Abstract
To develop excipient-free carrier-free nanoparticles (DE NPs) via self-assembly of doxorubicin (DOX) and emodin (EMO) for enhanced combination chemotherapy against non-small cell lung cancer (NSCLC). DE NPs were fabricated through non-covalent interactions between DOX and EMO using a self-assembly method. The nanoparticles were characterized for morphology, stability, encapsulation efficiency, and cellular uptake. In vitro cytotoxicity and in vivo antitumor efficacy of free drugs (EMO, DOX), DOX/EMO mixture, and DE NPs were evaluated on A549 NSCLC cells. Mechanistic studies explored the role of DE NPs in suppressing efflux pump proteins via modulation of p65 activation and nuclear translocation. DE NPs formed rod-like nanostructures with high encapsulation efficiency (> 90%) and stability. They exhibited enhanced cellular uptake and intracellular drug accumulation compared to free drugs or the mixture. In vitro studies demonstrated significantly stronger cytotoxicity in A549 cells (IC50 reduction of ~ 50%) and a synergistic antitumor effect (combination index < 0.5). In vivo studies further confirmed the superior antitumor efficacy of DE NPs over individual DOX or EMO monotherapy (72.67 ± 7.20% tumor inhibition rate). Mechanistically, DE NPs inhibited p65 activation and nuclear translocation, downregulating efflux pump protein expression and prolonging drug retention in cancer cells. The excipient-free DE NPs provide a promising carrier-free nanomedicine platform for NSCLC treatment, achieving synergistic antitumor efficacy through enhanced drug delivery and suppression of drug resistance mechanisms. This strategy highlights the potential of self-assembled dual-drug nanoparticles to improve therapeutic outcomes in combination.
Similar content being viewed by others
Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide, with non-small cell lung cancer (NSCLC) accounting for over 80% of cases1,2. Systemic combination chemotherapy is a standard first-line treatment, with doxorubicin (DOX) being a key agent3. However, DOX’s efficacy is hampered by severe toxicity (e.g., cardiotoxicity) and the induction of drug resistance, often mediated by efflux pump proteins like ABCB1 and ABCC14,5,6,7. Therefore, the development of novel, less toxic, and more effective therapeutic strategies is urgently needed to improve treatment outcomes for NSCLC patients.
Combination chemotherapy can enhance efficacy but faces challenges in controlling the pharmacokinetics and ratio of individual drugs8,9. Nanoscale co-delivery systems (e.g., liposomes, micelles) have been developed to address this, yet their clinical translation is often hindered by the complexity and potential toxicity of excipient materials. For example, brush-arm star polymer nanoparticles (NPs), created using ring-opening metathesis polymerization, were engineered to deliver doxorubicin (DOX), camptothecin, and cisplatin in precise molar ratios10,11,12,13,14,15,16.
Despite the development of numerous nanomedicine drug delivery systems, only a small fraction has successfully transitioned to clinical application. A significant barrier to clinical adoption lies in the reliance on large quantities of excipient materials and the complexity of production processes17. The inclusion of excipients often necessitates extensive clinical trials and additional regulatory approvals from bodies such as the FDA18. Furthermore, the intricate nature of nanomedicine production complicates scaling up for industrial manufacturing19. To address these challenges, there is a pressing need for “green” approaches to nanomedicine design that eliminate the use of potentially toxic excipients20,21. One promising strategy is the development of excipient-free nanodispersions through nano-assembly, which can be utilized for nanoparticulate anticancer agents to improve therapeutic efficacy—a fundamental principle of green pharmaceuticals. These formulations and processes have been optimized to minimize both the use and generation of hazardous substances22. Recently, Kushwah et al. introduced a carrier-free dual-drug delivery system based on drug self-nanocrystallization23. In this approach, DOX was utilized as a stabilizing agent to create uniformly sized spherical particles, improving the water solubility of 10-hydroxycamptothecin (HCPT)24. However, most studies have primarily focused on enhancing the solubility of poorly soluble drugs while overlooking drug compatibility. Therefore, excipient-free “pure” nanodrugs represent a promising direction for the next generation of nanomedicine.
Currently, an increasing number of natural compounds derived from traditional Chinese medicines (TCMs) are being discovered for their potential in treating various diseases. TCM, often regarded as a valuable cultural treasure of China, has been widely utilized across East Asia for thousands of years due to its therapeutic effects25. Emodin, a natural bioactive compound extracted from the roots and barks of several oriental herbs such as Polygonum cuspidatum and Rheum officinale, possesses a broad range of pharmacological activities. These include anti-tumor, anti-bacterial, anti-inflammatory, immunosuppressive, anti-osteoporotic, anti-diabetic, hepatoprotective, and anti-allergic properties26. Among these, emodin’s anti-cancer effects have garnered significant research attention, demonstrating its inhibitory impact on liver, breast, lung, colon, and renal cancers27. The anti-cancer mechanisms of emodin are linked to the TRIB3 pathway and endoplasmic reticulum (ER) stress28. Moreover, emodin has been shown to enhance the anti-tumor efficacy of paclitaxel in non-small cell lung cancer (NSCLC) by inhibiting multidrug resistance-associated proteins (MRPs)29. However, emodin’s hydrophobic nature poses significant challenges for vascular administration, limiting its clinical application30. These drawbacks necessitate the development of a nano-assembly strategy to overcome delivery obstacles and optimize therapeutic regimens.
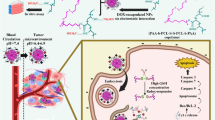
The structure of DOX exhibits characteristics similar to those of surfactants, with its unsaturated anthracycline rings and saturated ring system functioning as the hydrophobic and hydrophilic segments of the molecule, respectively. Additionally, DOX contains abundant hydroxyl groups adjacent to the amino sugar. Given that emodin is hydrophobic, DOX may potentially be employed to solubilize and nanosize emodin. In this study, we aim to develop excipient-free nanoparticles (DE NPs) by facilitating the self-assembly of emodin and DOX molecules, driven by non-covalent interaction forces such as hydrogen bonding, π-π stacking, and hydrophobic interactions, without the use of organic solvents. The strong interactions between DOX and emodin are expected to form a hydrophobic core, leaving the hydrophilic daunosamine moieties of DOX exposed, rendering the nanoparticles dispersible in aqueous solutions. As illustrated in Fig. 1, after DE NPs enter the cells, they could inhibit the expression of efflux pump proteins, thereby increasing intracellular drug accumulation and enhancing the cytotoxic effect against NSCLC cells.
Materials and methods
Materials
DOX (purity: 98%) and Emodin (EMO, purity: 98%) was applied by Aladdin Reagent Database Inc. (Shanghai, China). Cell counting kit-8 (CCK-8) was purchased from by Do Jindo Laboratories (Japan). The Annexin V-FITC/PI apoptosis detection kit and TUNEL assay kit were all obtained from Beyotime Biotechnology (Beyotime, China). All other reagents or chemicals used were analytical grade reagents and used without further purification.
The lung cancer cell line A549 was obtained from National Collection of Authenticated Cell Cultures and cultured in RPMI-1640 medium containing 10% Fetal Bovine Serum (FBS), 100 U/mL penicillin and 100 µg/mL streptomycin. The cells were cultured in an incubator at 37 °C in 5% CO2 atmosphere.
Preparation of nanoparticles
The carrier-free nanoparticles via the self-assembly of DOX and EMO molecules, named DOX/EMO nanoparticles (DE NPs) were fabricated using a nano-precipitation approach31,32. Briefly, 50 µL of the DOX (10 mM, DMSO) and 100 µL of the EMO (10 mM, DMSO) solution was gradually dispersed into 1 mL of deionized water (40 °C) under ultrasonication for 1 h to prepare the DE NPs nanoparticles. Subsequently, the nano-dispersions were purified using a dialysis bag (MWCO): 3500 Da), the resulting nanodispersions were further ultrafiltered to completely remove the free drugs. Finally, the DE NPs were collected and stored at 4 °C for further use. As previously described, additional formulations with varying molar (feeding) ratios of DOX to EMO were prepared. To prepare the DOX/EMO mixture, 100 µL of a 10 mM EMO solution in 1 mL of 20% polyethylene glycol (PEG 400) containing 5% NaHCO3, was mixed with 50 µL of a 10 mM DOX solution in DMSO.
Characterization
The morphology of DE NPs was examined using transmission electron microscopy (TEM). The size distribution and zeta potential of different nanoparticles were investigated using a Malvern Zetasizer 3000 system. UV-Vis spectrophotometer was used to obtain the absorption spectra, and a fluorescence spectrometer was used to obtain the fluorescence emission spectra of free EMO, free DOX and DE NPs (λex (EMO) = 360 nm; λex (DOX) = 475 nm). The concentrations of DOX and EMO within the nanoparticles were quantified using high-performance liquid chromatography (HPLC) method equipped with a Diamonsil C18 column (250 mm × 4.6 mm, 5 μm), with a mobile phase of acetonitrile and sodium dodecyl sulfate solution at a flow rate of 1 mL/min. The encapsulation efficiency (EE) was determined using the following equations:
Stability of nanoparticles
Particle size and polydispersity index (PDI) were used to evaluate the storage and serum stability of DE NPs in distilled water at 4 °C for 96 h and in fetal bovine serum (FBS), respectively. For storage stability, samples were collected and measured at predetermined intervals (0, 12, 24, 48, 72 and 96 h). For serum stability, samples were incubated with FBS at a 1:1 volume ratio and maintained at 37 °C with agitation at 100 rpm and measured at predetermined time points (0, 1, 4, 8, 12 and 24 h).
Molecular dynamics (MD) simulation
The geometries of DOX and EMO were refined through optimization employing the B3LYP exchange-correlation functional in conjunction with the 6-31G* basis set, utilizing the Gaussian09 computational chemistry package. Thereafter, the electrostatic potential (ESP) was determined employing the B3LYP/6-311G(d, p) computational method and basis set, which facilitated the derivation of the restrained ESP (RESP). The General Amber Force Field (GAFF) parameters were utilized for the parametrization of the molecular models of the two pharmaceuticals. Initially, a simulation box with a 40 Å edge length contained a random distribution of 24 EMO and 12 DOX molecules. To ensure charge neutrality, sodium and chlorine ions were incorporated, and the system was hydrated with TIP3P water molecules within a cuboid box, preserving a 10 Å buffer of solvent around the solute and the box boundaries. Molecular dynamics (MD) simulations were executed with GROMACS2021. The complex was positioned in the center of a simulation box, maintaining a 10 Å clearance, and solvated with the TIP3P water model. Periodic boundary conditions (PBC) were imposed to allow unrestricted movement within the cubic simulation space. Nonbonded interactions were calculated using the Lennard-Jones 12 − 6 potential with a truncation distance of 10 Å, while long-range electrostatics were handled by the Particle Mesh Ewald (PME) method. Constraints on hydrogen-containing bonds were implemented using the SHAKE algorithm. An energy minimization procedure employing the steepest descent method was conducted for 500,000 iterations to eliminate any steric clashes. The system was gradually warmed to 300 K over a period of 50 ps. Subsequently, a two-stage equilibration was conducted: initially under isochoric (NVT) ensemble, followed by isobaric-isothermal (NPT) ensemble, each lasting 100 ps at 300 K, employing Langevin thermostat with a collision frequency of 1.0 ps−1. The production MD simulation was then carried out for 50 ns with a time step of 2.0 fs for integration.
In vitro drug release profile
The release profiles of DOX, EMO, and DE NPs were studied in phosphate-buffered saline (PBS) at varying pH levels (5.0 and 7.4). Briefly, samples were placed in a dialysis bag (MWCO: 3500 Da) and immersed in 50 mL of PBS. The pH values were adjusted to 5.0 by adding diluted HCl. The samples were maintained at 37 °C with agitation at 100 rpm. At predetermined time points, 1 mL aliquots were collected, and the concentrations of DOX and EMO were determined using HPLC.
Cellular uptake
To investigate the cellular uptake of DE NPs, A549 cells were seeded in 24-well plates and cultured for 24 h. The cells were then treated with DOX, EMO, DOX/EMO, and DE NPs (1 µM DOX and 2 µM EMO) for 4 h, respectively. The fluorescence intensity of the different formulations was analyzed using flow cytometry (BD, Franklin Lakes, NJ).
The cellular internalization process was further observed using a Thunder Imaging System microscope (Leica). Briefly, A549 cells were cultured in a confocal laser scanning microscopy (CLSM) dish and incubated with DOX, EMO, DOX/EMO, and DE NPs (1 µM DOX and 2 µM EMO) for 4 h. After incubation, the medium was discarded, and the cells were washed with PBS to remove residual nanoparticles. The cells were then fixed with 4% paraformaldehyde and visualized using Thunder Imaging System microscope.
In vitro synergistic antitumor assay
The cytotoxicity of DE NPs was assessed with a CCK-8 assay kit. Briefly, cells were seeded in 96-well plates at a density of 5 × 103 cells/well and incubated for 24 h. Then cells were cultured with various concentrations of DOX, EMO, DOX/EMO, and DE NPs. After a 48 h incubation, the medium was replaced, and the cell viability was assessed using the CCK-8 kit. The absorbance was measured at 450 nm by a microplate reader. Untreated cells were used as controls. IC50 values were determined by CompuSyn 1.0 software. IC₅₀ values for combination groups were calculated based on total drug concentration at a fixed molar ratio, following the Chou-Talalay method for fixed-ratio designs.
The combination index (CI) of EMO and DOX was calculated using the Chou-Talalay equation:
In this equation, Dx denotes the dose of EMO or DOX required to achieve an x% inhibition rate, while (D)1and (D)2 represent the respective doses of EMO and DOX required to achieve the same level of inhibitory effect when administered in combination. CI values less than 1 indicate a synergistic interaction, CI values equal to 1 suggest an additive effect, and CI values greater than 1 reflect an antagonistic interaction.
Apoptosis assay
Annexin V-FITC/PI apoptosis detection kit (Beyotime, China) was used to evaluate the apoptosis-inducing capability of nanoparticles. Briefly, cells were cultured in 12-well plates, and then treated with DOX, EMO, DOX/EMO, and DE NPs (0.5 µM DOX and 1 µM EMO) for 12 h. Later, the cells were collected and stained with AnnexinV FITC and PI in accordance with the manufacturer’s protocol and monitored by flow cytometry.
TUNEL assay
TUNEL (terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling) is a popular method for identifying apoptotic cells in situ by detecting DNA fragmentation. Briefly, A549 cells were cultured in a 12-well plate and incubated overnight. Next, the cells were incubated with DOX, EMO, DOX/EMO, and DE NPs (0.5 µM DOX and 1 µM EMO) for 8 h. Subsequently, cells were fixed with 4% paraformaldehyde for 20 min, washed with PBS three times and treated with 20 µg/mL proteinase K at 37 °C for 5 min. Next, the cells were washed with 1×Equilibration Buffer and incubated with CF488-dUTP Labeling Mix. The TUNEL assay was then performed according to the instructions from the manufacturer (Servicebio, China). Next, the stained cells were further stained with DAPI for 10 min, and then washed with PBS twice and imaged under the microscope (Thunder Imaging Systems, Leica). DAPI stains both apoptotic and non-apoptotic cells blue, while the incorporation of CF488-dUTP produces green fluorescence that is specifically localized in the nuclei of apoptotic cells (CF488 excitation wavelength: 490 nm; emission wavelength: 515 nm).
In vivo antitumor efficacy
All experiments involving animals were performed in accordance with ARRIVE guidelines. Male BALB/c nude mice (18–22 g) were obtained from the Shanghai Laboratory Animal Center. All procedures complied with the Nanjing University of Chinese Medicine’s Guidelines for Animal Experimentation and received approval from the institutional Animal Ethics Committee. To establish orthotopic A549 tumor models, cell suspensions were injected subcutaneously into the right lower back region. Tumor volume (V) was monitored by caliper measurement of the length (a) and width (b), followed by calculation (V = a×b2/2). Mice bearing tumors reaching approximately 100 mm³ were randomized into 4 groups (n = 6) for evaluating DE NPs antitumor efficacy. Treatments (saline, DOX, DOX/EMO, or DE NPs containing 5 mg/kg DOX) were administered intravenously every other day. Tumor dimensions and body weights were recorded every other day, and the tumor inhibition rate was determined. On day 21, mice were euthanized by CO2 asphyxiation and tumors excised.
Western-bloting
Cellular proteins were extracted using a lysis buffer (R0030-100, Solarbi). Nuclear extracts were obtained employing the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo). Protein concentration was determined using the BCA protein assay kit (Thermo Fisher Scientific). Samples were separated by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, and blocked. Primary antibodies, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (YIFEIXUE Bio Tech), were used for detection. Enzymatic signals were visualized with an ECL detection reagent (Biosharp). The antibodies used included BAX, BCL-2, ABCB1, ABCC1, ABCC5, p65, p-p65, GAPDH and Lamin B (1:10,000, Bioworld).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from tissue samples or cultured cells using TRIzol reagent (Invitrogen). cDNA synthesis was carried out using 500 ng of total RNA and the Hiscript® II QRTSuperMix (Vazyme, China). qRT-PCR was conducted with the SYBR Green Master Mix (Bio-Rad, USA) following the manufacturer’s guidelines. Relative gene expression levels were quantified using the comparative cycle threshold (Ct) method (2−ΔΔCt). The specific primers employed for qRT-PCR are listed below: ABCB1: 5’-CTTCAGGGTTTCACATTTGGC-3’ (Foward) and 5’-GGTAGTCAATGCTCCAGTGG-3’ (Reverse); ABCC1: 5’-GACCCTCCCACACTGAATG-3’ (Foward) and 5’-CCTCCACTTTGTCCATCTCAG-3’ (Reverse); ABCC5: 5’-AGGCTTGTTTTGCTGCAGGG-3’ (Foward) and 5’-TGGGTGCTGGTGTTTGGAAG-3’ (Reverse); β-actin: 5’-TGACGGGGTCACCCACACTGTGCCCATCTA-3’ (Foward) and 5’-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3’ (Reverse).
Statistical analysis
Data are presented as the mean ± standard deviation (SD). Group differences were analyzed using one-way ANOVA, with statistical significance set at P < 0.05.
Results and discussion
Preparation and characterization of DE NPs
Based on the interactions between the free EMO and DOX in the aqueous phase, the carrier-free DE NPs were fabricated using a nano-precipitation approach. The morphology of DE NPs was observed by TEM. As illustrated in Fig. 2A, upon the addition of DOX to the EMO suspension at a DOX/EMO molar ratio of 1:2, the DOX molecules co-assembled with EMO through interactive forces, resulting in the formation of rod shaped DE NPs. The particle size, polydispersity index (PDI), and zeta potentials of DE NPs (Fig. 2B,C) were measured using dynamic light scattering (DLS) and found to be 257.9 ± 3.6 nm, 0.16 ± 0.05, and 27.4 ± 1.2 mV, respectively. These values fall within the accepted range for effective enhanced permeability and retention (EPR) effect, and enabling the nanoparticles to avoid filtration by the kidney, minimize specific sequestration by the splenic sinusoids, and pass through the hepatic fenestra21,33. The ultraviolet-visible (UV-Vis) spectroscopy and fluorescence emission spectra of DE NPs were subsequently characterized. As illustrated in Fig. 2D, DE NPs displayed the characteristic absorption peaks corresponding to DOX at 240 nm, 300 nm, 485 nm and EMO at 252 nm and 308 nm, consistent with the individual absorption profiles of DOX and EMO. The fluorescence emission spectrum of DE NPs exhibited slight variations compared to that of the free drugs, as depicted in Fig. 2E. Moreover, DOX was clear and transparent in aqueous solution, while the DE NPs solution exhibited a pronounced Tyndall light scattering effect (Fig. 2F). Due to the absence of any auxiliary materials during the preparation of the nanoparticles, both the encapsulation efficiency and drug loading capacity were extraordinarily high. The encapsulation efficiency (EE) of EMO and DOX, as determined by high-performance liquid chromatography, was 85.31 ± 0.26% and 79.57 ± 2.18%, respectively. Then, the loading efficiency (DL) of EMO and DOX was 64.28 ± 2.9% and 35.72 ± 3.2%, respectively.
The stability of DE NPs
Stability is a critical factor for the clinical use of drug delivery systems, as it is essential to prevent drug leakage and prolong blood circulation time for effective drug delivery34. Consequently, the storage stability of DE NPs was evaluated. As depicted in Fig. 3A, C, no significant changes were observed in particle size, PDI and zeta potentials over 96 h, indicating that the nanoparticles remained stable at 4 °C for up to four days. Additionally, the consistently low PDI value suggested a uniform particle size distribution. Next, the stability of DE NPs was further tested in fetal bovine serum (FBS). The results indicated that the presence of 10% FBS had a negligible effect on the particle size, PDI and zeta potentials of DE NPs, demonstrating good serum stability (Fig. 3B, D). Despite the absence of any auxiliary materials, the DE NPs exhibited desirable stability without any precipitation, which is advantageous for systemic circulation.
(A) The particle size and PDI of DE NPs during incubation at 4 °C for 96 h. (B) The particle size and PDI of DE NPs after incubation with fetal bovine serum for 24 h. (C) The zeta potentials of DE NPs during incubation at 4 °C for 96 h. (D) The zeta potentials of DE NPs after incubation with fetal bovine serum for 24 h. Results are reported as mean ± SD (n = 3).
The self-assembly mechanisms between DOX and EMO using molecular dynamics simulation
To further understand the self-assembly mechanism of DE NPs, the dynamics process and the interactions between drug molecules were investigated. 36 molecules (EMO: DOX = 2:1) were randomly included in a cubic box with a length of 40 Å and subjected to 50 ns of MD simulations. As shown in Fig. 4A, DOX and EMO self-organized and formed assemblies after 50 ns of simulations. We chose one snapshot of the aggregate to illustrate the stacking manner (including hydrogen bonds, π-π stacking and π-Alkyl bonds) of the two drugs and the representative interactions are shown in Fig. 4B–D. Thus, the amphiphilic nature of DOX (hydrophilic daunosamine vs. hydrophobic anthracycline ring) enables it to function as a ‘molecular scaffold’ that co-assembles with EMO via synergistic π-π stacking, H-bonding, and hydrophobic interactions. This mechanism bypasses the need for polymeric carriers, consistent with carrier-free nanodrug design principles. Collectively, these results elucidate how inter-molecular interactions stabilize the nanosystem.
In vitro drug release profile
The release performances of EMO, DOX and DE NPs were evaluated in buffer solutions with pH 7.4 and 5.0 over 24 h at 37 °C. As illustrated in Fig. 5, the release profiles of free EMO and DOX showed no significant differences under different pH conditions, with both exhibiting a rapid initial release, releasing approximately 85% of the drug within the first 10 h. In contrast, the release rates of DE NPs under varying pH conditions were lower than those of free EMO and DOX. In a medium with pH 7.4, the cumulative release rates of EMO and DOX from DE NPs within 2 h were 35.7% and 41.5%, respectively, with both drugs reaching a cumulative release rate of nearly 85% after 24 h. Additionally, the overall release rate of EMO enhanced upon lowering the pH, with a cumulative release of about 88% at 24 h. Under the same conditions, the release rate of DOX also increased, with a cumulative release of approximately 90% at 24 h. Under low pH conditions, the release of drugs from the nanoparticles was accelerated, which may be attributed to the reduced stability of the nanoparticles in acidic environments.
Cellular uptake
Efficient cellular internalization of nanoparticles is crucial for effective drug delivery and therapy. To investigate whether DE NPs can be efficiently internalized by A549 cells, the cellular uptake of DOX, EMO, DOX/EMO, and DE NPs was analyzed using flow cytometry. As shown in Fig. 6A, B, all four formulations were effectively taken up by the cells. Notably, the DE NPs group exhibited the highest uptake, with the strongest fluorescence intensity observed in both red and green channels. This indicates that the carrier-free nanoparticles can enhance drug delivery to tumor cells. Additionally, the Thunder Imaging System microscope was utilized to further investigate the cellular uptake and distribution of the formulations. The cellular fluorescence intensity of EMO and DOX in the DE NPs group was significantly higher than in the EMO, DOX, or EMO/DOX groups (Fig. 6C). The colocalization of red and green fluorescence confirmed that DOX and EMO were co-delivered to the cells. This enhanced cellular uptake is crucial for achieving a high antitumor effect. Nanoparticles of this size range (~ 260 nm) typically enter cells via energy-dependent endocytic pathways, such as clathrin- or caveolae-mediated processes. However, the exact route for DE NPs remains to be experimentally validated. Future studies will delineate specific endocytic mechanisms using pharmacological inhibitors (e.g., chlorpromazine for clathrin) or genetic knockdown approaches.
In vitro antitumor efficacy
The in vitro cytotoxicity of DOX, EMO, DOX/EMO, and DE NPs were tested by CCK-8 assay to evaluate the antitumor efficacy in A549 cell lines. The experimental results demonstrated a gradual increase in cytotoxicity with higher concentrations of the chemotherapeutics (Fig. 7). In the free DOX and free EMO groups, the half-maximal inhibitory concentration (IC50) for A549 cells was 0.65 ± 0.13 µM and 16.55 ± 1.27 µM, respectively (Table 1). Compared to single-drug treatments, combination therapy with DOX and EMO resulted in significantly lower cell viability. The IC50 of DOX decreased to 0.24 ± 0.17 µM when combined with EMO in the mixture. Notably, the DE NPs group demonstrated a further reduction in cell viability, with an IC50 of 0.05 ± 0.09 µM. In A549 cells, the IC50 of DE NPs was 13-fold lower than that of free DOX and 2.8-fold lower than that of free EMO, indicating a significantly enhanced therapeutic effect. In addition, we also calculated and found that the CI values for DOX/EMO and DE NPs were 0.85 and 0.43, respectively, suggesting a more profound synergistic antitumor effects of DE NPs. This superior antitumor efficacy of the DE NPs can be attributed to two primary factors. First, compared to free drugs, the nanoparticle formulation achieves more efficient cellular internalization, resulting in higher intracellular drug concentrations and enhanced therapeutic outcomes. Second, once internalized, the two drugs not only exert their respective therapeutic effects but also synergistically enhance the antitumor activity, leading to a more potent treatment response. While this study focused on A549 cells as a representative model to dissect the carrier-free synergy and mechanism, future work will comprehensively evaluate DE NPs across a panel of NSCLC cell lines, including KRAS-mutant (e.g., H460) and EGFR-mutant (e.g., PC-9) models, as well as in vivo models, to fully assess its broad-spectrum potential.
Cell apoptosis
The apoptosis rate is a crucial index for evaluating the therapeutic effects of antineoplastic agents. To further demonstrate the synergistic apoptosis-inducing efficacy of DE nanoparticles (DE NPs) on A549 cell lines, the Annexin V-FITC/PI method was used to quantitatively analyze apoptosis, with results assessed by flow cytometry. As shown in Fig. 8, the apoptotic rate in the control group was negligible, whereas the percentage of early and late apoptotic cells increased significantly after treatment with various formulations. The total apoptosis rates for the EMO, DOX, DOX/EMO, and DE NPs groups were 25.44%, 42.61%, 53.48%, and 71.84%, respectively. These results may be attributed to the enhanced cellular uptake of DE NPs compared to free drugs and the synergistic antineoplastic effects of the combined therapy.
TUNEL assay
The TUNEL assay is a widely utilized method for identifying apoptotic cells in situ by detecting DNA fragmentation. In apoptotic cells, the activation of Ca/Mg-dependent endonucleases leads to DNA degradation, resulting in strand breaks within the DNA. Terminal deoxynucleotidyl transferase (TdT) catalyzes the addition of biotin-dUTP to these cleaved DNA strand breaks, allowing for the identification of these sites. Therefore, the TUNEL apoptosis detection kit can be used to detect the DNA breaks in the nuclei of cells during the late stages of apoptosis. The underlying principle involves the incorporation of CF488-dUTP at the exposed 3’-OH ends of fragmented genomic DNA under the action of TdT, which can then be detected with green fluorescence using a fluorescence microscope. As shown in Fig. 9, almost no green fluorescence was observed in the control group, while all four other formulations exhibited the presence of green fluorescence, with the nanoparticle group displaying the highest intensity. This indicates the presence of the most apoptotic cells in this group. These findings are consistent with the apoptosis results from the Annexin V-FITC/PI assay, confirming the enhanced efficacy of the DE NPs formulation in inducing apoptosis.
In vivo antitumor efficacy of DE NPs
To evaluate the benefits of DE NP combination therapy, orthotopic A549 tumor models were established. As depicted in Fig. 10B, the model group exhibited rapid tumor growth, whereas DOX or DOX/EMO therapy only moderately suppressed progression. In contrast, DE NPs induced significant tumor growth inhibition (72.67 ± 7.20% tumor inhibition rate on day 21; Fig. 10C), further corroborated by tumor xenograft images (Fig. 10A). Post-treatment, the DE NP cohort showed minimal tumor volume, demonstrating a synergistic antitumor effect between DOX and EMO.
DE NPs significantly decreased the expressions of efflux pump proteins by inhibiting activation and nuclear translocatio of p65
Lower expressions of efflux pump proteins (such as ABCC1, ABCB1 and ABCC5) were positively correlated with high intracellular drug accumulation. In order to elucidate the intracellular accumulation of drugs and better anti-NSCLC efficacy of DE NPs, western bloting assay was used to detect the expression levels of the above-mentioned proteins. As shown in Fig. 11A-C, compared to the untreated control group, both the protein and mRNA levels of ABCB1, ABCC1 and ABCC5 were markedly decreased after the free EMO treatment. However, free DOX does not show similar efficacy. In addition, compared to free EMO treatments, combination therapy with DOX could not be further inhibited the expression of efflux pump proteins. Excitingly, DE NPs can further significantly inhibit ABCB1, ABCC1 and ABCC5 expression compared to free EMO (p<0.05). A similar trend was observed for apoptosis-related proteins (BAX and BCL2). A deeper investigation into the upstream mechanisms revealed that, in contrast to DOX free, EMO free notably suppressed the activation and nuclear translocation of p65, which is the main transcription factor of efflux pump proteins. Moreover, DE NPs exhibited the most pronounced regulatory effect on p65 (Fig. 11A, B).
Conclusions
In summary, EMO was successfully nanosized with the aid of DOX to create carrier-free nanoparticles (DE NPs) through a straightforward self-assembly method. EMO and DOX molecules co-assembled via multiple interaction forces, forming rod-like structures with good stability. Cellular uptake studies demonstrated a significant increase in intracellular drug accumulation following treatment with DE NPs. In vitro cytotoxicity assays revealed a significant enhancement in potency against A549 cells, with a profound synergistic interaction. Critically, in vivo validation in xenograft models demonstrated superior antitumor efficacy of DE NPs, achieving higher tumor inhibition versus DOX or EMO monotherapy, thereby confirming therapeutic synergy beyond additive effects. Mechanistic investigations demonstrated that DE NPs facilitated increased drug accumulation in tumor cells by downregulating the expression of multiple efflux pump proteins. This effect was mediated through the inhibition of p65 activation and its subsequent nuclear translocation. Collectively, the development of excipient-free nanomedicines represents a promising strategy, offering inspiration for the design of novel carrier-free nanodrugs for theranostics, particularly in achieving synergistic combination therapy for cancer.
Data availability
All data generated or analysed during this study are included in this published article.
References
Ganti, A. K., Klein, A. B., Cotarla, I., Seal, B. & Chou, E. Update of Incidence, Prevalence, Survival, and initial treatment in patients with Non-Small cell lung cancer in the US. JAMA Oncol. 7(12), 1824–1832 (2021).
Chen, P., Liu, Y., Wen, Y. & Zhou, C. Non-small cell lung cancer in China. Cancer Commun. (London England). 42(10), 937–970 (2022).
Riely, G. J. et al. Non-Small cell lung Cancer, version 4.2024, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Network: JNCCN. 22(4), 249–274 (2024).
Shin, D. H., Choi, Y. J. & Park, J. W. SIRT1 and AMPK mediate hypoxia-induced resistance of non-small cell lung cancers to cisplatin and doxorubicin. Cancer Res. 74(1), 298–308 (2014).
Mozaffari, S. et al. Design and application of hybrid cyclic-linear peptide-doxorubicin conjugates as a strategy to overcome doxorubicin resistance and toxicity. Eur. J. Med. Chem. 226, 113836 (2021).
Mi, Y. J. et al. Apatinib (YN968D1) reverses multidrug resistance by inhibiting the efflux function of multiple ATP-binding cassette transporters. Cancer Res. 70(20), 7981–7991 (2010).
Guo, L. et al. Gene expression profiling of drug-resistant small cell lung cancer cells by combining MicroRNA and cDNA expression analysis. Eur. J. Cancer (Oxford England: 1990). 46(9), 1692–1702 (2010).
Mörth, C. & Valachis, A. Single-agent versus combination chemotherapy as first-line treatment for patients with advanced non-small cell lung cancer and performance status 2: a literature-based meta-analysis of randomized studies. Lung Cancer (Amsterdam Netherlands). 84(3), 209–214 (2014).
Shi, C. et al. A drug-specific nanocarrier design for efficient anticancer therapy. Nat. Commun. 6(1), 7449 (2015).
Gu, Y. et al. Nanomicelles loaded with doxorubicin and Curcumin for alleviating multidrug resistance in lung cancer. Int. J. Nanomed. 11, 5757–5770 (2016).
Zhu, Y. X. et al. Photosensitizer-Doped and plasma Membrane-Responsive liposomes for nuclear drug delivery and multidrug resistance reversal. ACS Appl. Mater. Interfaces. 12(33), 36882–36894 (2020).
Wang, L. et al. Assessment of reversibility in pulmonary hypertension related to congenital heart disease by using biomarkers and clinical features. Congenit. Heart. Dis. 17(1), 87–97 (2022).
Cao, J., Zhu, C., Cao, Z. & Ke, X. CPPs-modified Chitosan as permeability-enhancing chemotherapeutic combined with gene therapy nanosystem by thermosensitive hydrogel for the treatment of osteosarcoma. Int. J. Biol. Macromol. 267(Pt 2), 130915 (2024).
Gatti, S. et al. Hydrazone linked doxorubicin-PLA prodrug nanoparticles with high drug loading. Nanotechnology 29(30), 305602 (2018).
Yin, Q. et al. Drug-initiated ring-opening polymerization of O-carboxyanhydrides for the Preparation of anticancer drug-poly(O-carboxyanhydride) nanoconjugates. Biomacromolecules 14(3), 920–929 (2013).
Zhang, B. et al. Development and evaluation of oxaliplatin and Irinotecan co-loaded liposomes for enhanced colorectal cancer therapy. J. Controll. Release 238, 10–21 (2016).
Su, C. et al. Absorption, distribution, metabolism and excretion of the biomaterials used in nanocarrier drug delivery systems. Adv. Drug Deliv. Rev. 143, 97–114 (2019).
Min, Y., Caster, J. M., Eblan, M. J. & Wang, A. Z. Clinical translation of nanomedicine. Chem. Rev. 115(19), 11147–11190 (2015).
Mei, H. et al. Carrier-free nanodrugs with efficient drug delivery and release for cancer therapy: from intrinsic physicochemical properties to external modification. Bioactive Mater. 8, 220–240 (2022).
Shim, M. K. et al. Carrier-free nanoparticles of cathepsin B-cleavable peptide-conjugated doxorubicin prodrug for cancer targeting therapy. J. Controlled Release. 294, 376–389 (2019).
Shim, M. K. et al. Cancer-specific drug-drug nanoparticles of pro-apoptotic and cathepsin B-cleavable peptide-conjugated doxorubicin for drug-resistant cancer therapy. Biomaterials 261, 120347 (2020).
Fu, S. et al. Pure drug nano-assemblies: A facile carrier-free nanoplatform for efficient cancer therapy. Acta Pharm. Sin. B. 12(1), 92–106 (2022).
Kushwah, V., Katiyar, S. S., Agrawal, A. K., Gupta, R. C. & Jain, S. Co-delivery of docetaxel and gemcitabine using pegylated self-assembled stealth nanoparticles for improved breast cancer therapy. Nanomedicine 14(5), 1629–1641 (2018).
Chen, F. et al. Synergistically enhanced therapeutic effect of a Carrier-Free HCPT/DOX nanodrug on breast cancer cells through improved cellular drug accumulation. Mol. Pharm. 12(7), 2237–2244 (2015).
Mijatović, S. et al. Naturally occurring compounds in differentiation based therapy of cancer. Biotechnol. Adv. 36(6), 1622–1632 (2018).
Dong, X. et al. A review of its Pharmacology, toxicity and pharmacokinetics. Phytother. Res. 30(8), 1207–1218 (2016).
Shrimali, D. et al. Targeted abrogation of diverse signal transduction cascades by Emodin for the treatment of inflammatory disorders and cancer. Cancer Lett. 341(2), 139–149 (2013).
Correia da Silva, D., Valentão, P. & Pereira, D. M. A survey of naturally occurring molecules as new Endoplasmic reticulum stress activators with selective anticancer activity. Cancers 15(1) (2022).
Chen, S., Zhang, Z. & Zhang, J. Emodin enhances antitumor effect of Paclitaxel on human non-small-cell lung cancer cells in vitro and in vivo. Drug. Des. Devel. Ther. 13, 1145–1153 (2019).
Wei, W. et al. Antitumor effects of Self-Assembling Peptide-Emodin in situ hydrogels in vitro and in vivo. Int. J. Nanomed. 16, 47–60 (2021).
Wang, R. et al. Synergistic Inhibition of metastatic breast cancer by dual-chemotherapy with excipient-free rhein/DOX nanodispersions. J. Nanobiotechnol. 18, 116 (2020).
Yuan, M. et al. Shi, L. A carrier-free supramolecular nano-twin-drug for overcoming irinotecan-resistance and enhancing efficacy against colorectal cancer. J. Nanobiotechnol. 21, 393 (2023).
Duan, X. et al. Smart pH-sensitive and temporal-controlled polymeric micelles for effective combination therapy of doxorubicin and Disulfiram. ACS Nano. 7(7), 5858–5869 (2013).
Zhu, J. et al. Dual-responsive polyplexes with enhanced disassembly and endosomal escape for efficient delivery of SiRNA. Biomaterials 162, 47–59 (2018).
Acknowledgements
This study was financially supported by the Foundation for Jiangsu Commission of Health (LKM2023011), The Project for the Construction of “Flagship” Departments of Integrated Traditional Chinese and Western Medicine([2024]221), and Foundation for Nanjing famous Chinese medicine studio construction project ([2023]9).The authors thank the members of their laboratory and their collaborators.
Author information
Authors and Affiliations
Contributions
Conceptualization, Cheng Tang, Yangyang Zou, Yaoyao Guo, and Chaojuan Li; methodology, Cheng Tang , Hao Meng, Jin Xu, and Chaojuan Li; experiment performance and validation, Cheng Tang, Hao Meng, Yaoyao Guo , and Fang Long ; formal analysis, Cheng Tang, Yangyang Zou, Hao Meng, and Xiaoli Chen ; data curation, Yangyang Zou; writing, original draft preparation, Cheng Tang, Yangyang Zou, and Hao Meng ; writing review and editing, Cheng Tang, Yangyang Zou, Hao Meng, Chaojuan Li, and Qingling Xiao; supervision, Cheng Tang, Yangyang Zou, Zhang Nan, and Kelei Su; project administration, Chaojuan Li, and Yaoyao Guo ; funding acquisition, Cheng Tang, Yangyang, Zou, Hao Meng, and Chaojuan Li.All authors reviewed the manuscript.Cheng Tang, Yangyang Zou, and Hao Meng contributed equally to this study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, C., Zou, Y., Meng, H. et al. Synergistic anticancer effects of carrier-free self-assembled doxorubicin and emodin nanoparticles in non-small cell lung cancer. Sci Rep 15, 40205 (2025). https://doi.org/10.1038/s41598-025-24027-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24027-2