The hexbin graph (right), which is created by dividing the plot area into a grid of hexagons and coloring each one based on the number of data points it contains, reveals the joint structure of the variables. The resulting map shows data concentrated at low insulin and moderate glucose levels. This suggests a weak, possibly nonlinear relationship between the two variables.
Ethics review and registration
This work is part of an observation study, and the study protocol was approved by Medical Ethics Committee of Sichuan Provincial People’s Hospital and Sichuan Academy of Medical Sciences (approval number: Yan-2022-137). The written informed consents were obtained from the patients or their representatives if the patient was unconscious. The study protocol was registered prior to the beginning of the study at Chinese Clinical Trial Registry (ChiCTR), registration number: ChiCTR2300077594.
Methods
All methods along with necessary datasets were used in accordance with the relevant guidelines and regulations. This study explores two modeling approaches:
-
Single-Variable Model: Here, glucose levels serve both as the input and the output for the forecasting model. This setup is designed to predict future glucose levels based solely on past glucose measurements in a lookback window.
-
Dual-Variable Model: This model incorporates both glucose and insulin levels as inputs, with glucose level prediction as the output. This approach aims to improve the predictive power of the models by accounting for the potential interaction between insulin and glucose.
For both models, the predicted sequence lengths or prediction horizons are defined as follows. For the Single-Variable Model:
-
\(Gl\_15\) for a 15-minute prediction horizon,
-
\(Gl\_30\) for a 30-minute prediction horizon, and
-
\(Gl\_60\) for a 60-minute prediction horizon.
For the Dual-Variable Model:
\(Gl2\_15\) indicates a model with both glucose and insulin as inputs for a 15-minute prediction horizon. This is the only model tested in this case.
All models are tested with varying lookback windows consisting of 30, 60, 90, 120 data points (minutes), respectively, to determine the optimal duration of retrospective data required for accurate forecasting. These configurations enable a systematic evaluation of the models’ performance across various prediction horizons and input complexities, facilitating a comprehensive understanding of how insulin data along with glucose levels influence the predictive accuracy.
Models
The time series dataset, \(D=\left\{ X_i\right\} _{i=1}^L\), is defined over interval [0, T], where each data point \(X_i \in \mathbb {R}^N\) consists of N variables sampled at regular time points \(t_1, t_2, \ldots , t_L \in [0, T]\). In this work, the number of variables is \(N\le 2\). The objective of the time series forecasting is to predict the next H time steps, \(X_{t+1}, \ldots , X_{t+H}\), based on the retrospective data at the past L time steps, \(X_{t-L+1}, \ldots , X_t\). This is achieved using a trainable machine learning model \(F_\theta\), which can be expressed as follows,
A comprehensive evaluation of the length of look-back windows (L) ranging from 30 to 120 and prediction horizons (H) ranging from 15 to 60 was conducted, and, in the meantime, the influence of the covariate insulin on forecasting performance was analysed as well. Six predictive models were evaluated, including four Transformer-based architectures, a linear-based model, and a novel ensemble zero-shot approach using a large language model (ChatGPT-4).
-
iTransformer: Introduced by Liu et al.35, this model adopts a novel approach by inverting dimensions and embedding the entire time series of each variable independently into a single temporal token. Specifically, for a multivariate time series \(X \in \mathbb {R}^{L \times N}\), the iTransformer converts the input into \(Z \in \mathbb {R}^{N \times L}\), embedding each variable into a single representation. The transformed output is subsequently fed into a standard Transformer architecture. This technique aims to capture multivariate correlations more effectively, representing an advanced case of segmentation in time series analysis. The core steps of the iTransformer include the following:
Embedding Variables: Each variable’s time series is embedded into a dense representation:
$$\begin{aligned} H_0 = \text {Embedding}(Z)\in \mathbb {R}^{N \times D}, \end{aligned}$$(4.2)where D is the dimension of the token.
Self-Attention for Variable Correlations: Self-attention calculates weights to capture relationships between variables:
$$\begin{aligned} \operatorname {Attention}(Q, K, V)=\operatorname {softmax}\left( \frac{Q K^T}{\sqrt{d_k}}\right) V, \end{aligned}$$(4.3)where Q, K, V are the query, key, and value matrices projected from \(H_0\) and \(d_k\) is the scaling factor.
Feed-Forward Network for Temporal Features: A two-layer neural network processes each variable token independently to extract temporal features:
$$\begin{aligned} H_{\text {FFN}} = \text {ReLU}(H_{\text {attn}} W_1 + b_1) W_2 + b_2, \end{aligned}$$(4.4)where \(W_{1,2}\) are weights and \(b_{1,2}\) are biases, respectively.
Output Prediction: The model projects the processed tokens back into the forecasting space:
$$\begin{aligned} \hat{Y} = \text {Projection}(H) \in \mathbb {R}^{N \times S}, \end{aligned}$$(4.5)where S is the forecast horizon.
-
PatchTST: Developed by Nie et al.40, PatchTST efficiently segments time series data into patches, processing each channel independently.
Patch Embedding: Consider a multivariate time series \(X\in \mathbb {R}^{L \times N}\), where the i-th univariate component is \(X^{(i)} = \bigl (x^{(i)}_{1},x^{(i)}_{2},\dots ,x^{(i)}_{L}\bigr )^{\top }, \quad i = 1, \dots , N.\) With patch length P and stride S (usually \(S=P\) for non-overlapping patches), each series is segmented into \(M=\lfloor (L-P)/S \rfloor + 2\) fixed-length patches
$$\begin{aligned} \textbf{p}^{(i)}_{j} =\bigl (x^{(i)}_{(j-1)S+1},\dots , x^{(i)}_{(j-1)S+P}\bigr )^{\top }\in \mathbb {R}^{P}, \quad j=1,\dots ,M. \end{aligned}$$(4.6)Each patch \(\textbf{p}^{(i)}_{j}\) is then mapped to a D-dimensional token via a learnable projection \(\textbf{W}_{p}\in \mathbb {R}^{D\times P}\), and positional vector \(\textbf{e}_{j}\):
$$\begin{aligned} \textbf{z}^{(i)}_{j} = \textbf{W}_{p}\textbf{p}^{(i)}_{j} + \textbf{e}_{j} \in \mathbb {R}^{D}. \end{aligned}$$(4.7)Channel-Independent Transformer Encoder: Tokens from all variables are stacked to a batch and processed by Transformer backbone, as defined in Equations (4.2)-(4.5). Denote the encoded sequence for variable i by \(\textbf{Z}^{(i)} =\bigl (\textbf{z}^{(i)}_{1},\textbf{z}^{(i)}_{2},\dots ,\textbf{z}^{(i)}_{M}\bigr ) \in \mathbb {R}^{M \times D}.\) By applying a flattening layer with a linear head to \(\textbf{Z}^{(i)}\), we obtain the H-step forecast:
$$\begin{aligned} \widehat{\varvec{x}}^{(i)} =\bigl (\widehat{x}^{(i)}_{L+1},\widehat{x}^{(i)}_{L+2},\dots ,\widehat{x}^{(i)}_{L+H}\bigr ) \in \mathbb {R}^{1 \times H}. \end{aligned}$$(4.8) -
Crossformer: Zhang et al.64 proposed Crossformer, which differentiates itself by addressing both temporal and cross-dimensional dependencies. It leverages Dimension-Segment-Wise (DSW) embedding coupled with a Two-Stage Attention (TSA) mechanism to adeptly capture dependencies.
Dimension-Segment-Wise (DSW) Embedding: DSW embedding processes each dimension of the time series independently by partitioning it into segments of length \(L_{seg}\).
$$\begin{aligned} X_{i, d}^{(s)} =\left\{ X_{t, d} \mid (i-1) \times L_{seg }<t \le i \times L_{seg }\right\} \end{aligned}$$(4.9)The embedded input is \(X \in \mathbb {R}^{L \times N}\), where L represents the lookback window length and N denotes the number of variables. \(X_{i, d}^{(s)} \in \mathbb {R}^{L_{seg}}\) represents the d-th variable in the i-th segment with a length of \(L_{seg}\). Each segment is then projected into a latent space with positional embeddings:
$$\begin{aligned} H_{i, d}=E X _{i, d}^{(s)}+E_{i, d}^{(p o s)} \end{aligned}$$(4.10)where \(E \in \mathbb {R}^{d_{model} \times L_{seg}}\) is the projection matrix, and \(E_{i, d}^{(pos)} \in \mathbb {R}^{d_{model }}\) encodes positional information.
Two-Stage Attention (TSA) Layer: The TSA layer captures both temporal and cross-dimension dependencies efficiently: Cross-Time Stage: Captures temporal dependencies within each dimension using Multi-Head Self-Attention (MSA):
$$\begin{aligned} \begin{aligned}&\hat{Z}_{:,d}^{\text {time}} = \text {LayerNorm}\left( Z_{:,d} + \text {MSA}_{\text {time}}(Z_{:,d}, Z_{:,d}, Z_{:,d})\right) \\&Z^{\text {time}} = \text {LayerNorm}\left( \hat{Z}^{\text {time}} + \text {MLP}(\hat{Z}^{\text {time}})\right) \end{aligned} \end{aligned}$$(4.11)where \(Z_{:,d}\) is the input embedding for all time segments of the d-th dimension, LayerNorm denotes layer normalization and MLP represents a two-layer neural network. Cross-Dimension Stage: Uses a router mechanism to aggregate and distribute information across dimensions:
$$\begin{aligned} \begin{aligned} B_{i,:}&=\text{ MSA}_1^{\operatorname {dim}}\left( R_{i,:}, Z_{i,:}^{\text{ time } }, Z_{i,:}^{\text{ time } }\right) \\ \overline{Z}_{i,:}^{\text{ dim } }&=\text {MSA}_2^{\operatorname {dim}}\left( Z_{i,:}^{\text{ time }}, B_{i,:}, B_{i,:}\right) \\ \hat{Z}^{\text{ dim }}&=\operatorname {LayerNorm}\left( Z^{\text{ time }}+\overline{Z}^{\text{ dim } }\right) \\ Z^{\text{ dim }}&=\text{ LayerNorm }\left( \hat{Z}^{\text{ dim } }+\operatorname {MLP}(\hat{Z}^{\text{ dim }})\right) \end{aligned} \end{aligned}$$(4.12)where \(Z_{i,:}\) is the input of all dimensions at time step i and \(R_{i,:}\) is a learnable set of vectors acting as routers.
-
FEDformer: Zhou et al.65 designed FEDformer to bolster long-term forecasting by merging the transformer architecture with seasonal-trend decomposition and Fourier transformations. This innovative combination yields improved forecasting accuracy.
Seasonal-Trend Decomposition: FEDformer incorporates seasonal-trend decomposition using a Mixture of Experts (MOE) mechanism to separate and refine seasonal and trend components dynamically:
$$\begin{aligned} X_{\text{ trend } }=\operatorname {Softmax}(L(X)) \cdot (F(X)), \end{aligned}$$(4.13)where \(F(\cdot )\) is a set of average pooling filters, and Softmax(L(X)) generates weights for combining these extracted trends.
Frequency Enhanced Attention: FEDformer replaces traditional self-attention by operating in the frequency domain using either Fourier Transform or Wavelet Transform. Fourier Transform converts input queries (Q), keys (K), and values (V), into its frequency representation:
$$\begin{aligned} \begin{aligned} \tilde{Q}&=\operatorname {Select}(\mathcal {F}(Q)) \\ \tilde{K}&=\operatorname {Select}(\mathcal {F}(K)) \\ \tilde{V}&=\operatorname {Select}(\mathcal {F}(V)) \end{aligned} \end{aligned}$$(4.14)where \(\mathcal {F}(\cdot )\) denotes the Fourier Transform. To optimize computational efficiency, a random subset of Fourier modes is selected. The attention computation in the frequency domain is given by \(Y =\sigma (\tilde{Q} \cdot \tilde{K}^{\top }) \cdot \tilde{V}\), where \(\sigma\) is an activation function (e.g., softmax or tanh). The output is then zero-padded to match the original sequence length L and transformed back to the time domain using the Inverse Fourier Transform:
$$\begin{aligned} \operatorname {FEA-f}(Q, K, V)=\mathcal {F}^{-1}(\operatorname {Padding}(\sigma (\tilde{Q} \cdot \tilde{K}^{\top }) \cdot \tilde{V})) \end{aligned}$$(4.15)
Linear-based Model:
-
DLinear: Introduced by Zeng et al.63, DLinear surpasses several existing Transformer-based models by utilizing straightforward linear methods alongside seasonal-trend decomposition techniques. It performs a weighted sum of historical inputs to predict future values:
$$\begin{aligned} \hat{X} = W X, \end{aligned}$$(4.16)where \(W \in \mathbb {R}^{H \times L}\) is the learnable weight matrix, L is the look-back window size, and H is the forecast horizon. Its simplicity and effectiveness highlight its competitive edge in time series forecasting.
Large Language Models (LLMs):
-
ChatGPT is a large language model developed by OpenAI. It’s an AI-powered chatbot that uses natural language processing to engage in conversational interactions with users. It was initially released in November 2022 and quickly gained widespread attention. ChatGPT is based on the GPT (Generative Pre-trained Transformer) architecture. It can understand and generate human-like text on a wide range of topics, answer questions, assist with tasks like writing and coding, and engage in creative exercises. The model is trained on a vast amount of text data from various sources, allowing it to draw upon a broad knowledge base. ChatGPT has been noted for its ability to maintain context in conversations and provide detailed, coherent responses. Since its initial release, OpenAI has continued to update and improve the model, including releasing GPT-4, GPT-4o, GPT-4.5,and the o3 series, which has even more advanced capabilities. ChatGPTs based on the zero-shot inference paradigm have recently been applied to time series prediction with the aid of prompt engineering55. Unlike traditional methods, zero-shot forecasting requires no task-specific training. This methodology leverages a pre-trained large language model (e.g., GPT-4 in this study) via prompt engineering, where a time-series segment is converted into a carefully designed textual prompt, which is then fed to the LLM to obtain a forecast. Because GPT-4 has been trained on vast scientific and biomedical data, it inherently encodes qualitative clinical relationships that are difficult to learn from small datasets, which is invaluable for short-horizon prediction. This advantage is particularly useful in bedside settings, where clinicians often have only a few glucose measurements and traditional deep learning forecasters are limited due to the data sparsity. In addition, the workflow in this zero-shot inference application is highly user-friendly. Clinicians can obtain reliable forecasts by interacting with a chat interface, without any requirements for specialized machine learning knowledge.
This study employs the above models to forecast glucose levels in a prescribed time horizon, aiming to benchmark their performance on the same dataset comparatively to produce a toolbox useful in ICU settings. The comparative analysis not only sheds light on the strength and weakness of each model but also provides insights into their robustness and applicability in real-world scenarios involving glucose data prediction.
To benchmark the results, six metrics are used below, among which three measure absolute errors while the other three relative errors.
MSE (Mean Squared Error),
where \(\hat{y}_{i}\) is the predicted glucose value, \(y_{i}\) is the value of the ground truth, and H is the length of the predicted time horizon.
MAE (Mean Absolute Error),
MME (Mean Maximum Error),
MSPE (Mean Squared Percentage Error),
MAPE (Mean Absolute Percentage Error),
and MMPE (Mean Maximum Percentage Error),
The transformer-based models and the DLinear model were trained following the machine learning protocol while the LLM-based zero-shot inference method was implemented using prompt engineering and ensemble average. The trained models were systematically applied to the holdout test set to benchmark their predictive accuracy across various forecasting horizons.
Results
The comparative forecasting performance of the five machine learning models and the ensemble zero-shot LLM approach is presented in this section. Since they use different approaches to make predictions, the predictive results obtained from the machine learning models and the LLM are discussed respectively.
Forecasting with machine learning models
After the models are trained on the training set (70% of the dataset) and validation set (10% of the dataset), the trained models are applied to the testing set to benchmark their predictive performance. The forecasting results on the testing dataset (20% of the overall data collected) are summarized in Table 2. The reported errors in the table are averaged errors calculated across all test samples for each model. For the 30+15 prediction, where 30 consecutive points are in the lookback window and 15 in the horizon, the number of samples tested is 3910, where each sample consists of 45 consecutive data points; for the 30+60 prediction, the number of samples in the test is 3865, where each sample consists of 90 consecutive data points. Specifically, given a sample of 30 data points in the lookback window in the testing dataset of the 30+15 prediction, the next lookback window sample is obtained by shifting the current 30 data point window in the testing dataset to the right by one position. By exhausting all the datapoints in the testing set, the testing phase of the study is completed and the averaged error is recorded as an entry in Table 2.
The best-performing results in Table 2 are highlighted in bold, and the second-best results are underlined. Overall, DLinear excels on average in all cases, closely followed by PatchTST. In the 15 minutes prediction, the average relative error in MMPE is slightly above 3% in both models. The error climbs to 7.46% in the 30 minutes prediction and deteriorates to 14.41% in the 60 minutes prediction horizon using the DLinear model. As shown in Fig. 4, with a 120-minute lookback window and a 15-minute prediction horizon, PatchTST and DLinear clearly outperformed other models, consistently achieving lower errors across all six metrics.
This comprehensive study examines the relationship between the length of the lookback window and the prediction horizon, uncovering an intriguing pattern. As the lookback window is expanded, the DLinear model’s error decreases, while other models experience increased errors. This finding suggests that the lookback window length may not be a universally sensitive hyperparameter for all machine learning models, provided a minimum threshold is met. The finding indicates that a minimum length of the lookback window consisting of approximately 30 data points yields satisfactory performance across all models. Consequently, the results are presented using only these 30 prior points in the lookback window for the horizon predictions.
However, if the length of the prediction horizon for a fixed-length lookback window is increased, the error escalates rapidly. For instance, the MSE of DLinear increases from 0.054 to 0.319 when the prediction horizon increases from 15 to 30 steps ahead. Furthermore, the analysis reveals that the inclusion of additional variables, such as insulin data, does not significantly improve the forecasting results among the models. For example, the MSE of PatchTST only decreases from 0.052 to 0.051 when insulin data is concatenated with glucose data as a combined input feature. This change is insignificant, which suggests that incorporating additional variables may not necessarily enhance the model’s predictive power in this particular data-driven modeling case significantly.
Comparison of performance metrics for the five forecasting models: iTransformer, Crossformer, PatchTST, DLinear, and FEDformer with a lookback window length = 120 and prediction length = 15. PatchTST and DLinear achieve significantly better results, with consistently lower errors across all metrics compared to other models.
Visualization of different models’ predictions with an input sequence length of 30 and a prediction horizon of 15. (a, b) Two examples of predictions from the supervised machine learning models vs. the ground truth. (c, d) Two examples of predictions from ChatGPT-4 (based on 20 samples) vs. the ground truth, showing the median prediction and the \(90\%\) prediction interval.
To illustrate the detailed performance of each model, Fig. 5 visualizes the predictions for an input sequence length of 30 and a prediction length of 15. As demonstrated in Fig. 5a , all models capture the correct trend of the data’s evolution in the horizon. The predictions of PatchTST, DLinear, and FEDformer closely follow the ground truth values. However, in Fig. 5b , Crossformer appears to perform better than the other models in this particular run. Considering the overall performance on all testing samples from the testing dataset, measured by average errors, DLinear and PatchTST excel. For practical applications, a weighted average of the results from DLinear and PatchTST may be the safest approach to adopt, as it leverages the strengths of both models. This is implemented as an option in the toolbox.
While varying the prediction horizon from 15 to 30 and then to 60 steps, and keeping the length of prediction horizon fixed at 30, the impact of the length of the prediction horizon on the forecasting errors was evaluated. As illustrated in Fig. 6, all models exhibit a similar scaling pattern in their errors. As the length of the prediction horizon increases, errors grow super-linearly. Particularly in Fig. 6b,e, it is clearly observed that MAE and MAPE increase linearly with the length of the prediction horizon. At lookback window length 30 and prediction horizon 30 steps, the overall average errors across all metrics remain clinically acceptable, with the largest error not exceeding 8.7%.
Considering a short-term prediction scenario, where the models forecast 30 steps (half an hour) ahead using only the preceding half-hour data, the predictions are reasonably accurate. For septic patients in an ICU, such half-hour predictions could provide attending physicians with much-needed information to make informed decisions. Moreover, when integrated into a digital twin framework as a forward-rolling forecaster, these predictive models could play a crucial role in projecting patients’ treatment outcomes. We next assess the performance of the zero-shot inference model based on an LLM.
Ensemble zero-shot forecasting with an LLM (ChatGPT-4)
Large language Models (LLMs) have been widely used in various domains, such as writing, encyclopedia, education, healthcare, finance, medicine, etc. With the help of Prompt Engineering, LLMs have shown potential in many other non-natural language processing functions such as mathematical optimization, mathematical proofs, time series forecasting, etc.1,28,57,66. Unlike traditional methods which use numerical time series as input and output, an LLM directly treats numerical series as textual data so that one can devise suitable prompts for LLMs to perform an ensemble zero-shot inference/forecasting55. For example, PromptCast61 transforms the numerical input and output into text prompts and forecasts time series in a sentence-to-sentence manner. LLMTime23 proposes an effective tokenization for time series data and a rescaling strategy, which are shown to be crucial for achieving good performance. Figure 7 illustrates the workflow for a question-answer-based ensemble zero-shot inference/prediction approach. In this method, the user queries the next values of a time series, and ChatGPT generates predictions based on its domain knowledge.
This work follows LLMTime’s approach and composes prompt templates as follows:
-
system:“You are a helpful assistant that performs time series predictions. The user will provide a sequence and you will predict the remaining sequence. The sequence is represented by decimal strings separated by commas.”
-
users:“Please continue the following sequence without producing any additional text. Do not say anything like ’the next terms in the sequence are’, just return the numbers. The values in the Glucose for the past {seq_len} time steps are {input_str}. What will the values for the next {pred_len} time steps be? The values for the next {pred_len} time steps will be.”
ChatGPT-4 is used as the base LLM as it was one of the most powerful large language models by the time this study conducted. Due to the cost of computations, 10 test samples are randomly chosen from the raw test dataset. To test the performance of the prompt-based ensemble zero-shot forecasting, results are compared under different sampling numbers, i.e. 5, 10 and 20 samples, noted as GPT4TS_5, GPT4TS_10 and GPT4TS_20, respectively. The median of the predictions from these samples are used as the final prediction. All results are reported in Table 3. Note that this method can only be applied to univariate time series, so Gl2_15 dataset is not used.
It is observed that in Table 3, GPT4TS_10 achieves the best performance in most cases while GPT4TS_20 is also competitive. The main reason for the difference between the two is the random error. The result of 20 samples is more stable while the result of 10 samples is relatively sparse. Overall, the ensemble zero-shot forecasting results yield the same predicative quality as the supervised learning models. As the length of the prediction horizon increases, the predictive error also increases. 30 minutes of lookback window is sufficient for the short-term forecasting since longer input sequence length seems to have little or no effect on reducing the error, which is consistent with the findings from the machine learning models.
The qualitative performance of the ensemble zero-shot inference approach with GPT-4 is visualized in Fig. 5c,d , using 20 prediction samples. The length of the lookback window is set at 30 and prediction horizon 15. As illustrated in Fig. 5(c), the median predicted trend aligns closely with the ground truth. Figure 5d highlights the importance of collecting multiple samples, as the 90% confidence interval covers a wide range while the median prediction remains close to the actual value.
Finally, the ensemble zero-shot forecasting results are compared with supervised learning results in Table 4. Top 2 supervised learning models, PatchTST and DLinear, are tested over the same test dataset. It can be seen that there is still a gap between the predictive results of the ensemble zero-shot inference with GPT-4 and state-of-the-art machine learning models since GPT-4 model is an ensemble zero-shot learning method without training. Figure 8 presents a comprehensive performance comparison between PatchTST and GPT4TS_10 based on MMPE metric across various configurations, such as 120_GI_60 and 90_GI_60, where “120_GI_60” represents a lookback window length of 120 and a prediction horizon of 60. PatchTST consistently outperforms GPT4TS_10, achieving lower error values across all scenarios, particularly in settings with longer input sequences, such as 120_GI_60 and 90_GI_60.
Nevertheless the ensemble zero-shot forecasting result is also competitive. It should be noted that this method is very convenient for clinicians without specialized training in AI/machine learning methods. However, care must be given when interpreting and presenting the results. It’s better off using the same sample multiple times to generate the predictions and then adopt the median of the predictive results in the ensemble as demonstrated. This resembles an ensemble method commonly used in machine learning22,43. More precisely, the ensemble zero-shot forecasting is an ensemble of zero-shot inferences.
Discussion
This comprehensive study explores the efficacy of various machine learning models and a novel ensemble zero-shot approach using GPT-4 for glucose level forecasting for septic patients. The findings offer valuable insights into the performance and potential clinical applications of these models. These models can be used as support tools for physicians to forecast patient outcomes and make quick clinical decision in ICU settings.
The comprehensive comparative study was conducted in the context a statistical mean of a large number of tests. The DLinear and PatchTST models demonstrated superior performance across different prediction horizons, particularly excelling in short-term forecasts. PatchTST’s remarkable accuracy for 15-minute predictions (MMPE \(\approx 3\)%) could prove invaluable for immediate interventions, while DLinear’s strong performance in 30-minute (MMPE \(\approx 7.46\)%) and 60-minute (MMPE \(\approx 14.41\)%) forecasts provides a wider window for treatment planning. These results suggest that these models could significantly enhance real-time patient monitoring, forecasting, and decision-making in intensive care settings.
A key finding is that a 30-minute lookback window suffices for short-term forecasting, indicating that recent glucose history is more relevant than long-term trends in the glucose dynamics. This could streamline data collection and model implementation in clinical environments. However, the super-linear increase in error rates for longer prediction horizons underscores the challenges of extended glucose forecasting and emphasizes the need for frequent model updates (forward-rolling) in prolonged monitoring scenarios.
The ensemble zero-shot forecasting approach using ChatGPT-4, while not outperforming the best machine learning models, showed competitive results and a cheap alternative for healthcare professionals to adopt in the environment with limited computational resources. This showcases the potential of large language models in medical time series monitoring and forecasting, particularly in situations prioritizing ease of use and rapid deployment over maximum accuracy.
Interestingly, the inclusion of additional variables, such as insulin data, did not show any significantly improvements in the forecasting results using the data-driven models. This implies that glucose levels alone may be sufficient for short-term predictions, potentially simplifying data collection and model inputs in clinical applications. Another reason why insulin appears to have limited influence on glucose forecasting is its reactive nature, as it is typically administered by physicians in response to elevated glucose levels rather than preemptively.
The results highlight the potential of machine learning models in predictive analytics in critical care settings. While existing methods primarily support the medical decision-making by focusing on early disease diagnosis, they often fall short in managing ongoing critical conditions. In contrast, this approach leverages continuous glucose data and advanced machine learning models to predict glucose levels, providing real-time feedback and actionable insights into the patient’s condition. This innovation enhances the potential for dynamic, data-driven patient care in critical care settings. In addition, continuous-time recurrent neural networks (CTRNNs) have shown promise in glucose forecasting by effectively handling irregular time series data. However, CTRNNs often face challenges related to computational complexity. This approach utilizes more efficient models, such as DLinear and PatchTST, ensuring greater scalability and performance in real-world applications.
From a clinical perspective, the ability to provide accurate glucose forecasts within a short time-frame, such as 15 to 30 minutes, can significantly impact decision-making in ICUs. Physicians can use these forecasts to anticipate glycemic fluctuations and administer timely interventions, potentially reducing the risk of complications associated with hypo- or hyperglycemia. The findings of this study suggest that models like DLinear and PatchTST are well-suited for integration into bedside monitoring systems, enabling real-time decision support for managing septic patients.
The current practice of insulin administration at ICUs is reactive. After physicians notice an increase of glucose beyond a certain threshold levels, they would administer insulin injection afterwards following the guideline tabulated in Table 5. The administration of insulin depends on the time point physicians notice the glucose increase and their response time. These models can provide the forecasted glucose level 30 minutes or 1 hour ahead of time, an insulin administration regimen can then be crafted and tested accordingly. This consists of the clinical implementation of the models developed in this study, which will be reported after a comprehensive clinical trial.
Looking forward, the potential application of these models in digital twin for health frameworks represents a promising venue for personalized medicine. By leveraging patient-specific data, digital twins can monitor and predict disease progression, offering clinicians a powerful tool to optimize treatment strategies. This study provides a foundation for incorporating glucose forecasting models into such frameworks, further advancing their utility in critical care settings.
This study highlights the reliability of DLinear and PatchTST for short-term predictions, noting their potential to complement existing clinical workflows. However, physicians/users might have concerns about the interpretability of transformer-based models, which may hinder their adoption in clinical settings. The ensemble zero-shot forecasting approach using GPT-4 is acknowledged for its ease of use and adaptability, but its relatively lower accuracy compared to DLinear and PatchTST limits its priority in the unsupervised, stand alone clinical utility. Physicians among the authors suggest that GPT-4 can serve as a supplementary tool, particularly in resource-limited settings.
This study presents promising findings for monitoring and forecasting glucose levels via bedside continuous data-collecting equipment for ICU-bound septic patients, but several limitations and challenges warrant additional considerations. First, the dataset was derived from a single patient, raising concerns about the generalizability of the results to broader populations. Transfer-learning would be used to re-tune the model parameters for other patients while retaining the hyper-parameters and the architecture of the model. Validation across diverse patient cohorts and conditions is essential for the safe use of the toolbox. Additionally, while this study focused on monitoring and forecasting of glucose levels, incorporating other physiological variables, such as hemodynamic data, could enhance the accuracy and applicability of the models, in particular, in understanding of the detailed glucose-insulin dynamical interaction.
Despite their strong predictive performance, transformer-based architectures, including GPT-4, remain inherently opaque, leaving clinicians unable to see how specific inputs influence the final results. The lack of interpretability in these machine learning models remains a critical concern, as healthcare providers often require transparent and explainable tools to ensure trust and informed decision-making. To address this limitation, several explainable techniques are proposed. First, SHapley Additive exPlanations (SHAP)36 leverages Shapley values from game theory to quantify the contribution of each feature to model predictions. Shen et al.47 used SHAP analysis to rank structural variables in their CFST capacity model, showing that section width and column height dominated predictive importance. However, SHAP is designed for multivariate settings, which limits its application to a single variable model like the models for glucose studied. Incorporating additional physiological variables, such as insulin levels, is crucial to fully exploit its explanatory power.
Furthermore, hybrid modeling frameworks integrate first-principles dynamical equations with machine learning components46 could shed lights on interpretability of the underlying models. This approach uses first-principles models as a backbone, while the data-driven component is employed to capture uncertain parameters, states, or uncertainties that are difficult to derive from physical laws alone. This fusion not only embeds domain knowledge into the model structure, but also helps reveal latent mechanisms that might be missed. In future work, these two approaches will be applied to identify the hidden glucose dynamics, thus strengthening the transparency and reliability of the decision-support system. Finally, prospective clinical studies will be required to integrate these explainable models into routine workflows and quantify their impact on patient care and treatment outcomes.
Conclusion
This study has demonstrated the potential use of machine learning and large language models for glucose forecasting in individual septic patients and produced a toolbox to support clinical decision making in the ICU. A comprehensive comparison of the models in the toolbox revealed that both DLinear and PatchTST exhibit high predictive accuracy across short- to medium-term forecasting horizons. The ensemble zero-shot approach using GPT-4 also emerged as a cost-effective alternative, delivering reasonably accurate predictions with minimal computational effort and no task-specific training.
These findings hold significant promise for clinical applications. The robust performance of the proposed models highlights their potential for being integrated into bedside monitoring and decision support systems, offering a pathway to enhance glycemic control and patient care in critical care settings. As AI technologies continue to evolve, their integration into clinical workflows could mark a paradigm shift in the management of critically ill patients. Additionally, the ensemble zero-shot GPT-4 approach provides a distinct application of AI tools for rapid and easy deployment, especially in resource-limited settings.
However, several limitations of this study warrant further investigations. While PatchTST and other transformer-based models achieve high accuracy, their complexity and lack of interpretability may hinder adoption and trust in clinical practices. The ensemble zero-shot LLM approach, though convenient, is less accurate than supervised methods, limiting its stand-alone utility in high-risk scenarios. Moreover, the study’s reliance on data from a single patient raises concerns about generalizability, underscoring the need for validation across diverse patient populations through transfer learning and model re-tuning.
Future research will focus on addressing these limitations. Primary priorities will include improving model interpretability, transfer-learning the machine learning models, validating performance across diverse patient cohorts, conducting prospective studies to assess the real-world clinical impact of these technologies, and developing innovative strategies for the seamless integration of these predictive tools into existing healthcare systems. These combined efforts are essential to advance this technology from a promising concept into a robust clinical tool, ultimately achieving personalized and adaptive glycemic control in septic patients.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AI :
-
Artificial intelligence
- APACHE II :
-
Acute physiology and chronic health evaluation II
- CGM :
-
Continuous glucose monitoring
- ICU :
-
Intensive care unit
- EICU :
-
Emergency intensive care unit
- DT :
-
Digital twin
- CTRNN :
-
Continuous time recurrent neural network
- LLM :
-
Large language model
- iTransformer :
-
Inverted-dimension transformer
- Crossformer :
-
Cross-dimension transformer
- PatchTST :
-
Patch time-series transformer
- DLinear :
-
Decomposition-linear model
- FEDformer :
-
Frequency-enhanced decomposition transformer
- GPT4TS_5 :
-
Zero-shot ChatGPT-4 forecaster using 5 repeated samples
- GPT4TS_10 :
-
Zero-shot ChatGPT-4 forecaster using 10 repeated samples
- GPT4TS_20 :
-
Zero-shot ChatGPT-4 forecaster using 20 repeated samples
- Gl_15 :
-
Glucose-only forecast, 15-min horizon
- Gl_30 :
-
Glucose-only forecast, 30-min horizon
- Gl_60 :
-
Glucose-only forecast, 60-min horizon
- Gl2_15 :
-
Dual-variable (glucose + insulin) forecast, 15-min horizon
- MSE :
-
Mean squared error
- MAE :
-
Mean absolute error
- MME :
-
Mean maximum error
- MSPE :
-
Mean squared percentage error
- MAPE :
-
Mean absolute percentage error
- MMPE :
-
Mean maximum percentage error
References
AhmadiTeshnizi, A., Gao, W., & Udell, M. Optimus: Optimization modeling using mip solvers and large language models. arXiv preprint. arXiv:2310.06116 (2023).
Aleman, L. & Guerrero, J. Sepsis hyperglycemia in the icu: From the mechanism to the clinic. Rev. Med. Chil. 146(4), 502–510 (2018).
Alsaadawi, M. M., Elshaarawy, M. K. & Hamed, A. K. Concrete compressive strength classification using hybrid machine learning models and interactive gui. Innov. Infrastruct. Solutions 10(5), 1–29 (2025).
Angus, D. C. & Poll, T. Severe sepsis and septic shock. N. Engl. J. Med. 369(9), 840–851 (2013).
Armanuos, A. M. & Elshaarawy, M. K. Estimating saltwater wedge length in sloping coastal aquifers using explainable machine learning models. Earth Sci. Inf. 18(2), 405 (2025).
Bellomo, R., & Egi, M. Glycemic control in the intensive care unit: Why we should wait for nice-sugar. In Mayo Clinic Proceedings vol 80, 1546–1548 (Elsevier, 2005).
Bergman, R. N. Toward physiological understanding of glucose tolerance: Minimal-model approach. Diabetes 38(12), 1512–1527 (1989).
Bondia, J., Romero-Vivo, S., Ricarte, B. & Diez, J. L. Insulin estimation and prediction: A review of the estimation and prediction of subcutaneous insulin pharmacokinetics in closed-loop glucose control. IEEE Control Syst. Mag. 38(1), 47–66 (2018).
Capes, S. E., Hunt, D., Malmberg, K. & Gerstein, H. C. Stress hyperglycaemia and increased risk of death after myocardial infarction in patients with and without diabetes: a systematic overview. The Lancet 355(9206), 773–778 (2000).
Capes, S. E., Hunt, D., Malmberg, K., Pathak, P. & Gerstein, H. C. Stress hyperglycemia and prognosis of stroke in nondiabetic and diabetic patients: A systematic overview. Stroke 32(10), 2426–2432 (2001).
Chadaga, K. et al. Artificial intelligence for diagnosis of mild-moderate covid-19 using haematological markers. Ann. Med. 55(1), 2233541 (2023).
DeMerle, K. M. et al. Sepsis subclasses: A framework for development and interpretation. Crit. Care Med. 49(5), 748–759 (2021).
Deng, H.-F. et al. Evaluating machine learning models for sepsis prediction: A systematic review of methodologies. iScience 25, 103651 (2022).
Dickson, J. L., Pretty, C. G., Alsweiler, J., Lynn, A. & Chase, J. G. Insulin kinetics and the neonatal intensive care insulin-nutrition-glucose (nicing) model. Math. Biosci. 284, 61–70 (2017).
Eichacker, P. Q. et al. Risk and the efficacy of antiinflammatory agents: Retrospective and confirmatory studies of sepsis. Am. J. Respir. Crit. Care Med. 166(9), 1197–1205 (2002).
Elmasry, N. H. & Elshaarawy, M. K. Hybrid metaheuristic optimized catboost models for construction cost estimation of concrete solid slabs. Sci. Rep. 15(1), 21612 (2025).
Elshaarawy, M. K., Zeleňáková, M. & Armanuos, A. M. Hydraulic performance modeling of inclined double cutoff walls beneath hydraulic structures using optimized ensemble machine learning. Sci. Rep. 15(1), 27592 (2025).
Stahlberg, E. et al. Exploring approaches for predictive cancer patient digital twins: Opportunities for collaboration and innovation. Front. Digit. Health 06, 10077084 (2022).
Finfer, S. Clinical controversies in the management of critically ill patients with severe sepsis: Resuscitation fluids and glucose control. Virulence 5(1), 200–205 (2014).
Fitzgerald, O. et al. Continuous time recurrent neural networks: Overview and benchmarking at forecasting blood glucose in the intensive care unit. J. Biomed. Inform. 146, 104498 (2023).
Ge, Y., Wang, Z., Ma, Y. & Zhang, C. Prognostic value of the glucose-to-albumin ratio in sepsis-related mortality: A retrospective icu study. Diabetes Res. Clin. Pract. 224, 112217 (2025).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016).
Gruver, N., Finzi, M., Qiu, S. & Wilson, A. G. Large language models are zero-shot time series forecasters. Adv. Neural Inf. Process. Syst. 36, 19622–19635 (2024).
Hovorka, R. et al. Five-compartment model of insulin kinetics and its use to investigate action of chloroquine in niddm. Am. J. Physiol.-Endocrinol. Metab. 265(1), E162–E175 (1993).
Hovorka, R., Chassin, L. J., Ellmerer, M., Plank, J. & Wilinska, M. E. A simulation model of glucose regulation in the critically ill. Physiol. Meas. 29(8), 959 (2008).
Ingels, C., Gunst, J. & Berghe, G. Endocrine and metabolic alterations in sepsis and implications for treatment. Crit. Care Clin. 34(1), 81–96 (2018).
Inzucchi, S. E. Management of hyperglycemia in the hospital setting. N. Engl. J. Med. 355(18), 1903–1911 (2006).
Jin, M., et al. Time-llm: Time series forecasting by reprogramming large language models. arXiv preprint. arXiv:2310.01728 (2023).
Kadri, S. S. et al. Estimating ten-year trends in septic shock incidence and mortality in united states academic medical centers using clinical data. Chest 151(2), 278–285 (2017).
Katsoulakis, E. et al. Digital twin for health: A review. NPJ Digit. Med. 7, 77 (2024).
Khanna, V. V. et al. A decision support system for osteoporosis risk prediction using machine learning and explainable artificial intelligence. Heliyon 9(12), e22456 (2023).
Krinsley, J. S. Association between hyperglycemia and increased hospital mortality in a heterogeneous population of critically ill patients. In Mayo Clinic Proceedings, vol 78 1471–1478 (Elsevier, 2003).
Lee, J. & Banerjee, D. Metabolomics and the microbiome as biomarkers in sepsis. Crit. Care Clin. 36(1), 105–113 (2020).
Lin, J. et al. A physiological intensive control insulin-nutrition-glucose (icing) model validated in critically ill patients. Comput. Methods Programs Biomed. 102(2), 192–205 (2011).
Liu, Y. et al. iTransformer: Inverted transformers are effective for time series forecasting. In International Conference on Learning Representations (2024).
Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 30, 4768–4777 (2017).
Mao, F. et al. Machine learning approach for sepsis risk assessment in ischemic stroke patients. J. Intensive Care Med. 40(6), 598–610 (2025).
Mifsud, S., Schembri, E. L. & Gruppetta, M. Stress-induced hyperglycaemia. Br. J. Hosp. Med. 79(11), 634–639 (2018).
Nayak, T. et al. Detection of monkeypox from skin lesion images using deep learning networks and explainable artificial intelligence. Appl. Math. Sci. Eng. 31(1), 2225698 (2023).
Nie, Y., Nguyen, N. H., Sinthong, P., & Kalagnanam, J. A time series is worth 64 words: Long-term forecasting with transformers. In International Conference on Learning Representations (2023).
Palumbo, P., Ditlevsen, S., Bertuzzi, A. & Gaetano, A. Mathematical modeling of the glucose-insulin system: A review. Math. Biosci. 244(2), 69–81 (2013).
Pieralli, F. et al. The classification of hospitalized patients with hyperglycemia and its implication on outcome: Results from a prospective observational study in internal medicine. Intern. Emerg. Med. 11, 649–656 (2016).
Simon, J. D. Understanding Deep Learning (MIT Press, 2023).
Puskarich, M. A. et al. Association between timing of antibiotic administration and mortality from septic shock in patients treated with a quantitative resuscitation protocol. Crit. Care Med. 39(9), 2066–2071 (2011).
Daniel Rittirsch, L., Hoesel, M. & Ward, P. A. The disconnect between animal models of sepsis and human sepsis. J. Leucocyte Biol. 81(1), 137–143 (2007).
Shah, P., Pahari, S., Bhavsar, R. & Sang-Il Kwon, J. Hybrid modeling of first-principles and machine learning: A step-by-step tutorial review for practical implementation. Comput. Chem. Eng. 194, 108926 (2024).
Shen, F. et al. Advanced predictive machine and deep learning models for round-ended cfst column. Sci. Rep. 15(1), 6194 (2025).
Shen, X., Shang, D., Sun, W. & Shuyan, R. Machine learning approach for the prediction of 30-day mortality in patients with sepsis-associated delirium. PLoS ONE 20(4), e0319519 (2025).
Singer, M. et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315(8), 801–810 (2016).
Sirlanci, M. et al. A simple modeling framework for prediction in the human glucose-insulin system. Chaos 33(7), 073150 (2023).
Berghe, G. et al. Intensive insulin therapy in the medical icu. N. Engl. J. Med. 354(5), 449–461 (2006).
Berghe, G. et al. Intensive insulin therapy in critically ill patients. N. Engl. J. Med. 345(19), 1359–1367 (2001).
Vught, L. A. et al. Admission hyperglycemia in critically ill sepsis patients: Association with outcome and host response. Crit. Care Med. 44(7), 1338–1346 (2016).
Wang, L. et al. Optimizing individualized energy delivery for septic patients using predictive deep learning models. Asian Pac. J. Clin. Nutr. 33(3), 348–361 (2024).
Wang, W., Zheng, V. W., Han, Yu. & Miao, C. A survey of zero-shot learning: Settings, methods, and applications. ACM Trans. Intell. Syst. Technol. 10(2), 1–37 (2019).
ZhanJin Wang, A. et al. Prediction of the mortality rate in the intensive care unit for early sepsis patients with combined hypoalbuminemia based on machine learning. Medicine 104(31), e43610 (2025).
Welleck, S., Liu, J., Ximing, L., Hajishirzi, H. & Choi, Y. Naturalprover: Grounded mathematical proof generation with language models. Adv. Neural. Inf. Process. Syst. 35, 4913–4927 (2022).
Williams, L. S. et al. Effects of admission hyperglycemia on mortality and costs in acute ischemic stroke. Neurology 59(1), 67–71 (2002).
Jianfeng, W. et al. Elevation of blood glucose level predicts worse outcomes in hospitalized patients with covid-19: A retrospective cohort study. BMJ Open Diabetes Res. Care 8(1), e001476 (2020).
Lin, X. et al. Association between the development of sepsis and the triglyceride-glucose index in acute pancreatitis patients: A retrospective investigation utilizing the mimic-iv database. BMC Gastroenterol. 25(1), 76 (2025).
Xue, H. & Salim, F. D. Promptcast: A new prompt-based learning paradigm for time series forecasting. IEEE Transactions on Knowledge and Data Engineering (2023).
Focai, Yu. et al. Predicting axial load capacity in elliptical fiber reinforced polymer concrete steel double skin columns using machine learning. Sci. Rep. 15(1), 12899 (2025).
Zeng, A., Chen, M., Zhang, L. & Qiang, X. Are transformers effective for time series forecasting?. In Proceedings of the AAAI Conference on Artificial Intelligence 37, 11121–11128 (2023).
Zhang, Y., & Yan, J. Crossformer: Transformer utilizing cross-dimension dependency for multivariate time series forecasting. In International Conference on Learning Representations (2023).
Zhou, T. et al. Fedformer: Frequency enhanced decomposed transformer for long-term series forecasting. In International Conference on Machine Learning 27268–27286 (PMLR, 2022).
Zhou, T. et al. One fits all: Power general time series analysis by pretrained lm. Adv. Neural. Inf. Process. Syst. 36, 43322–43355 (2023).
Funding
Hua Jiang’s research is supported by Sichuan Provincial Department of Science and Technology (No. 2021YFS0378). Xiang Cao, Jianguo Hou and Qi Wang’s research is partially supported by NSF awards DMS-2038080, OIA-2242812, and an SC GAIN-CRP award. The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
Xiang Cao implemented the basic methodologies, executed the analysis, performed the data analysis and data visualization, developed the concept for the article and wrote the initial draft. Dong Wang collected the clinical data, contributed to the development of the study protocol, provided comments and revisions. Jianguo Hou involved in the overall design of the project, provided comments and revisions. Jue-Xia He collected the clinical data, provided comments and revisions. Jun Li involved in the overall design of the project and provided comments and revisions. Hua Jiang initiated the project, developed the concept for the article, involved in the overall design of the project, contributed to the development of the study protocol, collected the data, contributed important ideas, participated in reviewing and editing of the text, supervised the team, and performed funding acquisition. Qi Wang initiated the project, developed the concept for the article, involved in the overall design of the project, performed the data analysis, drafted the article, contributed important ideas, participated in reviewing and editing of the text, supervised the team, and performed funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Code availability
The underlying code for this study and training/validation datasets is not publicly available but may be made available to researchers on reasonable requests from the corresponding authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, X., Wang, D., He, JX. et al. Machine learning glucose forecasting models for septic patients. Sci Rep 15, 40432 (2025). https://doi.org/10.1038/s41598-025-23057-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-23057-0