Abstract
Glioma is one of the most common and aggressive types of brain tumors, characterized by generally low survival rates. Circular RNAs (circRNAs) have been identified as key players in the development of glioma. Our study aims to investigate the effect of circNRIP1 on glioma progression and elucidate the underlying mechanisms involved. The mRNA expression of circNRIP1, miR-106a-5p, and GPR133 was determined using qRT-PCR. The protein expression of N-cadherin, E-cadherin, fibronectin, vimentin, and GPR133 in glioma cells and tissues was measured through Western blotting and IHC assays. Cell counting kit-8 (CCK-8), wound healing, and transwell assays were used to determine cell proliferation, migration, and invasion, respectively. The dual-luciferase reporter assay was utilized to elucidate the targeting interaction between miR-106a-5p and both circNRIP1 and GPR133. Finally, xenograft models were established to investigate the effects of circNRIP1 in vivo. The expression of circNRIP1 was significantly elevated in glioma cell lines and tissues, and this higher expression correlated with a reduced overall survival rate. Downregulation of circNRIP1 had been shown to inhibit the invasion, migration, proliferation, and epithelial-mesenchymal transition (EMT) of glioma cells and attenuate tumor growth in vivo. circNRIP1 functions as a molecular sponge for miR-106a-5p, consequently elevating the expression of GPR133. Additionally, upregulation of GPR133 could counteract the inhibition of the malignant phenotype of glioma cells caused by circNRIP1 knockdown. circNRIP1 knockdown suppresses glioma progression by elevating miR-106a-5p levels and simultaneously reducing GPR133 expression. The identified circNRIP1/miR-106a-5p/GPR133 axis could potentially be a therapeutic target for glioma.
Similar content being viewed by others
Introduction
Glioma is one of the most prevalent and aggressive primary malignant brain tumors, making up approximately 80% of all primary central nervous system (CNS) tumors1. The aggressive growth pattern and tendency to recur pose significant challenges for clinical treatment2. Although multimodal treatment strategies—including surgical resection, radiotherapy, chemotherapy, molecular targeted therapy, immunotherapy, and electric field therapy—have made notable advancements, effective management of gliomas remains elusive. The prognosis for glioma patients remains bleak, with median survival times being unacceptably brief and a less than 5% 5-year survival rate3,4,5. This highlights the urgent requirement to discover novel diagnostic biomarkers and dependable therapeutic targets for glioma.
Circular RNAs (circRNAs) are a form of non-coding RNA molecule characterized by their closed-loop structure created through back-splicing. These circRNAs exhibit remarkable stability due to their resistance to exonuclease activity, surpassing that of linear mRNA6,7. Functioning as microRNA (miRNA) sponges, they competitively bind miRNAs, thereby regulating the expression of oncogenes and tumor suppressor genes at both transcriptional and post-transcriptional levels. Consequently, circRNAs have emerged as key factors in cancer progression8,9. Recent studies have increasingly highlighted the significant role of circRNAs in tumor development, progression, metastasis, and prognosis across various cancer types, including hepatocellular carcinoma10, lung cancer11, colon cancer12, and glioma13. Specifically, their abnormal expression in gliomas has positioned them as a hopeful therapeutic target for gliomas. The circular RNA known as Hsa_circ_0002711, also referred to as circNRIP1, is formed through the head-to-tail splicing of exons 2 and 3 of the NRIP1 gene14. Previous research has demonstrated its role in regulating malignant biological activities in various types of cancer, including papillary thyroid carcinoma15, gastric cancer16, ovarian cancer17, esophageal squamous cell carcinoma18, and non-small cell lung cancer19. However, the specific role and mechanism of circNRIP1 in gliomas remain elusive.
MicroRNAs (miRNAs) are a class of non-coding RNAs capable of regulating gene expression post-transcriptionally. This regulation occurs through their binding to the 3ʹ-untranslated region (3ʹ-UTR) of target mRNAs, exhibiting either partial or complete complementarity. This interaction has the potential to hinder translation or induce degradation of the target mRNA, ultimately modulating gene expression20,21. New evidence indicates that circRNAs and miRNAs are crucial in the malignant progression of gliomas. GPR133 (ADGRD1) is part of the adhesion G-protein-coupled receptor (aGPCR) family22, and it has been shown that GPR133 is essential for the growth of tumors in glioblastoma multiforme (GBM). Silencing GPR133 results in significant inhibition of tumor growth in vivo23,24. Nevertheless, the upstream regulatory mechanisms that control GPR133 expression in gliomas remain elusive.
Interestingly, bioinformatics analyses have revealed potential binding sites between miR-106a-5p and both circNRIP1 and the 3ʹ-UTR region of GPR133. This observation hints at a possible binding interaction between miR-106a-5p and either circNRIP1 or GPR133. Our earlier research showed that miR-106a-5p can bind to the 3’-UTR of GPR133 in glioma cells. Considering this, we hypothesize that circNRIP1 might regulate GPR133 expression by serving as a sponge for miR-106a-5p.
In this study, it was discovered that circNRIP1 exerts an oncogenic role in both glioma tissues and cell lines. It functions as a natural sponge that interacts with miR-106a-5p, thereby affecting the expression of GPR133. These findings highlight the circNRIP1/miR-106a-5p/GPR133 pathway in glioma and indicate its potential as a therapeutic target for glioma treatment.
Materials and methods
Patients and tissue samples
Fifty tissue samples were collected from the Department of Neurosurgery of Suining Central Hospital (Sichuan, China), spanning from January 2019 to December 2021, including 40 glioma samples collected during surgical procedures, along with 10 samples of nontumorous brain tissues (NBTs) from patients undergoing surgery for severe traumatic brain injury. All samples were promptly frozen in liquid nitrogen and maintained at −80 °C. Histologic grading was performed according to the World Health Organization (WHO) classification (5th Edition)25. The group consisted of 5 patients with grade 2 gliomas, 11 with grade 3 gliomas, and 24 with grade 4 gliomas. Each patient was required to provide written informed consent for the use of their samples in the research. Approval from the Ethics Committee of Suining Central Hospital was also obtained before proceeding with the study.
Cell culture and transfection
The human glioma cell line LN229 (Cat# CRL-2611) was obtained from the American Type Culture Collection (ATCC, Manassas, VA). Additionally, the U251 MG (Cat# GDC0093) and U87 MG (Cat# GDC0628) cell lines were procured from the China Center for Type Culture Collection (CCTCC, Wuhan, China). Normal human astrocytes (NHAs) (Cat# HTX2408) were acquired from Otwo Biotech Inc. (Shenzhen, China). All cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) enriched with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY) and incubated at 37 °C with 5% carbon dioxide and 95% humidity. The overexpression plasmids for GPR133 (pEX1-GPR133) and the empty vector were all sourced from GenePharma (Shanghai, China). Small interfering RNA (siRNA) targeting circNRIP1, miR-106a-5p mimic, and their respective negative controls (si-NC, mimic-NC) were purchased from Sangon Biotech (Shanghai, China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The lentivirus vectors, either containing circNRIP1 short hairpin RNA (shRNA) or empty, were crafted and synthesized by Sangon Biotech (Shanghai, China). U87 cells were infected with a lentivirus vector and subsequently selected using 2 µg/ml puromycin (P8230-25, Solarbio, Beijing, China) over 2–3 weeks to generate stable cell lines.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells or snap-frozen glioma tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Subsequently, 2 µg of the extracted RNA underwent reverse transcription to generate complementary DNA (cDNA). For mRNA reverse transcription, we employed the ReverTra Ace® qPCR RT Master Mix with gDNA Remover kit (FSQ-301, TOYOBO, Osaka, Japan), while miRNA reverse transcription was achieved using the ReverTra Ace® qPCR RT Kit (FSQ-101, TOYOBO, Osaka, Japan). Next, qRT-PCR was conducted using SYBR® Green Real-time PCR Master Mix (QPK-201, TOYOBO, Osaka, Japan) on an Applied Biosystems device (QuantStudio 3, Thermo Fisher Scientific, Waltham, MA). β-actin served as the internal reference for normalizing circNRIP1 or GPR133 qRT-PCR results, and U6 small nuclear RNA was used as the internal reference for miR-106a-5p. The relative expression levels were determined via the 2−ΔΔCq method26. The primer sequences can be found in Table 1.
Immunohistochemistry assay
The tissue samples were embedded in paraffin, cut into 4-µm-thick sections, deparaffinized, and rehydrated. Microwave treatment in citrate buffer was utilized for antigen extraction. Subsequently, the endogenous peroxidase activity was blocked with a 30 g/l hydrogen peroxide solution for 5 min after cooling. The sections were subsequently sealed with 5% goat serum for 1 h. They were then stained using primary antibodies, specifically GPR133 (DF4947, Affinity Biosciences, Changzhou, China, 1:300), Ki67 (AB15580, Abcam, Boston, MA, 1:300), N-cadherin (AB76011, Abcam, Boston, MA, 1:200), Fibronectin (AF5335, Affinity Biosciences, Changzhou, China, 1:200) and Vimentin (AF7013, Affinity Biosciences, Changzhou, China, 1:200). These primary antibodies were incubated overnight at 4 °C. The sections were subsequently exposed to HRP-conjugated goat anti-rabbit IgG secondary antibody (H33654, Ventana, Export, PA, 1:200) for 30 min at room temperature. Afterward, the DAB Detection Kit (Ventana, Export, PA) was employed to stain the sections. Microscopic examination (EVOS M5000, Invitrogen, Carlsbad, CA) was conducted on five randomly chosen fields of view. Two pathologists independently reviewed all sections, without access to clinical information. They evaluated staining intensity and the proportion of stained cells within five randomly chosen fields. If more than 50% of the cells in a sample were stained, it was designated as a high-expression sample, otherwise, it was classified as a low-expression sample.
Dual-luciferase reporter assay
The wild-type (WT) sequence of circNRIP1, which contains the binding sites for miR-106a-5p (5’-CTATCTGTGTTCCTAGAGCACTTTT), and the mutant-type (MUT) sequence with the binding site mutated to 5’-GATTGACTGTACCATGTCGTGAAAT, were cloned into the pmirGLO dual-luciferase reporter vectors (GeneCreate Bioengineering, Wuhan, China) to generate the pmirGLO-WT-circNRIP1 and pmirGLO-MUT-circNRIP1 vectors, respectively. Similarly, fragments of the wild-type (WT) 3′-UTR of GPR133 containing the binding sites for miR-106a-5p (5’-GCACTTT) and the mutant-type (MUT) Fragments with the binding sites mutated to 5’-CGTGAAA were inserted into the pmirGLO vectors, resulting in the pmirGLO-WT-GPR133 and pmirGLO-MUT-GPR133 vectors, respectively. These recombinant reporter vectors were then co-transfected into U87 and LN229 cells, accompanied by either the miR-106a-5p mimic or the mimic-NC, employing Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) for each transfection process. Following forty-eight hours of transfection, the cells were subjected to luciferase activity analysis using a dual luciferase reporter kit (E1910, Promega, Madison, WI, USA). The relative light unit (RLU) values obtained reflected luciferase activity. Normalized luciferase activity was determined by calculating the ratio of firefly luciferase RLU value to Renilla luciferase RLU value.
Cell proliferation assay
Cell viability was assessed using the CCK-8 assay. In summary, transfected cells were seeded in 96-well plates (5 × 103/well) and incubated at 37 °C for 24, 48, and 72 h, respectively. Subsequently, cells were treated with a new medium containing 10% CCK-8 solution (KE1090-500T, Kingmorn, Shanghai, China) and further incubated at 37 °C for 4 h. The optical density (OD) of each sample was then measured at 450 nm.
Wound healing assay
After 24 h of transfection, the cells were seeded into 6-well plates. Scratches were created using 200-µl sterile pipette tips. The cells were then incubated in FBS-free DMEM at 37 °C for 36 h. Following this, five random fields of view were selected for microscopic examination, and the data were analyzed using ImageJ version 1.53k software (NIH, Bethesda, MD).
Transwell assay
Transwell membranes in the upper chambers (3422, Corning, New York, NY) were pre-coated with Matrigel (356234, Corning, New York, NY). After transfection for 48 h, cells were seeded into the upper chamber at a density of 5 × 104 cells/well, using serum-free DMEM. The lower chamber was then filled with DMEM supplemented with 10% FBS. Following an incubation period at 37 °C for 24 h, cells that had invaded the lower surface were immobilized with 4% paraformaldehyde and colored with 0.2% crystal violet. Images of 5 randomly selected fields were captured using a microscope (EVOS M5000, Invitrogen, Carlsbad, CA) for each sample. The total number of invasive cells was quantified using ImageJ version 1.53k software (NIH, Bethesda, MD).
Western blotting assay
Firstly, total protein was extracted from the cells using ice-cold RIPA lysis buffer (R0010, Solarbio, Beijing, China) and subsequently isolated using 10% polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer to polyvinylidene difluoride (PVDF) membranes. After being blocked with 5% bovine serum albumin (Kingmorn, Shanghai, China) for 1 h, the membranes were incubated overnight at 4 °C with the specified primary antibodies: GPR133 (DF4947, Affinity Biosciences, Changzhou, China, 1:1000), E-cadherin (AF0131, Affinity Biosciences, Changzhou, China, 1:1000), N-cadherin (AB76011, Abcam, Boston, MA, 1:1000), Vimentin (AF7013, Affinity Biosciences, Changzhou, China, 1:1000), Fibronectin (AF5335, Affinity Biosciences, Changzhou, China, 1:1000), and GAPDH (T0004, Affinity Biosciences, Changzhou, China, 1:10000). Subsequently, the membranes were washed three times and incubated for 1 h with HRP-conjugated anti-mouse (SA00001-1, Proteintech, Wuhan, China, 1:2000) or anti-rabbit (SA00001-2, Proteintech, Wuhan, China, 1:2000) secondary antibodies. Finally, an enhanced chemiluminescence system (Amersham Imager 680, Cytiva, USA) was utilized for visualizing the protein bands, and their grayscale was analyzed using ImageJ version 1.53k software (NIH, Bethesda, MD).
Xenograft assay
Twelve male BALB/c nude mice (five weeks old, 16.8 ± 1.2 g) were purchased from Cavens Model Animal Co., Ltd. (Changzhou, China) and housed in SPF-class barrier feeding conditions. To conduct xenograft experiments, U87 cells stably transfected with sh-circNRIP1 or sh-NC were subcutaneously injected into the armpits of mice (2 × 106 cells/mouse, 6 mice/group). Tumor growth was tracked and measured using a vernier caliper every two days. The tumor volume (V) was calculated by applying the formula: volume (mm3) = (length × width2)/2. At the end of 28 days, mice were humanely euthanized using a combination of 3% sodium pentobarbital solution and high-concentration carbon dioxide, and subcutaneous tumors were dissected and weighed. Subsequently, immunohistochemistry (IHC) analysis was performed on the tumor samples. The Suining Central Hospital Ethics Committee approved the animal experiments, and all experiments were performed in accordance with relevant guidelines and regulations.
Statistical analysis
Each experiment was conducted three times, and the data were presented as mean ± SD. Statistical analysis was performed using SPSS version 26.0 software (IBM, Armonk, NY). An unpaired t-test was utilized to compare the differences between the two groups. One-way analysis of variance (ANOVA) and Tukey’s post hoc test were employed to analyze differences among multiple groups. Kaplan-Meier survival curves were used to assess overall survival rates, while log-rank tests were conducted for analysis. Linear regression was applied to examine the correlation between the expression levels of miR-106a-5p and circNRIP1 in glioma tissues. Results were considered statistically significant at p < 0.05.
Results
circNRIP1 is upregulated in glioma tissues and cell lines
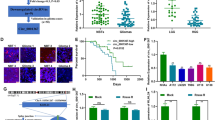
First, we conducted qRT-PCR to analyze the expression level of circNRIP1 in glioma tissues and NBTs. The results revealed that circNRIP1 expression was significantly upregulated in glioma tissues compared to NBTs (Fig. 1A). Subsequently, the expression of circNRIP1 in glioma cell lines (U251, U87 and LN229) were markedly increased compared with NHAs (Fig. 1B). Furthermore, evaluation of circNRIP1 expression across glioma samples of different histological grades revealed significantly higher expression in grade 3/4 samples relative to grade 2 samples (Fig. 1C). In addition, we assessed the correlation between circNRIP1 expression and patient prognosis. Kaplan-Meier survival analysis indicated that patients with elevated circNRIP1 levels had a poorer overall survival (Fig. 1D). Collectively, these findings implicated that circNRIP1 was associated with the malignant progression of glioma.
Expression level of circNRIP1 in glioma and its relationship with the survival duration of glioma patients. (A) qRT-PCR was conducted to compare the expression of circNRIP1 between glioma tissues and NBT samples. (B) The level of circNRIP1 in U251, U87, LN229, and NHA cells was detected by qRT-PCR. (C) The level of circNRIP1 in glioma tissues of different grades was measured using qRT-PCR. (D) Kaplan–Meier survival analysis revealed the relationship between circNRIP1 expression levels and the OS of glioma patients. The results are presented as the mean ± SD of three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
circNRIP1 knockdown suppresses the proliferation of glioma cells
First, we sought to verify the efficiency of circNRIP1 knockdown. We utilized three siRNAs targeting circNRIP1 to diminish its expression in glioma cells. Our findings revealed that si-circNRIP1-1 exhibited the highest knockdown efficiency (Fig. 2A), therefore, it was selected for further experiments. Following this, we performed a CCK-8 assay to evaluate the effect of circNRIP1 on cell proliferation. The data revealed that reducing circNRIP1 significantly hampered the growth of U87 and LN229 cell lines in comparison to the control group (Fig. 2F).
Downregulation of circNRIP1 suppresses the proliferation, migration, and invasion of glioma cells. (A) The knockdown effectiveness of circNRIP1 was determined by qRT–PCR. (B and C) Cell migration was determined by wound healing assay in U87 and LN229 cells. The scale bars are 600 μm. (D and E) Cell invasion was determined by transwell assay in U87 and LN229 cells. The scale bars are 400 μm. (F) Cell proliferation was detected by CCK-8 assay in U87 and LN229 cells. (G and H) Protein levels of E-cadherin, N-cadherin, Fibronectin, and Vimentin were detected by western blotting. Data are presented as the mean ± SD of three independent experiments. (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
circNRIP1 knockdown suppresses the invasion and migration of glioma cells
We performed wound healing, transwell, and western blotting assays to investigate the impact of circNRIP1 on the migration and invasion of glioma cells. Our results, as depicted in Fig. 2B and C, reveal that decreasing circNRIP1 significantly hinders cell migration in both cell lines when compared to the control group. Likewise, the elimination of circNRIP1 led to a marked decrease in the invasive cell count in both cell lines, as illustrated in Fig. 2D and E.
According to previous research, epithelial-to-mesenchymal transition (EMT) is a pivotal molecular event in tumor invasion, specifically including glioblastoma (GBM)27. During EMT, tumor cells change adhesion properties and harness developmental mechanisms to gain migratory and invasive capabilities28. Therefore, we conducted a western blotting assay to investigate whether circNRIP1 affects the EMT-like process in glioma cells. Our findings revealed that downregulating circNRIP1 significantly reduced the protein levels of mesenchymal markers (N-cadherin, Fibronectin, and Vimentin) while increasing the level of the epithelial marker E-cadherin (Fig. 2G and H). Collectively, these observations suggested that circNRIP1 knockdown diminished the migratory and invasive capabilities of glioma cell lines by modulating the EMT-like pathway.
circNRIP1 functions as a sponge for miR-106a-5p
Firstly, we employed the online prediction database ENCORI (https://rnasysu.com/encori/index.php)29 to identify potential miRNAs targeted by circNRIP1. Our findings indicated that miR-106a-5p emerged as the primary candidate target for circNRIP1 (Fig. 3A). The luciferase reporter assay was conducted to verify the direct binding between miR-106a-5p and circNRIP1. The results revealed that the miR-106a-5p mimic significantly reduced the relative luciferase activity of circNRIP1-WT in both U87 and LN229 cells, whereas circNRIP1-MUT remained unaffected (Fig. 3C). Following this, qRT-PCR analysis demonstrated that the downregulation of circNRIP1 markedly increased the expression of miR-106a-5p in these cell lines (Fig. 3D). Moreover, Pearson’s correlation analysis indicated a negative correlation between the expression levels of circNRIP1 and miR-106a-5p in glioma tissues (Fig. 3E). Taken together, these results indicated that circNRIP1 acted as a sponge to bind miR-106a-5p at its “seed” region, thereby suppressing its expression and function.
The binding relationship between circNRIP1 and miR-106a-5p, as well as GPR133 and miR-106a-5p. (A and B) The predicted binding sites between circNRIP1 and miR-106a-5p, as well as GPR133 3′UTR and miR-106a-5p were exhibited. (C) The interaction between circNRIP1 and miR-106a-5p was confirmed by dual-luciferase reporter assay. (D) The expression of miR-106a-5p in U87 and LN229 cells transfected with si-circNRIP1 or si-NC was detected by qRT-PCR. (E) The correlation between miR-106a-5p and circNRIP1 expression in glioma tissues. (F) The interaction between GPR133 and miR-106a-5p was confirmed by dual-luciferase reporter assay. (G) The mRNA level of GPR133 in U87 and LN229 cells transfected with miR-106a-5p mimic or mimic-NC was detected by qRT-PCR. (H and I) The protein level of GPR133 in U87 and LN229 cells transfected with miR-106a-5p mimic or mimic-NC was detected by western blotting. Data are presented as the mean ± SD of three independent experiments. (**p < 0.01, ***p < 0.001 and ****p < 0.0001).
GPR133 is targeted by miR-106a-5p
To delve deeper into the mechanism by which miR-106a-5p affects glioma cells, we initially utilized the online gene prediction database TargetScan (http://www.targetscan.org/vert_72/)30 to pinpoint potential downstream genes regulated by miR-106a-5p. In our previous study, we discovered that GPR133 is directly targeted by miR-106a-5p and contributes to enhancing the malignant phenotype of glioma cells31. Curiously, both the 3ʹ-UTR of GPR133 and circNRIP1 share a comparable “seed” region for miR-106a-5p binding (Fig. 3B). This prompted us to focus our further investigations on GPR133. Using luciferase reporter assays, we observed that miR-106a-5p mimic notably reduced the luciferase activity of GPR133-WT in U87 and LN229 cells, but had no impact on GPR133-MUT luciferase activity (Fig. 3F). Subsequent qRT-PCR and western blotting assays demonstrated that the overexpression of miR-106a-5p resulted in a clear reduction in both the mRNA (Fig. 3G) and protein levels (Fig. 3H and I) of GPR133 in U87 and LN229 cells.
circNRIP1 exerts its effect on malignant phenotypes of glioma cells via the miR‑106a‑5p/GPR133 axis
To investigate the impact of circNRIP1 on glioma cells through the regulation of GPR133, we performed rescue experiments. Results from wound healing (Fig. 4A and B) and transwell assays (Fig. 4C and D) demonstrated that the reduced migration and invasion abilities of U87 and LN229 cells resulting from circNRIP1 knockdown were notably restored by the upregulation of GPR133. Next, the CCK-8 assay (Fig. 4E) confirmed that the proliferative capacity of these cells decreased with the downregulation of circNRIP1, an effect reversed by GPR133 overexpression. Western blotting assay further confirmed that the protein levels of EMT markers, significantly regulated by circNRIP1 downregulation, could be notably reversed through GPR133 overexpression (Fig. 4F and G). These results implicated that circNRIP1 exerted its oncogenic functions in glioma cells through the miR-106a-5p/GPR133 axis.
Overexpression of GPR133 reversed the effect of circNRIP1 knockdown on proliferation and invasion of glioma cells. (A and B) Cell migration was detected by wound healing assay in U87 and LN229 cells. The scale bars are 600 μm. (C and D) Cell invasion was detected by transwell assay in U87 and LN229 cells. The scale bars are 400 μm. (E) Cell proliferation was detected by CCK-8 assay in U87 and LN229 cells. (F and G) Protein levels of GPR133, N-cadherin, Fibronectin, Vimentin, and E-cadherin were detected by western blotting. Data are presented as the mean ± SD of three independent experiments. (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
circNRIP1 knockdown suppresses glioma cell growth in vivo
To investigate the impact of circNRIP1 on the proliferation and migration capabilities of glioma cells in vivo, we subcutaneously injected nude mice with U87 cells stably transfected with either sh-circNRIP1 or sh-NC. Our findings revealed that the tumors developed in the sh-circNRIP1 group were notably smaller (Fig. 5A and B) and lighter (Fig. 5C) than those in the control group. This confirmed that suppressing circNRIP1 expression substantially impeded glioma cell growth in vivo. Moreover, the immunohistochemistry (IHC) results demonstrated that the expression levels of Fibronectin, N-cadherin, Vimentin, GPR133, and Ki67 were significantly reduced in tumors where circNRIP1 was downregulated (Fig. 5D). These findings indicated that inhibiting circNRIP1 could obstruct both the proliferation and EMT of glioma cells in vivo.
Downregulation of circNRIP1 inhibits glioma cell growth in vivo. (A) Image of subcutaneous xenograft tumors (n = 6 for each group). (B) Tumor volume was measured every 2 days. (C) Tumor weight was examined 28 days after injection. (D) Ki67, Fibronectin, N-cadherin, Vimentin, GPR133 IHC staining of xenograft tumors. The scale bars are 100 μm. (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
Discussion
Previous studies have indicated that dysregulated circRNA expression is a crucial factor in cancer progression. Specifically, Fu et al. reported that circNRIP1 was prominently expressed in papillary thyroid cancer, and its augmented levels in vitro facilitated the aggressive growth of the disease32. Likewise, Li et al. noted high levels of circNRIP1 in cervical cancer, demonstrating that its upregulation markedly boosted the invasion and migration abilities of cervical cancer cells33. Furthermore, Zhou et al. identified that circNRIP1 promoted the malignant progression of esophageal squamous cell carcinoma in vitro18. However, the specific biological role and mode of action of circNRIP1 in gliomas have not been determined yet. Our study has discovered that circNRIP1 is significantly upregulated in both glioma tissues and cell lines, and its elevated expression is correlated with a poorer prognosis for glioma patients. Functional experiments have shown that reducing circNRIP1 expression significantly restrains glioma cell proliferation, invasion, migration, and EMT. Additionally, xenograft tumor experiments have indicated that suppressing circNRIP1 impedes the growth of glioma cells in vivo. These findings implicate circNRIP1 as an oncogenic factor in glioma progression, highlighting its potential as a valuable diagnostic and prognostic biomarker for glioma patients.
Many studies have demonstrated that circRNAs play crucial biological roles in the development of gliomas by functioning as miRNA sponges. Specifically, circ-HIPK3 has been found to enhance the proliferation and invasion of glioma cells via the miR-654/IGF2BP3 pathway34, while circ_0008344 could promote glioma advancement and angiogenesis by modulating the miR-638/SZRD1 signaling pathway35. Additionally, Guo et al. have revealed that circEGFR could restrict the malignant progression of gliomas through the regulation of the miR-183-5p/TUSC2 signaling pathway36. In the present study, it was noted that miR-106a-5p was a target miRNA regulated by the circNRIP1 sponge mechanism. The glioma cells displayed a notable decrease in the expression of miR-106a-5p upon an increase in circNRIP1 levels. Furthermore, qRT-PCR analysis revealed a negative relationship between circNRIP1 and miR-106a-5p.
MiRNAs are crucial in the advancement of gliomas. In recent years, several miRNAs have been recognized as either oncogenes or anti-oncogenes in gliomas. Specifically, elevated levels of miR-10b, miR-17 ~ 92 cluster, miR-125b, and miR-145 have been linked to enhanced glioma proliferation, invasion, and angiogenesis. Conversely, heightened expression of miR-29b, miR-124, miR-125a, miR-218, and miR-195 have been shown to suppress the malignant phenotype of glioma cells37. Previous research has demonstrated that miR-106a-5p could hinder the advancement of renal cell carcinoma38, hepatocellular carcinoma39, and glioma40. In a similar vein, our previous study has further supported these findings, revealing that miR-106a-5p can impede the growth, invasion, migration, and EMT of glioma cells by modulating GPR133 expression31. In line with these earlier results, our current study has confirmed that miR-106a-5p specifically binds to GPR133 and negatively regulates its expression level.
Our previous research has indicated that GPR133 is elevated in gliomas, enhancing the proliferation, invasion, migration, and EMT of glioma cells. In our latest study, we discovered that GPR133 competes with circNRIP1 for the same binding site of miR-106a-5p. Through rescue experiments, we demonstrated that the overexpression of GPR133 significantly counteracted the inhibitory effects of circNRIP1 knockdown on the proliferation, migration, invasion, and EMT of glioma cells. These findings suggest that circNRIP1 facilitates the malignant progression of gliomas via the miR-106a-5p/GPR133 axis. Nevertheless, there are some limitations in this study. The clinical sample size is relatively modest. Expanding the cohort or using public data sets would enhance the confidence of the findings. Second, using orthotopic xenograft models that more closely mimic the microenvironment of glioma growth would make the results more convincing. In addition, the specific mechanism by which circNRIP1 promotes glioma progression may involve more miRNAs or proteins, and its downstream signaling pathways need to be further explored.
Conclusion
Here, we have discovered that circNRIP1 is a newly identified circRNA that is elevated in glioma. It contributes to the aggressive progression of glioma by modulating the miR-106a-5p/GPR133 axis. The possibility of targeting circNRIP1 emerges as a potential anti-tumor strategy in gliomas, offering potential benefits for patients with advanced gliomas.
Data availability
The data generated in the present study may be requested from the corresponding author.
References
Schwartzbaum, J. A. et al. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2, 494–503. https://doi.org/10.1038/ncpneuro0289 (2006). quiz 1 p following 516.
Koo, S. et al. Serial selection for invasiveness increases expression of miR-143/miR-145 in glioblastoma cell lines. BMC Cancer. 12, 143. https://doi.org/10.1186/1471-2407-12-143 (2012).
Stephan, G., Ravn-Boess, N. & Placantonakis, D. G. Adhesion G protein-coupled receptors in glioblastoma. Neurooncol Adv. 3, vdab046. https://doi.org/10.1093/noajnl/vdab046 (2021).
Aggarwal, P. et al. Pediatric versus adult high grade glioma: immunotherapeutic and genomic considerations. Front. Immunol. 13, 1038096. https://doi.org/10.3389/fimmu.2022.1038096 (2022).
Binder, D. C., Davis, A. A. & Wainwright, D. A. Immunotherapy for cancer in the central nervous system: current and future directions. Oncoimmunology 5, e1082027. https://doi.org/10.1080/2162402X.2015.1082027 (2015).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. https://doi.org/10.1038/s41576-019-0158-7 (2019).
Qu, S. et al. Circular RNA. A new star of noncoding RNAs. Cancer Lett. 365, 141–148. https://doi.org/10.1016/j.canlet.2015.06.003 (2015).
Yang, G., Zhang, Y. & Yang, J. Identifcation of potentially functional CircRNA-miRNA-mRNA regulatory network in gastric carcinoma using bioinformatics analysis. Med. Sci. Monit. 25, 8777–8796. https://doi.org/10.12659/MSM.916902 (2019).
Su, M. et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer. 18, 90. https://doi.org/10.1186/s12943-019-1002-6 (2019).
Zhang, X. Y. et al. CircPIAS1 promotes hepatocellular carcinoma progression by inhibiting ferroptosis via the miR-455-3p/NUPR1/FTH1 axis. Mol. Cancer. 23, 113. https://doi.org/10.1186/s12943-024-02030-x (2024).
Li, H. et al. circRNA-CPA4 regulates cell proliferation and apoptosis of non-small cell lung cancer via the miR-1183/PDPK1 axis. Biochem. Genet. 62, 4087–4102. https://doi.org/10.1007/s10528-023-10641-0 (2024).
Lun, J. et al. Circular RNA circHIPK2 inhibits colon cancer cells through miR-373-3p/RGMA axis. Cancer Lett. 593, 216957. https:// (2024).
Pan, Z. et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer. 21, 16. https://doi.org/10.1186/s12943-021-01485-6 (2022).
Du, Z. CircNRIP1: an emerging star in multiple cancers. Pathol. Res. Pract. 241, 154281. https://doi.org/10.1016/j.prp.2022.154281 (2023).
Fu, L. et al. CircNRIP1 exerts oncogenic functions in papillary thyroid carcinoma by sponging miR-653-5p and regulating PBX3 expression. J Oncol 2022: 2081501. (2022). https://doi.org/10.1155/2022/2081501
Liang, L. & Li, L. Down-regulation of circNRIP1 promotes the apoptosis and inhibits the migration and invasion of gastric cancer cells by miR-182/ROCK1 axis. Onco Targets Ther. 13, 6279–6288. https://doi.org/10.2147/OTT.S221633 (2020).
Li, M., Cai, J., Han, X. & Ren, Y. Downregulation of circNRIP1 suppresses the Paclitaxel resistance of ovarian cancer via regulating the miR-211-5p/HOXC8 axis. Cancer Manag Res. 12, 9159–9171. https://doi.org/10.2147/CMAR.S268872 (2020).
Zhou, S. et al. circ_NRIP1 is oncogenic in malignant development of esophageal squamous cell carcinoma (ESCC) via miR-595/SEMA4D axis and PI3K/AKT pathway. Cancer Cell. Int. 21, 250. https://doi.org/10.1186/s12935-021-01907-x (2021).
D’Ambrosi, S. et al. The analysis of Platelet-Derived circrna repertoire as potential diagnostic biomarker for non-small cell lung cancer. Cancers (Basel). 13, 4644. https://doi.org/10.3390/cancers13184644 (2021).
Pu, M. et al. Regulatory network of MiRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 76, 441–451. https://doi.org/10.1007/s00018-018-2940-7 (2019).
O’Brien, J., Hayder, H., Zayed, Y. & Peng, C. Overview of MicroRNA Biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne). 9, 402. https://doi.org/10.3389/fendo.2018.00402 (2018).
Hamann, J. et al. International union of basic and clinical pharmacology. XCIV. Adhesion g protein-coupled receptors. Pharmacol. Rev. 67, 338–367. https://doi.org/10.1124/pr.114.009647 (2015).
Bayin, N. S. et al. GPR133 (ADGRD1), an adhesion G-protein-coupled receptor, is necessary for glioblastoma growth. Oncogenesis 5, e263. https://doi.org/10.1038/oncsis.2016.63 (2016).
Frenster, J. D. et al. Expression profiling of the adhesion G protein-coupled receptor GPR133 (ADGRD1) in glioma subtypes. Neurooncol Adv. 2 (1), vdaa053. https://doi.org/10.1093/noajnl/vdaa053 (2020).
WHO Classification of Tumours Editorial Board. World Health Organization Classification of Tumours of the Central Nervous System 5th edn (International Agency for Research on Cancer, 2021).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods 25 (4), 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Ma, Y. S. et al. DRR1 promotes glioblastoma cell invasion and epithelial-mesenchymal transition via regulating AKT activation. Cancer Lett. 423, 86–94. https://doi.org/10.1016/j.canlet.2018.03.015 (2018).
Yilmaz, M. & Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 28, 15–33. https://doi.org/10.1007/s10555-008-9169-0 (2009).
Li, J. H. et al. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–97. https://doi.org/10.1093/nar/gkt1248 (2014).
Agarwal, V. et al. Predicting effective MicroRNA target sites in mammalian mRNAs. Elife 4, e05005. https://doi.org/10.7554/eLife.05005 (2015).
Zhang, S. Y., Zhang, Y. & Sun, X. Targeting GPR133 via miR-106a-5p inhibits the proliferation, invasion, migration and epithelial-mesenchymal transition (EMT) of glioma cells. Int. J. Neurosci. 134, 991–1002. https://doi.org/10.1080/00207454.2023.2201873 (2024).
Fu, L. et al. CircNRIP1 exerts oncogenic functions in papillary thyroid carcinoma by sponging miR-653-5p and regulating PBX3 expression. J. Oncol. 2022, 2081501. https://doi.org/10.1155/2022/2081501 (2022).
Li, X. et al. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell. Death Dis. 11, 399. https://doi.org/10.1038/s41419-020-2607-9 (2020).
Jin, P. et al. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem. Biophys. Res. Commun. 503, 1570–1574. https://doi.org/10.1016/j.bbrc.2018.07.081 (2018).
Deng, L., Gong, K. & Wang, G. Hsa_circ_0008344 promotes glioma tumor progression and angiogenesis presumably by regulating miR-638/SZRD1 pathway. Neurotox. Res. 40, 825–836. https://doi.org/10.1007/s12640-022-00504-8 (2022).
Guo, Q. et al. Circ-EGFR functions as an inhibitory factor in the malignant progression of glioma by regulating the miR-183-5p/TUSC2 axis. Cell. Mol. Neurobiol. 42, 2245–2256. https://doi.org/10.1007/s10571-021-01099-y (2022).
Møller, H. G. et al. A systematic review of MicroRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Mol. Neurobiol. 47, 131–144. https://doi.org/10.1007/s12035-012-8349-7 (2013).
Ma, J. et al. miR-106a-5p functions as a tumor suppressor by targeting VEGFAin renal cell carcinoma. Dis Markers 2020:8837941. (2020). https://doi.org/10.1155/2020/8837941
Xu, Y. et al. ROS-Related MiRNAs regulate immune response and chemoradiotherapy sensitivity in hepatocellular carcinoma by comprehensive analysis and experiment. Oxid. Med. Cell. Longev. 2022, 4713518. https://doi.org/10.1155/2022/4713518 (2022).
Zhou, H. et al. The MicroRNA-106a/20b strongly enhances the antitumour immune responses of dendritic cells pulsed with glioma stem cells by targeting STAT3. J Immunol Res 2022:9721028. (2022). https://doi.org/10.1155/2022/9721028
Acknowledgements
We want to acknowledge the service provided by the Basic Laboratory of Suining Central Hospital.
Author information
Authors and Affiliations
Contributions
Haiping Shi and Shiyuan Zhang conceived and designed the experiments. Shiyuan Zhang, Wei Huang, Jimin He, and Changchen Cai conducted a series of experiments, including Dual-luciferase reporter assay, IHC, qRT-PCR, Western blotting, cell experiments, and xenograft assay. Shiyuan Zhang and Jimin He performed the statistical analysis and wrote the manuscript text. Haiping Shi revised the manuscript. All authors confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Ethics Committee of the Suining Central Hospital (Suining, China) (Grant No. LLSNCH20220019). All experiments on tissue samples were conducted in strict accordance with the Declaration of Helsinki’s ethical standards. Patients signed the informed consent form before their participation. The animal experiment was conducted following the ARRIVE guidelines and was approved by the Animal Ethics Committee of Suining Central Hospital.
Consent to participate
Written informed consent for the use of tumor samples in this study was obtained from all participants.
Consent for publication
All the patients provided written informed consent for the publication of any identifying information (data or accompanying images).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, S., Huang, W., He, J. et al. Circular RNA circNRIP1 promotes glioma progression by regulating the miR-106a-5p/GPR133 pathway. Sci Rep 15, 39666 (2025). https://doi.org/10.1038/s41598-025-23343-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-23343-x