Abstract
Ecotropic viral integration site 5 (EVI5), a member of the Tre-2/Bub2/Cdc16 (TBC) domain-containing protein family, has been implicated in the initiation of various cancers. However, its precise role in lung adenocarcinoma (LUAD) remains unclear. This study aimed to elucidate the function of EVI5 in LUAD, with a focus on its regulation of the immune checkpoint molecule PD-L1. In the present study, we found that EVI5, Rab11 and PD-L1 were significantly overexpressed in LUAD tissues compared to adjacent normal tissues. In vitro experiments demonstrated that EVI5 knockout suppressed the expression of Rab11 and PD-L1 in LUAD cells. Notably, EVI5 overexpression promotes the expression of Rab11 and PD-L1 in LUAD cells. EVI5 was also shown to interact with Rab11 to upregulate PD-L1 expression in LUAD cells. Mechanistically, our findings identify a novel EVI5-Rab11-PD-L1 axis and suggest that EVI5 is a potential regulator of the tumor immune microenvironment, which may have implications for the efficacy of immunotherapy.
Similar content being viewed by others
Background
Lung adenocarcinoma (LUAD), a major histological subtype of non-small cell lung cancer (NSCLC), represents a leading cause of cancer-related mortality worldwide1,2,3,4. While the advent of immune checkpoint blockade (ICB) targeting the PD-1/PD-L1 axis has revolutionized LUAD treatment, primary and acquired resistance remain significant clinical hurdles5,6,7,8,9. Objective response rates to anti-PD-1/PD-L1 monotherapy are limited, underscoring the urgent need to elucidate the molecular mechanisms governing PD-L1 regulation and immune checkpoint activity10,11,12.
The identification of novel regulators of PD-L1 requires looking beyond transcriptional control to post-translational mechanisms such as intracellular trafficking13. Recent studies have highlighted the small GTPase Rab11 as a critical player in recycling cargoes like PD-L1 to the plasma membrane. For instance, amlodipine was shown to induce PD-L1 degradation in a Rab11-dependent manner, and TRAPPC4 was found to regulate PD-L1 trafficking via Rab1114. These findings position Rab11 as a central node in PD-L1 regulation. The activity of Rab GTPases is tightly controlled by GTPase-activating proteins (GAPs)15. Notably, ecotropic viral integration site 5 (EVI5) encodes a protein containing a TBC domain, which confers GAP activity specifically toward Rab1116.
EVI5 is not only an oncogene implicated in non-small cell lung cancer and other malignancies but has also been genetically associated with immune-mediated diseases like multiple sclerosis (MS)17,18. In MS, dysregulated T-cell activation is paramount, and Rab11 facilitates the transport of lymphocyte-specific protein tyrosine kinase (LCK) to the immune synapse19,20,21. This independent link between EVI5 and immune cell regulation strongly suggests its potential involvement in immunomodulatory pathways.
Based on these converging lines of evidence, we hypothesized that EVI5, through its regulation of Rab11, influences the cell surface expression of PD-L1 in LUAD, thereby impacting the tumor’s capacity to modulate the immune microenvironment. This study aims to investigate this novel EVI5-Rab11-PD-L1 axis and assess its potential as a therapeutic target for overcoming resistance to immune checkpoint inhibitors in LUAD.
Methods
Tissue samples
Paired LUAD and adjacent non-cancerous lung tissue samples (n = 20 each) were obtained with informed consent from patients at the Third Affiliated Hospital of Soochow University between 2020 and 2024. All patients were diagnosed with LUAD based on histological and pathological criteria in accordance with the Revised International System for Staging Lung Cancer. None of the patients had undergone chemotherapy or radiotherapy before tissue collection. Upon collection, Samples are immediately fixed in 10% neutral buffered formalin. Following fixation, the samples are dehydrated and subsequently embedded in paraffin blocks. The study protocol was approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University. We confirming that all experiments were performed in accordance with relevant guidelines and regulations.
Immunohistochemical assay
The tissue samples were fixed in 10% neutral buffered formalin, dehydrated through a graded ethanol series, and embedded in paraffin blocks. The embedded tissues were sectioned at a thickness of 4 μm and mounted on glass slides. Following deparaffinization and rehydration, antigen retrieval was performed by heating the sections in sodium citrate buffer (10 mM, pH 6.0) using a microwave oven for 10 min, followed by cooling to room temperature naturally. Endogenous peroxidase activity was quenched by incubation with 3% hydrogen peroxide for 15 min. Subsequently, non-specific binding sites were blocked with 10% normal goat serum for 30 min at room temperature. After blocking, the sections were incubated with primary antibodies diluted in antibody diluent overnight at 4 °C. The primary antibodies used were as follows: anti-human EVI5 ( ABN194, Millipore, dilution 1:50), anti-human PD-L1 ( #13684, CST, dilution 1:100), anti-human Rab11 (Abcam, Ab242196, dilution 1:100). After washing with PBS, the sections were incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (EnVision™ Detection Kit, Dako) for 1 h at room temperature. The signal was visualized using a 3,3’-diaminobenzidine(DAB) substrate kit. The nuclei were counterstained with hematoxylin. Finally, the sections were dehydrated, cleared, and mounted. Appropriate positive controls(known positive tissue) and negative controls (replacement of the primary antibody with PBS) were included in each experiment to ensure the specificity and reliability of the staining. The stained sections were evaluated by pathologists who was blinded to the clinical and pathological data. A scoring system based on the intensity of staining and the percentage of positive cells was applied. The H-score was calculated using the formula: H-score = (% of weakly stained cells × 1) + (% of moderately stained cells × 2) + (% of strongly stained cells × 3), yielding a range of 0 to 300. Cases were then categorized into high and low expression groups based on the median H-score of all samples.
Cell lines and cultures
The LUAD cell line, A549, was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) and L-glutamine (Invitrogen, Carlsbad, CA, USA) at 37 °C in a humidified atmosphere with 5% CO₂.
Establishment of stable EVI5-overexpressing cell lines
To establish stable EVI5-overexpressing LUAD cell lines, a 2493-bp fragment of the EVI5 coding sequence was synthesized (GeneChem, Shanghai, China) and subcloned into the PLVX-IRES-Neo vector using EcoRI and XbaI. The construct was transfected into HEK293T cells, along with packaging plasmids, using Lipofectamine 3000 (Invitrogen). Packaged lentiviruses were collected and used to infect the A549 cells. Stable transfectants were selected using 400 µg/mL G418 (Amresco, Solon, OH, USA). An empty vector was used as a control.
Establishment of stable EVI5-knockout cell lines
EVI5 knockout was achieved using the CRISPR-Cas9 system. Guide RNA (gRNA) sequences targeting EVI5 were designed and subcloned into the LentiCRISPR v2 vector (GeneChem, Shanghai, China). The accuracy of the constructs was confirmed by sequencing. Lentiviral particles were packaged into HEK293T cells, collected, and used to infect A549 cells. Stable transfectants were selected using puromycin (0.4–2 µg/mL; Sigma-Aldrich, St. Louis, MO, USA).
Establishment of stable Rab11-knockdown cell lines
To establish stable A549 cell lines in which Rab11 was silenced, two DNA fragments (Rab11-shRNA-1:5′-GATCCGGAGTAGAGTTTGCAACAAGACTCGAGTCTTGTTGCAAACTCTACTCCTTTTTG-3′; Rab11-shRNA-2:5′-GATCCGGCCCTAGACTCTACAAATGTCTCGAGACATTTGTAGAGTCTAGGGCCTTTTTG-3′) were subcloned into the lentiviral vector pGMLV-SC5 (Genomeditech, Shanghai, China) with BamHI and EcoRI endonucleases. The scrambled sequence (underscored) of Rab11 shRNA, which served as a negative control, was as follows: 5ʹ-ATCGACTAGC CACTTAGACTTCAAGAGGTCTAAGTGGCTAGT CGATTTTTTTT-3’. The CD151-silenced construct or negative control was co-transfected with packs of aging plasmids into human embryonic kidney 293 T cells using Lipofectamine 3000 (Invitrogen). Forty-eight hours later, A549 cells were infected with the packaged lentiviruses and cultured for 2 days, and stable cell lines were selected using 2 µg/ml of puromycin.
RNA extraction, cDNA synthesis, and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using RNAiso Plus (Takara) and quantified using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized using M-MLV reverse transcriptase (Takara, Shiga, Japan). qRT-PCR was conducted using SYBR Premix Ex Taq ™ (Takara) on an ABI StepOne Plus system (Applied Biosystems). The primer sequences used were as follows:
EVI5, Forward: 5′-GCATCATCCTGGTTTCTGAC-3′ and reverse: 5′-AGCTTGTCTGGG ACACCATC-3′; β-actin, forward: 5′-CACAGAGCCTCGCCTTTGCC-3′ and reverse: 5′-ACCCATGCCCACCATCACG-3′; Rab11, Forward 5′-GGGACACAGCAGGGCAA-3′ and reverse: 5′-CTCAGTTCTTTCAGCCATCGC-3′; PD-L1, Forward: 5′-GGTGCCGACTACAAGCGAAT-3′ and reverse: 5′-GGTGACTGGATCCACAACCAA-3′; qRT-PCR was performed using SYBR Premix ExTaq™ (Takara) according to the manufacturer’s instructions with an ABI Step One Plus Real Time PCR system (Applied Biosystems). Relative mRNA expression was normalized to β-actin and calculated using the ΔΔCt method22.
Cell viability assay
Cell proliferation was examined using a Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China). Tumor cells were seeded in 96-well plates at a density of 3 × 103 cells/well and grown under normal culture conditions for 24, 48 and 72 h. Cell viability was measured according to the manufacturer’s instructions at several time points (24, 48, and 72 h).
Western blotting assay
Western blotting was performed as described previously23. The antibodies used were anti-EVI5 (Millipore, Billerica, MA, USA, dilution 1:500), anti-Rab11 (Abcam, USA, dilution 1:500), anti-PD-L1 (CST, Danvers, MA, USA, dilution 1:500), anti-β-actin (CST, Danvers, MA, USA, dilution 1:1000), and GAPDH (ABclonal, A19056, dilution 1:10000).
Immunofluorescence staining
Cells were rinsed with PBS buffer three times, fixed with 4% paraformaldehyde for 15 min at room temperature, and permeabilized with 0.5% Triton X-100 for an additional 15 min. Subsequently, cells were washed three times with PBS and blocked with an immunofluorescence staining blocking solution (Beyotime, Shanghai, China) for 30 min at 37℃. They were then incubated overnight at 4 °C with EVI5 primary antibody (Novus, USA, dilution 1:200), anti-Rab11 (Abcam, USA, dilution 1:200), anti-PD-L1 (CST, Danvers, MA, USA, dilution 1:100). Next, cells were washed three times with PBS and incubated with a secondary antibody at 37 °C for 2 h. Cell nuclei were stained with DAPI (Beyotime, Shanghai, China) for 10 min at room temperature. Finally, fluorescent microscopy was used for observation and image capture.
Gene expression and clinical data collection
The RNA sequencing data and corresponding clinical information of LUAD patients in the TCGA database were obtained from UCSC Xena (https://xenabrowser.net/). All RNA sequencing data were log2-transformed for further analysis. There were 513 samples in the RNA sequencing dataset.
Survival analysis
Survival analysis for EVI5 in lung adenocarcinoma (LUAD) was performed using the online Kaplan-Meier Plotter tool. The mRNA expression data and corresponding clinical survival information were retrieved from the database’s integrated TCGA and GEO datasets. Samples were automatically split into “high-expression” and “low-expression” groups based on the best-performing cutoff value as determined by the tool’s algorithm. The two groups were compared using a Kaplan-Meier survival curve, and the hazard ratio (HR) with 95% confidence intervals and log-rank P -value were calculated and displayed on the plot. A P -value of less than 0.05 was considered statistically significant.
Evaluation of immune microenvironment
RNA sequencing(RNA-seq) data and corresponding clinical information were obtained from The TCGA database, encompassing 535 lung adenocarcinoma (LUAD) samples and 59 adjacent non-tumor tissues. Immune cell infiltration was estimated using the immunedeconv R package. For this study, the CIBERSORT algorithm was employed to analyze the differences in the abundance of 22 immune cell subtypes between high and low EVI5 expression groups (stratified by the median expression level of EVI5). A significance threshold of P < 0.05 was applied to define statistically differential expression between the groups.
Proteomics sequencing and analysis
Our proteomic sequencing was commissioned by Shanghai Baispectrum. Protein was extracted from tissue samples using SDT lysis buffer (4% SDS, 100 mM DTT, 100 mM Tris-HCl pH 8.0). Samples were boiled for 3 min and further ultra-sonicated. Undissolved cellular debris were removed by centrifugation at 16 000 g for 15 min. The supernatant was collected and quantified with a BCA Protein Assay Kit (BeyoTime, China). Protein digestion was performed with FASP method described by Wisniewski, Zougman et al.24. Briefly, the detergent, DTT and IAA in UA buffer was added to block reduced cysteine. Finally, the protein suspension was digested with trypsin (Promega) at ratio 50:1 overnight at 37 °C. The peptide mixtures were collected by centrifugation at 16 000 g for 15 min and desalted with C18 StageTip for further LC-MS analysis. The concentrations of re-dissolved peptides were determined with OD280 by Nanodrop One device (Thermo, USA).
Statistical analysis
Data are presented as the mean ± SD. Statistical analyses were performed using unpaired or paired two-tailed t-tests as appropriate. Statistical significance was set at p < 0.05. Analyses were performed using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA).
Result
Correlation between EVI5, Rab11, and PD-L1 mRNA expression in public databases
Analysis of TCGA database ((http://xenabrowser.net/) revealed a significant positive correlation between EVI5 mRNA and Rab11 mRNA levels in LUAD samples (P < 0.001, Fig. 1a). Similarly, EVI5 mRNA expression positively correlated with PD-L1 mRNA levels in LUAD (P < 0.001, Fig. 1b). More importantly, dataset extracted from Kaplan-Meier Plotter (http://www.kmplot.com) indicated that mRNA expression level of EVI5 was significantly associated with poor survival of patients with LUAD.
Additionally, leveraging data from 535 lung adenocarcinoma samples in the TCGA database, this study investigated the differences in the abundance of 22 types of immune cells between the low and high EVI5 expression groups (stratified based on the median expression level of EVI5). The results presented in Supplementary Fig. 1 indicate that in the high EVI5 expression group, the abundance of resting CD4 + memory T cells, monocytes, M2 macrophages, mast cells, and myeloid dendritic cells was higher than that in the low expression group. Conversely, the abundance of CD8 + T cells, plasma cells, follicular helper T cells (TFH), regulatory T cells (Tregs), NK cells, and neutrophils was higher in the low EVI5 expression group compared to the high expression group. These findings collectively suggest that the aberrant expression of EVI5 is closely associated with PD-L1-mediated immune infiltration.
Correlation of EVI5 mRNA expression with Rab11 and PD-L1 mRNA levels in LUAD. (a) EVI5 mRNA is positively correlated with Rab11 mRNA in TCGA database, (b) EVI5 mRNA is positively correlated with PD-L1 mRNA in TCGA database. (c) Effect of the EVI5 mRNA expression level on overall survival in 1161 LUAD patients. Kaplan–Meier plots were generated using Kaplan–Meier Plotter (http://www.kmplot.com). Significant differences: *P < 0.05, **P < 0.01, ***P < 0.001.
High EVI5 expression in LUAD tissues
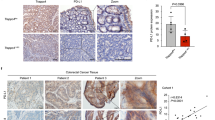
Immunohistochemistry (IHC) analysis of 20 paired LUAD and adjacent normal lung tissue samples revealed elevated expression of EVI5 in 20 LUAD cases (70%), Rab11 in 12 LUAD cases (60%), and PD-L1 in 11 LUAD cases (55%) (Fig. 2a). EVI5 protein expression was predominantly localized in the cytoplasm of tumor cells, As shown in Fig. 2b and d, we performed semi-quantitative analysis of EVI5, PD-L1, and Rab11. The results provided more objective evidence that the expression levels of EVI5, PD-L1, and Rab11 were significantly higher in LUAD tumor tissues compared to adjacent normal tissues. However, there was no correlation between the H-score expression of EVI5 and clinicopathological parameters (Supplementary Table 1). Collectively, our data showed that EVI5, Rab11 and PD-L1 were upregulated in LUAD tissues.
Rab11 and PD-L1 expression in EVI5-KO A549 cells. (a) EVI5 mRNA expression levels. (b) Rab11 mRNA expression levels. (c) PD-L1 mRNA expression levels. (d) Protein levels of EVI5, Rab11, and PD-L1. β-actin served as the internal control. Data represent mean ± SD of three independent experiments. Significant differences: *P < 0.05, **P < 0.01, ***P < 0.001.
Knockout of EVI5 inhibits Rab11 and PD-L1 expression in vitro
To explore the role of EVI5 in tumor immune escape, EVI5-knockout A549 cell lines (EVI5-KO) were established. Knockout was confirmed by the absence of EVI5 protein expression and unchanged EVI5 mRNA levels, indicating effective gene silencing (Fig. 3a, 3 d). In EVI5-KO cells, Rab11 and PD-L1 mRNA and protein levels were significantly reduced compared to those in vector-transfected control cells (Fig. 3b and d).
Immunohistochemical analysis of EVI5, Rab11, and PD-L1 expression in LUAD tissues. (a) Expression levels of EVI5, Rab11, and PD-L1 were assessed in LUAD tissues and compared to adjacent noncancerous lung tissues. (b) EVI5 expression was evaluated by IHC and semi-quantitatively assesed using an H-score methodology. (c) Rab11 expression was evaluated by IHC and semi-quantitatively assesed using an H-score methodology. (d) PD-L1 expression was evaluated by IHC and semi-quantitatively assesed using an H-score methodology.
Overexpression of EVI5 promotes Rab11 and PD-L1 expression in vitro
To further elucidate the function of EVI5, stable EVI5-overexpressing A549 cell lines were generated. Overexpression was confirmed by the increased EVI5 mRNA and protein levels (Fig. 4a and d). In these cells, Rab11 and PD-L1 mRNA and protein levels were significantly elevated, suggesting that EVI5 enhanced tumor immune escape by upregulating Rab11 and PD-L1 (Fig. 4b and d).
Rab11 and PD-L1 expression in EVI5-overexpressing A549 cells. (a) EVI5 mRNA expression levels. (b) Rab11 mRNA expression levels. (c) PD-L1 mRNA expression levels. (d) Protein levels of EVI5, Rab11, and PD-L1. β-actin served as the internal control. Data represent mean ± SD of three independent experiments. Significant differences: *P < 0.05, **P < 0.01, ***P < 0.001.
Inhibition of Rab11 can reduce Rab11 and PD-L1 expression caused by EVI5 overexpression in vitro
To validate the relationship of the EVI5/Rab11/PD-L1 axis, we further constructed a stable Rab11-knockdown in A549 plvx cell lines and EVI5-overexpressing cell lines, followed by detection of the expression levels of EVI5, Rab11, and PD-L1. As depicted in Fig. 5, in A549 cell lines overexpressing EVI5, Rab11 knockdown reduced the expression of Rab11 and PD-L1 induced by EVI5 overexpression.
Rab11 and PD-L1 expression in both EVI5-overexpression and Rab11-knockdown A549 cells. (a) Protein levels of EVI5, Rab11, and PD-L1. (b) EVI5 mRNA expression levels. (c) Rab11 mRNA expression levels. (d) PD-L1 mRNA expression levels. Data represent the mean ± SD of three independent experiments. Significant differences: *P < 0.05, **P < 0.01, ***P < 0.001.
Inhibition of PD-L1 can reduce Rab11 and PD-L1 expression caused by EVI5 overexpression in vitro
In order to validate the relationship between EVI5, Rab11, and PD-L1 in LUAD, we treated PLVX- and EVI5-overexpressing cells with a PD-L1 inhibitor and detected the expression levels of EVI5, Rab11, and PD-L1. First, we selected BMS-202, a selective PD-L1 functional inhibitor to suppress PD-L1 signaling. The IC50 of the drug was determined using the CCK-8 cytotoxicity assay (Fig. 6a and b), the control and experimental groups were treated with the PD-L1 inhibitor at concentrations of 20 µM and 12 µM, respectively.
Upon treatment with the PD-L1 inhibitor in both EVI5-overexpressing and PLVX cell lines, western blot and qRT-PCR analyses revealed that compared to the PLVX group, the EVI5-OE group exhibited significantly increased protein and mRNA expression of Rab11 and PD-L1, However, after adding the PD-L1 inhibitor, both Rab11 and PD-L1 expression levels were markedly reduced in the EVI5-OE group compared to the PLVX and untreated EVI5-OE groups (Fig. 6c-f). These results suggest that inhibition of PD-L1 function attenuates the high expression of Rab11 and PD-L1 in LUAD cells induced by EVI5 overexpression.
Rab11 and PD-L1 expression in PD-L1 inhibition A549 cells. (a) IC50 of PLVX cells in inhibiting PD-L1. (b) IC50 of EVI5-OE cells in inhibiting PD-L1. (c) Protein levels of EVI5, Rab11, and PD-L1. (d) EVI5 mRNA expression levels. (e) Rab11 mRNA expression levels. (f) PD-L1 mRNA expression levels. Data represent the mean ± SD of three independent experiments. Significant differences: *P < 0.05, **P < 0.01, ***P < 0.001.
EVI5 promotes PD-L1 expression via Rab11/PD-L1 signaling pathway in LUAD
EVI5 has been implicated in the regulation of immune-related diseases via its association with Rab1125. However, its precise role in LUAD remains unclear. To elucidate the underlying mechanisms, we investigated the interactions between EVI5, Rab11, and PD-L1 using Immunofluorescence assay. We observed a notable subcellular co-localization of EVI5, Rab11 and PD-L1, indicatign their spatial proximity and potential functional interplay within a regulatory complex (Fig. 7a and b, and 7c). Furthermore, we performed comprehensive Mender’s coefficient to strengthen the evidence base (Fig. 7d), thereby enhancing the reliability of our research findings.
To further explore the relationship between EVI5 and Rab11, we performed proteomic analysis of Rab11-associated proteins in EVI5-overexpressing LUAD cell lines using a Proteome Profiler Array, and Our mass spectrometry proteomics data have been deposited to the ProteomeX Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository26,27 with the dataset identifier PXD064761. The data confirmed a strong correlation between EVI5 and Rab11 expression (Supplement Fig. 2). Additionally, the overexpression of EVI5 significantly enhanced Rab11 expression, further supporting the regulatory role of EVI5 in Rab11-mediated processes.
Taken together, these findings provide compelling evidence that EVI5 interacts with and regulates the Rab11/PD-L1 signaling pathway, thereby promoting tumor immune escape in LUAD. The proposed mechanistic model suggests that EVI5 enhances immune evasion by modulating Rab11/PD-L1 signaling (Fig. 8).
EVI5, Rab11, and PD-L1 Are Highly Correlated in LUAD Cells. (a) Immunofluorescence staining showing co-expression of PD-L1 and Rab11 in EVI5-overexpressing cells compared to control cells (scale bar: 5 μm). (b) Analysis of PD-L1and Rab11 co-located Mander coefficients. M1 represents the proportion of co-localization in the red channel, and M2 represents the proportion of co-localization in the green channel. (c) Immunofluorescence staining showing co-expression of PD-L1 and EVI5 in EVI5-overexpressing cells compared to control cells (scale bar: 5 μm). (d) Analysis of PD-L1and EVI5 co-located Mander coefficients. M1 represents the proportion of co-localization in the red channel, and M2 represents the proportion of co-localization in the green channel. (e) Immunofluorescence staining showing co-expression of EVI5 and Rab11 in EVI5-overexpressing cells compared to control cells (scale bar: 5 μm). (f) Analysis of Rab11 and EVI5 co-located Mander coefficients. M1 represents the proportion of co-localization in the red channel, and M2 represents the proportion of co-localization in the green channel.
Discussion
The clinical success of immune checkpoint blockade (ICB) against the PD-1/PD-L1 axis has been a landmark advancement in lung adenocarcinoma (LUAD) therapy28,29,30. Nevertheless, the modest response rates in unselected patient populations underscore our incomplete understanding of the pathways that govern PD-L1 abundance and dynamics at the tumor cell surface31. Our study identifies the oncoprotein EVI5 as a novel regulator of PD-L1 in LUAD, operating through a mechanism involving the small GTPase Rab11. This finding extends the functional repertoire of EVI5 beyond cell cycle regulation and suggests its potential role in shaping the immunosuppressive tumor microenvironment.
To further investigate the role of EVI5 in LUAD, we constructed EVI5-knockout and EVI5-overexpressing A549 cell lines. EVI5 knockout resulted in a significant reduction in Rab11 and PD-L1 expression, whereas EVI5 overexpression led to their upregulation. Interestingly, the EVI5 knockout cell line we generated using CRISPR/Cas9 technology did not show significant changes at the mRNA level. This is a not uncommon but critically important phenomenon in CRISPR gene editing experiments. The specific reasons for this observation may include the following: (1) The frameshift mutation may not have triggered Nonsense-Mediated mRNA Decay (NMD). Although CRISPR/Cas9-induced indels often generate premature termination codons (PTCs), mRNAs with PTCs located in the last exon or near the natural stop codon may escape NMD-mediated degradation, resulting in stable truncated transcripts. (2) Potential isoform-specific knockout. The EVI5 gene may have multiple transcript isoforms. The designed gRNA might only target certain isoforms, while other unaffected isoforms continue to be expressed, leading to no significant change in overall mRNA levels as detected by qPCR. (3) Low editing efficiency. If the percentage of cells with successful biallelic knockout is low, the presence of a large number of wild-type cells may mask the reduction in mRNA levels, making the overall change undetectable. (4) Production of stable truncated proteins. In some cases, the truncated mRNA may still be translated into a truncated protein, but the mRNA itself remains stable despite the loss of function. (5) Compensatory mechanisms. After gene knockout, feedback mechanisms or compensatory overexpression of other genes in the same pathway may maintain mRNA expression levels, although this is less likely to directly affect the mRNA level of the target gene itself32,33,34,35. Furthermore, addition of a PD-L1 inhibitor or knockdown of Rab11 to EVI5-overexpressing cells markedly decreased the expression of related proteins, underscoring the regulatory role of EVI5 in the PD-L1 signaling axis. Mechanistically, Immunofluorescence assays demonstrated that EVI5, RAB11, and PD-L1 colocalize in the cytoplasm. Proteomic profiling using Proteome Profiler Array further confirmed the correlation between EVI5 and Rab11.
Our in vitro gain- and loss-of-function studies provided direct evidence that EVI5 is upstream of Rab11 and PD-L1 in the regulatory hierarchy, significantly modulating their protein expression. While the precise biochemical mechanism requires further dissection, our protein-protein interaction data, including proteomic profiling and observed co-localization, are consistent with a model wherein EVI5, potentially via its influences Rab11 expression to facilitate the expression of PD-L1, thereby enhancing its immunosuppressive function. This model is further supported by the reversal of the EVI5-overexpression phenotype upon Rab11 knockdown or PD-L1 inhibition, confirming the functional dependency of this axis.
Nevertheless, we fully acknowledge the following limitations in the current study: (1) The findings are primarily derived from in vitro cellular experiments, lacking validation through in vivo animal models to confirm the tumor-promoting effects of EVI5 in LUAD; (2) The precise molecular.
mechanisms underlying EVI5-mediated regulation of the Rab11/PD-L1 signaling axis remain to be elucidated. Future studies should focus on addressing these knowledge gaps. The present work establishes a foundational experimental framework for the preliminary investigation of LUAD progression mechanisms and facilitates subsequent in-depth research.
Conclusion
This study demonstrates that EVI5 promotes PD-L1 expression through Rab11 in LUAD, suggesting its potential role in tumor immune regulation. These findings provide new insights into the molecular mechanisms controlling PD-L1 expression and identify EVI5 as a potential therapeutic target for overcoming immunotherapy resistance in LUAD.
Data availability
Our mass spectrometry proteomics data have been deposited to the ProteomeX Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD064761. If anyone wants to request data from this study, please contact Tingting Cai.
References
Wei, X., Li, X., Hu, S., Cheng, J. & Cai, R. Regulation of ferroptosis in lung adenocarcinoma. Int. J. Mol. Sci. 24 (19), 14614. https://doi.org/10.3390/ijms241914614 (2023).
Liu, Y. et al. circ_0004140 promotes LUAD tumor progression and immune resistance through circ_0004140/miR-1184/CCL22 axis. Cell. Death Discov. 8 (1), 181. https://doi.org/10.1038/s41420-022-00983-w (2022).
Chen, P., Quan, Z., Song, X., Gao, Z. & Yuan, K. MDFI is a novel biomarker for poor prognosis in LUAD. Front. Oncol. 12, 1005962. https://doi.org/10.3389/fonc.2022.1005962 (2022).
Li, J., Gu, X., Gao, C. & Zhang, J. Six MicroRNA prognostic models for overall survival of lung adenocarcinoma. Genet. Res. (Camb). 5955052. https://doi.org/10.1155/2022/5955052 (2022).
Shao, L. et al. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell. Death Discov. 7 (1), 145. https://doi.org/10.1038/s41420-021-00527-8 (2021).
Boucherit, N., Gorvel, L. & Olive, D. 3D tumor models and their use for the testing of immunotherapies. Front. Immunol. 11, 603640. https://doi.org/10.3389/fimmu.2020.603640 (2020).
Liu, B. et al. Genetically engineered CD276-anchoring biomimetic nanovesicles target senescent escaped tumor cells to overcome chemoresistant and immunosuppressive breast cancer. Biomaterials313:122796. (2025). https://doi.org/10.1016/j.biomaterials.2024.122796
Zhang, C. et al. H3K18 lactylation potentiates immune escape of Non-Small cell lung cancer. Cancer Res. 84 (21), 3589–3601. https://doi.org/10.1158/0008-5472.CAN-23-3513 (2024).
Novoplansky, O. et al. Dual Inhibition of hers and PD-1 counteract resistance in KRASG12C-mutant head and neck cancer. J. Exp. Clin. Cancer Res. 43 (1), 308. https://doi.org/10.1186/s13046-024-03227-0 (2024).
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-Positive Non-Small-Cell lung cancer. N. Engl. J. Med. 375 (19), 1823–1833. https://doi.org/10.1056/NEJMoa1606774 (2016). & KEYNOTE-024 Investigators
Tony, S. K. et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. The Lancet. 393(10183):1819–1830. (2019). https://doi.org/10.1016/S0140-6736(18)32409-7
Mazieres, J. et al. Atezolizumab versus docetaxel in pretreated patients with NSCLC: final results from the randomized phase 2 POPLAR and phase 3 OAK clinical Trials. Journal of thoracic oncology: official publication of the international association for the study of lung cancer. J. Thorac. Oncol. 16 (1), 140–150. https://doi.org/10.1016/j.jtho.2020.09.022 (2021).
Ye, Z. et al. Manipulation of PD-L1 endosomal trafficking promotes anticancer immunity. Adv. Sci. (Weinheim Baden-Wurttemberg Germany). 10 (6), e2206411. https://doi.org/10.1002/advs.202206411 (2023).
Ren, Y. et al. TRAPPC4 regulates the intracellular trafficking of PD-L1 and antitumor immunity. Nat. Commun. 12 (1), 5405. https://doi.org/10.1038/s41467-021-25662-9 (2021).
Müller, M. P. & Goody, R. S. Molecular control of Rab activity by GEFs, gaps and GDI. Small GTPases. 9 (1–2), 5–21. https://doi.org/10.1080/21541248.2016.1276999 (2018).
Laflamme, C. (ed Emery, G.) In vitro and in vivo characterization of the Rab11-GAP activity of drosophila Evi5. Methods Mol. Biology (Clifton N J) 1298 187–194 https://doi.org/10.1007/978-1-4939-2569-8_16 (2015).
Cai, T. et al. EVI5 is an oncogene that regulates the proliferation and metastasis of NSCLC cells. J. Exp. Clin. Cancer Res. 39 (1), 84. https://doi.org/10.1186/s13046-020-01585-z (2020).
Yang, W., Liu, C., Li, Z. & Cui, M. Multi-omic biomarkers associated with multiple sclerosis: from Mendelian randomization to drug prediction. Sci. Rep. 15 (1), 9421. https://doi.org/10.1038/s41598-025-94303-8 (2025).
Mitra, J., Hegde, P. M. & Hegde, M. L. Loss of endosomal recycling factor RAB11 coupled with complex regulation of MAPK/ERK/AKT signaling in postmortem spinal cord specimens of sporadic amyotrophic lateral sclerosis patients. Mol. Brain. 12 (1), 55. https://doi.org/10.1186/s13041-019-0475-y (2019).
Soltani, S., Webb, S. M., Kroll, T. & King-Jones, K. Drosophila Evi5 is a critical regulator of intracellular iron transport via transferrin and ferritin interactions. Nat. Commun. 15 (1), 4045. https://doi.org/10.1038/s41467-024-48165-9 (2024).
Schmidt, T., Zörnig, M., Beneke, R. & Möröy, T. MoMuLV proviral integrations identified by Sup-F selection in tumors from infected myc/pim Bitransgenic mice correlate with activation of the gfi-1 gene. Nucleic Acids Res. 24 (13), 2528–2534. https://doi.org/10.1093/nar/24.13.2528 (1996).
Zhu, J. et al. CD73 promotes non-small cell lung cancer metastasis by regulating Axl signaling independent of GAS6. Proc. Natl. Acad. Sci. U S A. 121 (43), e2404709121. https://doi.org/10.1073/pnas.2404709121 (2024).
Fu, Y. et al. Abnormally activated OPN/integrin αVβ3/FAK signalling is responsible for EGFR-TKI resistance in EGFR mutant non-small-cell lung cancer. J. Hematol. Oncol. 13 (1), 169. https://doi.org/10.1186/s13045-020-01009-7 (2020).
Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample Preparation method for proteome analysis. Nat. Methods. 6 (5), 359–362. https://doi.org/10.1038/nmeth.1322 (2009).
Welz, T., Wellbourne-Wood, J. & Kerkhoff, E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 24 (7), 407–415. https://doi.org/10.1016/j.tcb.2014.02.004 (2014).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47 (D1), D1211–D1217. https://doi.org/10.1093/nar/gky869 (2019).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 50 (D1), D1522–D1527. https://doi.org/10.1093/nar/gkab1081 (2022).
Huang, D. et al. MAX transcriptionally enhances PD-L1 to inhibit CD8 + T cell-mediated killing of lung adenocarcinoma cells. Cell. Immunol. 386, 104706. https://doi.org/10.1016/j.cellimm.2023.104706 (2023).
Wang, Y. et al. MTSS1 curtails lung adenocarcinoma immune evasion by promoting AIP4-mediated PD-L1 monoubiquitination and lysosomal degradation. Cell. Discovery. 9 (1), 20. https://doi.org/10.1038/s41421-022-00507-x (2023).
Zheng, C. et al. Systems pharmacology: a combination strategy for improving efficacy of PD-1/PD-L1 Blockade. Brief. Bioinform. 22 (5), bbab130. https://doi.org/10.1093/bib/bbab130 (2021).
Matulonis, U. A. et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Annals Oncology: Official J. Eur. Soc. Med. Oncol. 30 (7), 1080–1087. https://doi.org/10.1093/annonc/mdz135 (2019).
Haapaniemi, E. et al. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage respone. Nature Medicine. 24(7), 927-–930 (2018). https://www.nature.com/articles/s41591-018-0049-z
Mathis, N. et al. Predicting prime editing efficiency and product purity by deep learning. Nat. Biotechnol. 41 (8), 1151–1159 (2023). https://www.nature.com/articles/s41587-020-0677-y
Roper, J. et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35 (6), 569–576. https://doi.org/10.1038/nbt.3836 (2017).
Lykke-Andersen, S. & Jensen, T. H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16 (11), 665–677 (2015). https://www.nature.com/articles/s41580-019-0128-0
Acknowledgements
We thank all patients who participated in this study for their cooperation, and we thank Dr. YongQiang Shi and Dr. Qing Li at The Third Affiliated Hospital of soochow University, Changzhou, China, for clinical sample collection.
Funding
This work was supported by grants from National Natural Science Foundation of China (No.81970080), Changzhou Municipal Health Commission project (No.QN202309), the Science and Technology Support Plan (Social Development) Project of Changzhou (CE20235057) and Changzhou Applied Basic Research Program (CJ20220091).
Author information
Authors and Affiliations
Contributions
CTT, XYY, ZJ and LC contributed to the conception and design; CTT, XYY, ZPP, ZXY, SYQ contributed to the acquisition of data; CTT, XJ, XYY, LH, HQ and QY contributed to the analysis and interpretation of data; CTT and LC contributed to the writing, review, and/or revision of the manuscript; CTT, XYY, ZPP, ZXY contributed to the administrative, technical, or material support; LQ, ZJ and LC supervised the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
20 paired NSCLC tissues and adjacent noncancerous lung tissues were collected, with the informed consent of the patients, from the Third Affiliated Hospital of Soochow University between 2020 and 2024. This study was approved by the Academic Advisory Board of Soochow University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cai, T., Xu, Y., Zhang, P. et al. EVI5 unveils a role of the oncogene in modulating the Rab11/PD-L1 pathway in lung adenocarcinoma. Sci Rep 15, 40882 (2025). https://doi.org/10.1038/s41598-025-24726-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24726-w