Abstract
Inflammatory bowel disease (IBD), including Crohn’s disease and Ulcerative colitis, often shows variable responses to biological therapies. Identifying the most significant variables for predicting the response to these therapies could help prioritize efforts in data collection and preprocessing. This study evaluated the predictive performance of machine learning models in forecasting remission and response to vedolizumab and ustekinumab in the treatment of IBD. The goal was not to compare the two therapies, but rather to identify the variables most influential in predicting response for each treatment. Data from 227 IBD patients treated at Virgen Macarena University Hospital (2015–2022) were analyzed. Clinical, demographic, and laboratory variables were used to develop Extreme gradient boosting (XGBoost) models to predict the clinical response at 26 and 52 weeks and remission at 52 weeks. Model performance was evaluated via F1 scores, accuracy, precision, and recall, with fairness analyses across sex and age groups. The models achieved F1 scores of 0.842, 0.869, and 0.649, respectively. The predictors included leukocyte count, FCP, CRP, and vitamin B12 levels, with higher inflammatory marker levels linked to poorer responses. Demographic subgroup analysis revealed variability in model performance due to small sample sizes. Machine learning models have potential as clinical decision support tools for personalizing IBD treatment. These findings underscore the value of leveraging real-world evidence in optimizing therapeutic strategies. Further multicenter studies are needed to validate these models and enhance their applicability across diverse populations. Taken together, these findings arise from an exploratory, hypothesis-generating study that constitutes an early step toward truly personalized biologic therapy in IBD.
Similar content being viewed by others
Introduction
Background
Inflammatory bowel disease (IBD) encompasses two main entities: Crohn’s disease (CD) and Ulcerative colitis (UC). These chronic, relapsing conditions are only partially understood in terms of cause and are characterized by an excessive immune response that leads to variable depths and extents of intestinal lesions. These diseases follow a chronic, recurrent and relapsing course, with unpredictable progression and no tendency toward spontaneous remission.
An improved understanding of this disease has led to the development of a broad therapeutic arsenal that reflects not only the complexity and heterogeneity of the disease but also the suboptimal efficacy of these drugs. Despite advancements in the introduction of anti-TNF medications, approximately one-third of patients do not respond to this therapy, and approximately 30–40% lose efficacy within the first year of treatment1. New biological agents that have emerged in recent years, such as ustekinumab and vedolizumab, help some patients who do not respond to anti-TNF therapy. However, these agents have also been found to be ineffective in a significant number of patients2, and the optimal sequence for using these new treatments relative to anti-TNF agents remains unclear.
The chronic and recurrent course of IBD, its complex and partially understood etiology and pathogenesis, as well as the cost of available treatments, have imposed an increasing burden on healthcare systems and present a challenging management landscape. The underlying mechanisms for primary nonresponse are multifactorial and include disease and drug characteristics as well as factors related to treatment strategy3. In this context, efforts have been directed toward identifying predictive factors for response to biological therapy that can help determine the likelihood of a patient responding to a specific drug before initiating treatment, representing a cost-effective approach to treatment individualization. However, this goal has not yet been achieved in this pathology, with results up to this point being variable and often contradictory, with less evidence available for vedolizumab and ustekinumab.
Artificial intelligence (AI) and its subdiscipline, machine learning (ML), have become promising tools in the field of IBD. Their ability to integrate and analyze large datasets allows the correlation of patient characteristics with disease susceptibility, activity, and response to specific therapies. Interest in using big data for personalized IBD treatment is growing. In particular, predicting therapeutic nonresponse highlights ML’s potential to uncover novel insights from complex and voluminous clinical data4.
Previous studies have used ML models to predict response to thiopurines5 and biological therapies such as infliximab, ustekinumab6,7, and vedolizumab8.
Other studies using large longitudinal claims datasets have demonstrated the feasibility of accurately predicting various adverse outcomes. These include hospitalizations, long-term steroid use, and initiation of biologics. Complex ML models such as Random Forests and LASSO regression were used in these analyses9. Our study uniquely focuses on this area and aims to further contribute to the growing body of real-world evidence. Specifically, we evaluate the predictive performance of XGBoost models for vedolizumab and ustekinumab in a real-world clinical practice setting. The goal is to identify key clinical and biochemical predictors of treatment response and remission. The results so far have been promising. Indeed, a systematic review of machine learning-based prediction models for diagnosis and prognosis in IBD—primarily utilizing routinely available clinical data—concluded that ML models generally outperform traditional statistical models in risk prediction10.
Objectives
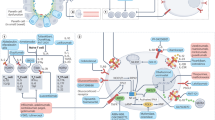
The identification of these factors could reduce the costs of assigning ineffective drugs with high economic impact, avoid hospitalizations, surgical procedures, complications, and achieve earlier symptomatic and biological remission. This goal remains unmet in this pathology, and all studies providing results in this regard should be considered to improve patient care. The main goal is to identify the most determinant variables for predicting treatment effectiveness in the overall patient population. Specifically, the aim of our study was to find factors to achieve remission or response with vedolizumab or ustekinumab. The aim was not to compare the two therapies, but rather to identify the variables most influential in predicting response for each treatment. Effectiveness will be assessed through clinical response at 26 and 52 weeks, as well as clinical remission at 52 weeks. Additionally, the project seeks to develop predictive models to determine the likelihood of achieving a clinical response or remission. A comprehensive overview of the study, including the population study, methodology, and key findings, is visually summarized in Fig. 1. Given that our work is exploratory in nature, demonstrating the potential of machine learning models as clinical decision support tools, the models represent and are intended to be an early first step to future confirmatory trials rather than to dictate immediate clinical practice.
Study population, key findings, and machine learning pipeline. Left: Overview of the 227 participants with Crohn’s disease (CD) or ulcerative colitis (UC), showing treatment distribution (vedolizumab or ustekinumab), drug proportions by disease type, and key clinical predictors of treatment response identified from SHAP analysis. Right: Machine learning pipeline illustrating dataset refinement to 29 clinical variables, XGBoost model training and validation, and evaluation with accuracy, precision, recall, F1 score, SHAP feature importance, and fairness analysis.
Methods
Data collection
Biological therapies such as vedolizumab and ustekinumab have proven effective in treating IBD. However, patient responses vary, prompting research to identify clinical, demographic, and biochemical predictors for treatment outcomes. Recognizing these factors is crucial for developing algorithms to optimize drug selection and enable personalized treatment decisions.
To identify key predictors, a dataset was compiled at Hospital Virgen Macarena University Hospital (Hospital Universitario Virgen Macarena—HUVM, Seville) from primary care records within the Andalusian Health Population Database (Base poblacional de salud de andalucía—BPS). This dataset includes electronic health records (EHR) from Diraya Primary Care, XXI prescription records, mortality data, and medical records from Diraya Specialized and hospital prescription programs, with laboratory variables obtained from the Analytical Testing Module (Módulo de pruebas analíticas—MPA).
This study was approved by the Coordinating Ethics Committee for Biomedical Research of Andalusia (CCEIBA, from the Spanish acronym), under protocol code FPS-EII-2021-04, version 1 dated September 14, 2021. The Committee confirmed that the study meets the ethical requirements established by Royal Decree 957/2020, the Declaration of Helsinki, and applicable regulations. Due to the retrospective nature of the study, the Coordinating Ethics Committee for Biomedical Research of Andalusia (CCEIBA) waived the need for obtaining informed consent. All research procedures were performed in accordance with relevant guidelines and regulations.
The dataset contains 111 variables, including five target variables. The key explanatory variables include active ingredient, sex, family history of IBD, disease location, prior treatments, smoking status, disease duration, and various laboratory tests. In accordance with the clinical criteria, the final dataset included 26 independent variables and 3 target variables. Three of the target variables were used in the analysis: clinical response at 26 and 52 weeks and clinical remission at 52 weeks. Clinical response was defined as a reduction of at least 3 points in the Harvey-Bradshaw Index for patients with Crohn’s disease (CD) and in the partial Mayo score for those with Ulcerative Colitis (UC) (dichotomous variable: yes/no). Clinical remission was defined as a dichotomous variable (yes/no), based on established clinical criteria. Although data points for clinical remission at week 104 (2 years) and clinical response at week 104 were collected, the predictive models in this study were specifically developed for the 26- and 52-week outcomes as detailed in the objectives. The dataset initially had 228 patient records, but after one patient was excluded because of missing data, the final cohort included 227 participants, with therapy initiation between January 2015 and January 2022. The independent variables are labelled X, and the target variables (clinical response at 26 and 52 weeks and clinical remission at 52 weeks) are y1, y2, and y3. A rigorous anonymization process was employed to ensure that specific patient data could not be identified, safeguarding privacy and maintaining compliance with ethical standards. The final cohort by disease and treatment is presented in Fig. 1.
Model development
The study followed the inclusion criteria (age ≥ 18 years, diagnosis of moderate-to-severe IBD, and at least three months of treatment with vedolizumab and/or ustekinumab) and exclusion criteria (lack of EHR data, registration of less than 50% of the study variables, or loss to follow-up before 26 weeks after the start of treatment). Given the retrospective, single-center nature of the dataset and the modest cohort size (n = 227), the resulting models may not generalize unaltered to external populations; all performance metrics should therefore be interpreted as center-specific.
The primary objective was to identify the variables that are most relevant for the predictive models of clinical response and remission using baseline and follow-up data. Three models predict the clinical response at 26 weeks, 52 weeks, and clinical remission at 52 weeks. Initially, eight models were considered for subpopulations defined by disease and therapy combinations, but these were excluded from the analysis.
For each model, training (75%) and test sets (25%) were created and stratified by the target variable to account for imbalance. The F1 score, which is ideal for imbalanced datasets, was used for optimization11. Unlike area under curve and receiver operating characteristic (AUC-ROC), which may overestimate performance in imbalanced datasets, the F1 score balances precision and recall. The XGBoost classifier12 was chosen for its performance, and a random search with fivefold cross-validation was used for hyperparameter tuning. The model was trained with the best parameters and then tested on the test set.
Three general models were trained based on the three target variables, excluding subpopulations with fewer than 163 samples. This threshold was determined via power analysis (Table S1) for the general linear model13, ensuring statistical robustness. Only the three general models are included in further analyses. Subpopulation models with insufficient sample sizes are noted in Online Resource 1.
To minimize optimistic bias typical of retrospective studies, we employed nested fivefold cross-validation for hyper-parameter tuning, regularization (L2) within XGBoost, early stopping on an unseen validation fold, and independent hold-out testing; these steps reduce performance inflation.
Fairness assessment
A fairness analysis will be conducted on each model’s F1 scores, stratified by sex and age groups (Adolescence: 10–19 years, Youth: 20–24 years, Adulthood: 25–64 years, and Old Age: 65 years and older). F1 scores for demographic subgroups with fewer than 10 test cases are not reported or interpreted due to the unreliability of performance metrics with such limited data. This approach supports the assessment of equitable performance across demographic groups and helps identify potential disparities for that may require corrective action.
Results
Study population
This section outlines key findings on treatment effectiveness and patient outcomes over time. Statistical analyses were used to evaluate the clinical response at 26 and 52 weeks and remission at 52 weeks.
The study population was stratified based on the presence or absence of three previously defined target variables. Such stratification enables a comprehensive evaluation of treatment effectiveness over time. As shown in Tables 1, 2 and 3, 74.4% (n = 169) of the participants were diagnosed with Crohn’s disease. Moreover, ustekinumab was administered to 59.5% (n = 135) of the participants. Statistical analysis employing P-values revealed no significant disparities in treatment outcomes between the two medications across the assessed time intervals and response criteria. The behavior of patients with Crohn’s disease indicated a statistically significant difference in the presence or absence of a clinical response at 52 weeks, with P = 0.002. The clinical response recovery rate at 52 weeks for patients with fistulizing behavior (90.6%, 29/32) was greater than that for those with stenosing behavior (65.6%, 40/61). The presence of comorbidity asthma at the start of treatment was associated with statistically significant differences between groups across all analyzed outcomes. Specifically, patients without asthma at the start of treatment had better response rates at 26 and 52 weeks, as well as a higher remission rate at 52 weeks, than did those with asthma. The C-reactive protein (CRP) level measured at the start of treatment was 7.5 [3.4, 26.6] mg/L in patients who did not achieve a clinical response at 26 weeks, whereas it was 4.7 [2.0, 13.9] mg/L in those who did, P = 0.03, indicating a statistically significant difference between these groups. However, for clinical response and remission outcomes at 52 weeks, CRP levels at the start of treatment were not significantly different. Therefore, CRP appears to be a more relevant predictor of short-term outcomes, which aligns with expectations, as CRP levels are measured at treatment initiation. The calprotectin levels exhibited a similar pattern in relation to the clinical response at 26 weeks, with P = 0.01. Notably, patients who do not achieve a clinical response at this time point have higher calprotectin levels, measuring 654.1 [141.6, 1333.6] µg/g, than those who achieve a response of 226.9 [86.6, 775.2] µg/g. However, the calprotectin level was not a statistically significant predictor of other outcomes. In patients assessed for clinical response at 26 weeks, the percentage of neutrophils was significantly greater in non-responders (64.5% [53.8, 70.8]) than in responders (58.6% [53.1, 65.3]) with P = 0.02. Similarly, at 52 weeks, non-responders presented an increased percentage of neutrophils (65.3% [54.3, 72.1]) compared to responders (58.3% [53.0, 65.2]), with P = 0.006, indicating statistical significance. However, no significant difference in the percentage of neutrophils was observed for patients in clinical remission at 52 weeks. In terms of clinical remission at 52 weeks, there was a significant difference related to the presence of extraintestinal manifestations (EIMs), P = 0.01. Specifically, 50.5% of patients with EIMs achieved remission, whereas 33.9% of those without EIMs achieved remission. Despite this, no significant differences were found in the clinical response at either 26 or 52 weeks based on the presence or absence of EIMs. Finally, prior exposure to vedolizumab was associated with a statistically significant difference in clinical remission at 52 weeks P = 0.04. Patients with previous vedolizumab treatment had a lower remission rate (5.6%) than did those without prior exposure (94.4%). However, no significant differences were observed for the clinical response at either 26 or 52 weeks.
Models’ performances and key predictors
Three XGBoost classifier models were developed to predict the clinical response at both 26 and 52 weeks and clinical remission at 52 weeks, and the influence of each variable was analyzed. All the models use the same set of explanatory variables, with the only difference being the target variable. The models were stratified by the respective outcome.
The model trained to predict the clinical response at 26 weeks demonstrated the following performance metrics on the test set: Accuracy = 0.737, F1 Score = 0.842, Precision = 0.800 and Recall = 0.889 (Table 4). Notably, the model tends to overpredict positive clinical responses at 26 weeks compared with negative responses. This tendency may be due to the imbalance in the dataset, with a greater number of positive clinical responses at 26 weeks (No = 49, Yes = 178).
The beeswarm plot provides a detailed view of how different variables contribute to the prediction of the clinical response at 26 weeks. Figure 2 displays the variables ranked in order of their importance. It is observed that low values of Leukocyte count (× 10^3/µL) are associated with a negative effect on the clinical target variable. The variable Creatinine (mg/dL) has a strong positive influence on the target variable for high values. High values of Fecal Calprotectin (FCP) (µg/g) and CRP (mg/L) also negatively impact the target variable. High levels of Total Protein (g/dL) and Vitamin B12 (pg/mL) were associated with a positive effect on the response variable. Additionally, the greater age of the patients at the start of the treatment is associated with a positive effect on the target variable; moreover, higher values of the variable Years with the disease (calculated as the difference between diagnosis and the start of treatment in this study) positively impact the target variable. For patients with Crohn’s disease, inflammatory behavior is associated with a positive effect on the target variable. Moreover, patients who took corticosteroids in the 12 months prior to the start of treatment were associated with a slightly negative effect on the target variable.
The same architecture was employed to predict the clinical response at 52 weeks, yielding the following performance metrics on the test set: Accuracy = 0.772, F1 Score = 0.869, Precision = 0.811 and Recall = 0.935 (Table 5). Like the previous model for 26-week predictions, this model tends to overpredict positive clinical responses outcomes at 52 weeks compared with negative responses. This inclination may be attributed to data imbalance, as the dataset comprises a greater proportion of positive clinical responses at 52 weeks (No = 44, Yes = 183).
The Shapley additive explanations (SHAP) beeswarm plot for this model is presented in Fig. 3. A lower lymphocytes count (× 10^3/µL) is associated with a negative effect on the clinical response at 52 weeks. Patients who initiate treatment at an older age and those with a longer disease duration have a positive association with the target variable. This effect is also observed at 26 weeks concerning these two variables, with the association remaining consistent across the results from 26 to 52 weeks. Additionally, high values of FCP (µg/L) and elevated levels of CRP (mg/L) negatively impact the target variable, which aligns with previous analyses. Furthermore, high levels of Creatinine (mg/dL) are correlated with a positive response at 52 weeks, which is consistent with earlier findings. Finally, elevated levels of Total protein (g/dL) and Vitamin B12 (pg/mL) are associated with a positive response of the target variable.
For the prediction of clinical remission at week 52, we obtained the following metrics in the test set: Accuracy = 0.526, F1 Score = 0.649, Precision = 0.543 and Recall = 0.806 (Table 6). This model demonstrated a greater tendency to predict positive clinical remission than negative outcomes did with the previous two models. This tendency may be attributed to data imbalance, as the dataset for clinical remission outcomes at 52 weeks contains a greater proportion of positive outcomes (Yes = 124) compared to negative outcomes (No = 103). While this distribution for clinical remission is more balanced than that for the clinical response outcomes at 26 and 52 weeks (where clinical response at 52 weeks had No = 44 and Yes = 183 outcomes), the model for clinical remission still shows a propensity to overpredict positive clinical outcomes.
Owing to the limited performance of the model, the insights from the SHAP beeswarm plot, displayed in Fig. 4, should be interpreted with caution. In line with previous analyses, high levels of Total Protein (g/dL) and Vitamin B12 are associated with a positive effect on the clinical remission at 52 weeks. The influences of high and low values of FCP (µg/g) and CRP (mg/L) are also ambiguous. With respect to the Age at start of drugs, older patients are associated with a positive effect on the target variable. The effect of Years with Disease is not clear. Notably, male biological sex is associated with a worse response than female biological sex. Furthermore, it should be noted that the X-axis in the SHAP beeswarm plot is scaled differently from that in the previous plots.
Fairness analysis
To obtain a more comprehensive understanding of the models’ performance across different demographic groups, a fairness analysis was conducted by evaluating the F1 score on the test dataset, stratified by sex (female and male) and age categories described in the Materials and Methods section. Unfortunately, data on race were not available for inclusion in this analysis. Fairness evaluation was performed for each of the trained models.
For the clinical response at 26 weeks Figs. 5 and 6 indicate that the F1 score on the test data is slightly higher for males than for females. When stratifying by age groups, differences are observed in adolescence, youth, and old age groups due to the small sample sizes; these results should be interpreted with caution and are unlikely to be reliable. The adulthood group had a lower F1 score than did the adolescence and youth groups but a higher F1 score than did the old age group.
Moreover, the Clinical Response at 52 weeks is presented in Figs. 7 and 8. The F1 score performance is balanced between the sexes. However, estimates for adolescence, youth and old-age strata are unstable (F1 range 0.0–0.92) because each contains < 15 test cases; any apparent performance gap in these groups should therefore be viewed as inconclusive rather than definitive evidence of bias. Stratified by age, extreme values are observed in the adolescence, youth, and old age groups, with F1 scores of 0.0, 0.91, and 0.92, respectively. These results should be interpreted cautiously because of the small sample sizes. The adulthood group shows more consistent performance because of their larger sample sizes.
Finally, for Clinical Remission at 52 weeks Figs. 9 and 10 show consistent performance across demographic groups. Males achieved a slightly higher F1 score than females did. The adolescence and youth groups displayed lower F1 scores due to small sample sizes, whereas the adulthood group performed better, and the old age group had an intermediate F1 score. Small sample sizes in the old age group make it difficult to predict the model’s performance in new individuals.
It is important to note that results for subgroups with fewer than 10 individuals should not be interpreted due to their unreliability, and the corresponding visualizations should be considered exploratory only. Where applicable, we recommend omitting subgroup results with n < 10 to avoid overinterpretation.
Discussion
Principal results
The extensive therapeutic arsenal developed for inflammatory bowel disease underscores the inherent heterogeneity and complexity of this disease. While the predictive models for clinical response at weeks 26 and 52 demonstrated satisfactory performance, the model for predicting clinical remission at week 52 showed comparatively lower accuracy. This difference may indicate that the current set of selected variables is insufficient for reliably predicting clinical remission at week 52. Furthermore, the stricter criteria required for clinical remission, along with the smaller number of patients achieving remission, may have contributed to the observed differences in model performance.
Comparison with prior work
These results are comparable to those obtained in previous studies6,7,8,14 in which machine learning methodologies were used to develop predictive models of response to ustekinumab and vedolizumab, most of which used data from multicenter cohort studies and clinical trials. In this study, assessing the weighting of variables within the algorithm has been indispensable, as it enables interpretation of the results from a medical and physiological perspective. This evaluation is essential for determining the clinical relevance of the model and for ruling out spurious correlations that could otherwise lead to misleading conclusions.
Among the characteristics included in the models, laboratory parameters are variables that seem to be relevant in the prediction of clinical response and remission. Our methodological approach further distinguishes this study. While previous machine learning studies in IBD have often used broad administrative claims data or a variety of ML algorithms9, our research takes a more focused approach. We specifically applied XGBoost models to detailed real-world clinical and biochemical parameters. This allowed us to generate interpretable insights into key predictors of treatment response, using SHAP analysis. In this sense, inflammatory parameters such as FCP, CRP and leukocyte count have been associated with a negative effect; thus, higher disease activity at the start of treatment, represented by elevated values of these markers, seems to be related to a worse response to treatment. These results have also been observed in previous studies using machine learning methodologies6,7,8. The Lymphocyte count is another parameter that seems to be related to a positive effect on treatment response and has also been described in previous studies15. Although the etiology of IBD has not been clearly elucidated, the available evidence describes the role of the adaptive immunity of the intestinal mucosa in the development and evolution of the disease. These results are in line with those obtained in previous studies and could suggest, as described in the study by Miyoshi et al.14, the existence of "lymphocyte-dominant" subgroups of patients, a characteristic that could confer patients an advantage in response to biologic therapy, although we believe that larger studies are needed to confirm these results.
Key predictive variables
Total protein, creatinine and vitamin B12 levels are analytical parameters that are related to the nutritional status of the patient and represent other variables that are positively associated with the response to biological therapy in models. Suboptimal nutritional status and low serum micronutrient levels can negatively affect both the induction and maintenance of disease remission and the quality of life of IBD patients16. Studies by Waljee et al.8 and Miyoshi et al.14 have described this association between protein and creatinine levels and the response to vedolizumab. On the other hand, the evidence on vitamin B12 as a potential predictor of response to biologic therapy is limited and has not been analyzed in the referred studies that use machine learning methodologies; thus, more studies are needed to support this association.
Among the clinical variables included in the models, biological sex, disease duration, age at treatment initiation, CD behavior and location, and the need for previous corticosteroid treatment are characteristics that appear to be related to the response to biologic therapy.
The influence of sex on the response to biologic therapy has been analyzed in some observational studies17,18, reporting a tendency toward a better response among women with CD treated with ustekinumab, which could be in line with the results of our study. With respect to disease duration, Chaparro et al.7 reported that disease duration and age at drug initiation were associated with a lower probability of clinical remission over time. However, the results obtained in our study were the opposite, with a positive association between disease duration and response to the drug; thus, these results should be interpreted with caution, and larger studies are needed to draw more robust conclusions in this regard.
Most published studies have not demonstrated a clear association between CD location and treatment response3. However, in an observational study of pediatric IBD patients treated with vedolizumab, patients with colonic involvement only (UC or isolated Crohn’s colitis without small bowel involvement) were more likely to achieve clinical remission19.
The behavior of CD constitutes another feature that appears to influence the response to biologic therapy. Patients with disease progression to a stenosing or perforating phenotype with established intestinal damage may be expected to be less likely to respond to medical treatment. In this context, some observational studies have reported a worse response to ustekinumab20 and anti-TNF drugs21 in patients with CD and a stenosing phenotype.
Regarding the influence of corticosteroid treatment, some observational studies have reported a negative influence of the concomitant use of corticosteroids on the response to vedolizumab22,23. Similarly, in the work published by Chaparro et al.7, in which the baseline characteristics of patients with active CD were used for the development of clinical response prediction models with ustekinumab via ML methodologies, the number of cycles of steroid treatment concomitant with the use of ustekinumab was related to a lower probability of clinical remission. These results are related to those observed in our study in the vedolizumab group, and could be explained if previous maintenance corticosteroid treatment was used in those patients with more severe or refractory inflammatory activity at the initiation of biologic therapy.
Limitations
To date, few published studies have used ML methodologies to determine predictors of response to ustekinumab and vedolizumab, and most of these studies use data from clinical trials. The use of data from patients seen in daily clinical practice is an advantage when these models are applied in the real world.
This study presents several limitations that affect the robustness and generalizability of the findings. The retrospective, single-center design introduces variability in data completeness, follow-up timing, and clinical documentation. These limitations—particularly the single-center, retrospective design and small sample size—indicate that the predictive models developed here are specific to the Virgen Macarena University Hospital context and cannot be directly generalized or applied to other patient populations or healthcare systems without rigorous external validation. A systematic review highlighted that, although machine learning models generally outperform traditional statistical models in IBD risk prediction, most models are at high risk of bias and often lack external validation. These issues frequently stem from the same design constraints10. Drug trough levels, which may be informative, were not included due to current monitoring guidelines24. Some relevant confounders may be absent from the structured EHR data, limiting model interpretability.
The small overall sample size, particularly in ulcerative colitis subgroups, restricted model specificity by disease or treatment type. This inherent limitation further underscored that the primary aim of our study was to identify predictors of remission and response for vedolizumab and ustekinumab separately, rather than to perform a direct head-to-head comparison of their efficacy. Subgroup predictions should be interpreted cautiously. In addition, data were limited to structured clinical and laboratory variables; excluding other modalities (e.g., imaging, patient-reported outcomes) reduces the dimensionality of disease characterization.
Biases inherent to real-world data—such as missingness and unmeasured confounding—may influence model outputs. Although fairness analyses across sex and age groups were conducted, limited representation in certain strata (e.g., adolescents, older adults) precludes strong conclusions about equity25.
Although changes in inflammatory markers such as CRP and fecal calprotectin (FCP) during induction may provide valuable information, we did not include delta values in the present analysis. This limitation is due, in part, to data governance constraints that currently restrict access to longitudinal laboratory series in our hospital’s information systems. Furthermore, our study was designed to support treatment decision-making at the point of initiation, before exposure to biologics. As such, only baseline values were considered appropriate for building clinically actionable predictive models. We acknowledge that future studies could benefit from incorporating dynamic biomarker trajectories as additional predictive features once data collection frameworks are adapted.
External validation was not possible due to the single-center design, and this remains a critical step to ensure model generalizability. Future prospective multicenter studies with harmonized definitions and standardized data collection are essential to support reproducibility, allow stratified modeling, and confirm performance across diverse populations.
Lastly, implementation of ML in clinical practice is challenged by technical and infrastructural barriers, including data interoperability, integration with workflows, and limited digital expertise. Addressing these issues will require multidisciplinary collaboration to enable safe and effective deployment in real-world settings.
Clinical implications
The results obtained in this study are satisfactory, supporting their potential use as a clinical decision support tool in the study area. However, the limited patient sample prevents these findings from being considered definitive and may have hindered significant outcomes for certain response factors, particularly those related to UC, owing to the lower representation of these patients in the cohort. Additionally, the small sample size has limited the development of precise predictive models for each drug and disease subtype. Larger multicenter studies are needed to achieve a broader cohort, enabling more robust and generalizable results and the development of predictive models tailored to patient subgroups, considering drug type and inflammatory bowel disease subtype.
While these models demonstrate promising potential as clinical decision support tools, it is crucial to emphasize that given the retrospective, single-center nature and modest cohort size of this study (n = 227), these findings are not generalizable to external populations and any extrapolation to other healthcare settings or diverse patient cohorts would be speculative and unsupported by the current data. Our models are intended as an early first step for future confirmatory trials, not for immediate clinical practice.
In clinical settings, predictive machine learning models can significantly enhance personalized treatment for patients with CD and UC. By analyzing clinical, demographic, and biochemical data, these models help clinicians select the most effective biologic treatment tailored to a patient’s specific profile.
These models also support timely treatment adjustments by predicting remission at key intervals. A low remission probability might lead to proactive changes in therapy, preventing complications that often result from prolonged ineffective treatments, such as unnecessary hospitalizations or surgeries. This data-driven approach not only improves patient outcomes but also optimizes healthcare resource use, ensuring cost-effective care for chronic, high-impact conditions.
Conclusions
Our study demonstrates the potential of machine learning models supported by SHAP-based interpretability. A key strength of this research is its reliance on real-world evidence from patients in daily clinical practice, offering a significant advantage for the practical application and external validity of these models compared to those derived solely from clinical trials. The use of SHAP-based interpretability is indispensable, as it provides a transparent framework that clarifies the influence of individual variables and enables the interpretation of results from a medical and physiological perspective, fostering greater confidence in model predictions. Furthermore, a comprehensive and proactive fairness analysis was conducted across sex and age groups, confirming performance within different patient subgroups and addressing an important aspect of model robustness, demonstrating a commitment to evaluating equitable model performance and identifying potential disparities, which is vital for responsible machine learning deployment. While our findings underscore the promise of machine learning in personalizing IBD treatment strategies, further validation across diverse, multicenter cohorts is necessary to enhance the robustness and generalizability of these models in clinical practice.
Data availability
The data supporting the findings of this study were collected from the Virgen Macarena University Hospital under the authority of the Andalusian Health Service (Servicio Andaluz de Salud, SAS). These data contain sensitive patient information and, in accordance with Spanish and European regulations, are not publicly available. Researchers may direct reasonable requests for data access to the Fundación Pública Andaluza Progreso y Salud at: [fpsdata.fps@juntadeandalucia.es] (mailto:fpsdata.fps@juntadeandalucia.es).
Code availability
Resources to ensure reproducibility, including code, Jupyter notebooks, and a data dictionary, are publicly available on GitHub through the following URL: https://github.com/Data-Science-Laboratory-FPS/IBD-DRTxML
References
Yanai, H. & Hanauer, S. B. Assessing response and loss of response to biological therapies in IBD. Am. J. Gastroenterol. 106(4), 685–698. https://doi.org/10.1038/ajg.2011.103 (2011).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383(27), 2603–2615. https://doi.org/10.1056/NEJMoa2034577 (2020).
Gisbert, J. P. & Chaparro, M. Predictors of primary response to biologic treatment [Anti-TNF, Vedolizumab, and Ustekinumab] in patients with inflammatory bowel disease: From basic science to clinical practice. J. Crohns Colitis 14(5), 694–709. https://doi.org/10.1093/ecco-jcc/jjz195 (2020).
Javaid, A. et al. Machine learning predictive outcomes modeling in inflammatory bowel diseases. Inflamm. Bowel Dis. 28(6), 819–829. https://doi.org/10.1093/ibd/izab187 (2021).
Waljee, A. K. et al. Machine learning algorithms for objective remission and clinical outcomes with Thiopurines. J. Crohns Colitis 11(7), 801–810. https://doi.org/10.1093/ecco-jcc/jjx014 (2017).
Waljee, A. K. et al. Development and validation of machine learning models in prediction of remission in patients with moderate to severe Crohn disease. JAMA Netw. Open 2(5), e193721. https://doi.org/10.1001/jamanetworkopen.2019.3721 (2019).
Chaparro, M. et al. Using interpretable machine learning to identify baseline predictive factors of remission and drug durability in Crohn’s disease patients on Ustekinumab. J. Clin. Med. 11(15), 4518. https://doi.org/10.3390/jcm11154518 (2022).
Waljee, A. K. et al. Predicting corticosteroid-free biologic remission with Vedolizumab in Crohn’s disease. Inflamm. Bowel Dis. 24(6), 1185–1192. https://doi.org/10.1093/ibd/izy031 (2018).
Zand, A., Stokes, Z., Sharma, A., van Deen, W. K. & Hommes, D. Artificial intelligence for inflammatory bowel diseases (IBD): accurately predicting adverse outcomes using machine learning. Dig. Dis. Sci. 67(10), 4874–4885. https://doi.org/10.1007/s10620-022-07506-8 (2022).
Nguyen, N. H. et al. Machine learning-based prediction models for diagnosis and prognosis in inflammatory bowel diseases: A systematic review. J. Crohns Colitis 16(3), 398–413. https://doi.org/10.1093/ecco-jcc/jjab155 (2022).
Anuj, ‘F1 Score in Machine Learning’, Machine Learning Architect. Accessed: 03 Dec 2024. [Online]. Available: https://machinelearningarchitect.com/machine-learning-basics/f1-score-in-machine-learning/
T. Chen and C. Guestrin, ‘XGBoost: A Scalable Tree Boosting System’, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, in KDD ’16. New York, NY, USA: Association for Computing Machinery, Aug. 2016, pp. 785–794. https://doi.org/10.1145/2939672.2939785.
S. Champely, ‘pwr: Basic Functions for Power Analysis’. p. 1.3–0, 2006. https://doi.org/10.32614/CRAN.package.pwr.
Miyoshi, J. et al. Machine learning using clinical data at baseline predicts the efficacy of vedolizumab at week 22 in patients with ulcerative colitis. Sci. Rep. 11(1), 16440. https://doi.org/10.1038/s41598-021-96019-x (2021).
Chang, J. T. Pathophysiology of Inflammatory Bowel Diseases. N. Engl. J. Med. 383(27), 2652–2664. https://doi.org/10.1056/NEJMra2002697 (2020).
Valvano, M. et al. Nutrition, nutritional status, micronutrients deficiency, and disease course of inflammatory bowel disease. Nutrients 15(17), 3824. https://doi.org/10.3390/nu15173824 (2023).
Hoffmann, P. et al. Ustekinumab: “Real-world” outcomes and potential predictors of nonresponse in treatment-refractory Crohn’s disease. World J. Gastroenterol. 25(31), 4481–4492. https://doi.org/10.3748/wjg.v25.i31.4481 (2019).
Kubesch, A. et al. Short and long-term effectiveness of Ustekinumab in patients with Crohn’s disease: Real-world data from a German IBD cohort. J. Clin. Med. 8(12), 2140. https://doi.org/10.3390/jcm8122140 (2019).
Singh, N. et al. Multi-center experience of vedolizumab effectiveness in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 22(9), 2121–2126. https://doi.org/10.1097/MIB.0000000000000865 (2016).
Ma, C. et al. Clinical, endoscopic and radiographic outcomes with ustekinumab in medically-refractory Crohn’s disease: Real world experience from a multicentre cohort. Aliment. Pharmacol. Ther. 45(9), 1232–1243. https://doi.org/10.1111/apt.14016 (2017).
Moran, G. W. et al. Phenotypic features of Crohn’s disease associated with failure of medical treatment. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 12(3), 434-442.e1. https://doi.org/10.1016/j.cgh.2013.08.026 (2014).
Amiot, A. et al. Effectiveness and safety of vedolizumab induction therapy for patients with inflammatory bowel disease. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 14(11), 1593-1601.e2. https://doi.org/10.1016/j.cgh.2016.02.016 (2016).
Amiot, A. et al. One-year effectiveness and safety of vedolizumab therapy for inflammatory bowel disease: A prospective multicentre cohort study. Aliment. Pharmacol. Ther. 46(3), 310–321. https://doi.org/10.1111/apt.14167 (2017).
Restellini, S. & Afif, W. Update on TDM (therapeutic drug monitoring) with ustekinumab, vedolizumab and tofacitinib in inflammatory bowel disease. J. Clin. Med. 10(6), 1242. https://doi.org/10.3390/jcm10061242 (2021).
Char, D. S., Shah, N. H. & Magnus, D. Implementing machine learning in health care—Addressing ethical challenges. N. Engl. J. Med. 378(11), 981–983. https://doi.org/10.1056/NEJMp1714229 (2018).
Acknowledgements
The authors would like to thank the healthcare professionals of Virgen Macarena University Hospital for their support, which was essential for this research. The authors also extend their sincere gratitude to the patients whose clinical data were instrumental in the findings of this study.
Funding
This work was supported by the EU–NextGenerationEU and the Recovery, Transformation, and Resilience Plan, granted to the Ministry of Employment, Training, and Autonomous Work of the Junta de Andalucía under the Investigo Program, Recovery and Resilience Mechanism [SE/INV/0008/2022 to J.L.G.R.]. Further support was provided by the Grupo Español de Trabajo en Enfermedad de Crohn y Colitis Ulcerosa (GETECCU) through the VII Beca GETECCU—Johnson & Johnson 2024 [to F.A.A.]. Additionally, research was supported by the Sociedad Andaluza de Patología Digestiva through the Beca del Grupo Andaluz de Trabajo de Enfermedad Inflamatoria Intestinal 2022 [to C.V.P.].
Author information
Authors and Affiliations
Contributions
J.C.R. and F.A.A. contributed to the conception and design of the study. J.C.R. collected the data, and J.L.G.R. performed the data analysis. The fairness analysis was conducted by J.L.G.R. and A.B. Data interpretation and results were developed by J.L.G.R., C.V.P., and J.C.R. The discussion was drafted by J.C.R., F.A.A., M.A.H., and C.V.P. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Coordinating Ethics Committee for Biomedical Research of Andalusia (CCEIBA, protocol code FPS-EII-2021-04, version 1 dated September 14, 2021). The Committee, acting under the authority of the Consejería de Salud y Consumo, Secretaría General de Investigación, Desarrollo e Innovación en Salud, Dirección Gerencia del Servicio Andaluz de Salud, confirmed that the study meets the ethical requirements established by Royal Decree 957/2020, the Declaration of Helsinki, and applicable regulations. Due to the retrospective nature of the study, the Coordinating Ethics Committee for Biomedical Research of Andalusia (CCEIBA) waived the need for obtaining informed consent. All research procedures were performed in accordance with relevant guidelines and regulations.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rodríguez, J.L.G., Parrado, C.V., Cordero-Ramos, J. et al. Identifying key clinical and biochemical predictors of treatment outcomes in inflammatory bowel disease: a real-world evidence study. Sci Rep 15, 41350 (2025). https://doi.org/10.1038/s41598-025-25370-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-25370-0