Abstract
The study of soft, amorphous fascia has been constrained by definitional ambiguities regarding its fine structure, constituent and precise function. Here, a previously unrecognized CD34+ membranous cell (CMC) population was identified by single-cell RNA sequencing, which displayed a distinct spatial distribution in UMAP coordinates in the subcutaneous fascia of rat abdominal midline. Constituting one of the predominant cell populations in the fascia, CMCs were further validated morphologically in situ and in vitro. They exhibited lamina shapes and distributed alternating with the collagen bundles. Equipped with mechanosensors (integrins, cadherins, PIEZOs), CMCs are intricately interwoven with matrix fibers, forming the tensile mechanical apparatuses in the fascia. Enriched with Cx43 + gap junctions, Cdh1 + adherens junctions, integrin β1 + focal adhesions, Tsg101 + extracellular vesicles, F-actin + cytoskeletons, and caveolae, CMCs with marginal process foaming possessed typical evidence of signal conversion and transduction. The extensive membrane architecture of CMCs easily enabled their interactions with nerves, blood vessels, immune cells and interstitial fluid, promoting the integrations among different systems. In addition, functioning as membranous boundaries and signaling hubs, sheeting-like CMCs incompletely encapsulated or separated some structures, creating unique microenvironments. Molecular function analyses aligned with the ultrastructural features, distinguishing CMCs from other flat cells like fibroblasts. This discovery highlights a previously overlooked cell subpopulation, advancing our understanding of the fascial biology.
Similar content being viewed by others
Introduction
Fascia, an uninterrupted viscoelastic tissue, envelops and permeates all bodily structures, forming a continuous network throughout the body1,2. Fascia connects dispersed muscles, bones, and joints into a synergistically functioning whole, maintaining body form and postural stability. Recent research indicates that fascia is not only an encapsulated structure, but also a dynamic force transmission system3. Fascia has garnered increasing attention due to its critical roles in sports medicine4. However, the definition of fascia remains ambiguous, presenting challenges in isolation and nomenclature standardization1,5,6. Historically, classical dissection techniques often disregarded the fascia, removing it to access deeper structures7,8. The study of the mechanical properties of fascia network contributes to the advancement of sports medicine, necessitating microscopic investigation to unravel its complexities9. Previous single-cell sequencing studies of fascial tissues have primarily focused on functional aspects such as wound healing10, stem cell therapy11, and angiogenesis12, with limited attention to the cellular identity and molecular characteristics of resident cell populations13. This study addresses this gap by employing morphological analysis combined with single-cell RNA sequencing to systematically characterize the cellular composition of subcutaneous fascia, potentially revealing previously overlooked cell populations.
Identifying novel cell types within the body is inherently challenging, as histology is traditionally viewed as a “closed” morphological science14. The diversity of interstitial cells in fascia further complicates accurate identification. Advances in technology, however, now enable the detection of previously overlooked cells through sophisticated methodologies15,16. Recent research has increasingly focused on the anatomical features of fascia, driven by the adage that “the fear of surgery is the fear of anatomy”—Ian Taylor17. Bioinformatics, combined with in situ stereoscopic observation, has proven indispensable for uncovering novel structures and hypothesizing their functions16. This study aims to identify previously unrecognized cells, characterize their physical attributes, and predict associated molecular functions within rat subcutaneous fascia of rat abdominal midline. By elucidating the molecular and ultrastructure characteristics of mechanoreception, cell communication, microenvironment homeostasis, and matrix remodeling, this work seeks to advance the understanding of fascial biology.
Results
A population of CD34 + cells was identified within the subcutaneous fascia from rat abdominal midline by single-cell RNA sequence (scRNA)
In this study, single-cell RNA sequencing of abdominal rat subcutaneous fascia yielded 13,224 high-quality cells, which were annotated into 9 distinct cell populations based on marker expression. These populations included macrophages (MPs; 25.7%), CD34 + cells (CCs; 20.0%), pericytes (PerCs; 15.8%), endothelial cells (ECs; 13.3%), myofibroblasts (MyoFs; 8.8%), lymphocytes (LPs; 8.8%), fibroblasts (FCs; 5.9%), dendritic cells (DCs; 1.2%) and granulocytes (GNCs; 0.5%) (Fig. 1A–D). Notably, CCs were identified as a previously unrecognized cell type, displaying a distinct spatial distribution in UMAP coordinates compared to other cell types including FCs and ECs (Fig. 1A), and constituting one of the predominant cell populations in the subcutaneous fascia (Fig. 1D). In addition, we found that CD34 and Pdgfra exhibited a certain degree of overlap in the single-cell sequencing results (Fig. 1C), which was further validated by the co-expression of CD34/Pdgfra on CCs by dual immunofluorescence staining (Supp Fig. G–G3).
Cell Types and Functional Characteristics of CCs in the Subcutaneous Fascia. A. UMAP visualization of cell types identified in the subcutaneous fascia from rat abdominal midline. B. Average expression levels of marker genes across distinct cell populations. C. Distribution of marker gene expression within each cell type. D. Proportional representation of cell populations in the subcutaneous fascia, highlighting CCs as a major constituent. E. Enriched biological functions associated with highly expressed genes in CCs, emphasizing roles in mechanosensation, extracellular matrix (ECM) assembly, and connective tissue development. F. Heatmap depicting the expression of genes linked to the biological functions shown in (E), demonstrating the pronounced upregulation of mechanoresponsive genes in CCs compared to other cell types.
Differential gene expression analysis revealed that CCs exhibit significant upregulation of mechanosensitive genes including Piezo2, as well as genes associated with extracellular matrix (ECM) assembly and mechanoresponsiveness, such as Eln (encoding elastin) and Itgb3 (encoding integrin β3) (Fig. 1F, Supplementary Table 1). Gene set enrichment analysis (GSEA) further highlighted the functional enrichment of CCs in biological processes related to mechanosensation, ECM assembly, cell junction organization, and connective tissue development (Fig. 1E). The expression of mechanosensitive genes including Piezo2, elastin, and Itgb3 in CCs suggested potential roles in mediating mechanical signaling and ECM dynamics within the subcutaneous fascia.
CD34 + cells established a complex microenvironmental signaling network in the subcutaneous fascia
Cell communication analysis performed by CellCall18 tool demonstrated that CCs engaged in extensive signaling interactions with other cell types within the fascia (Fig. 2A, B). They secreted diverse signaling molecules, including members of the WNT, BMP, and FGF families (Fig. 2C, D), critical for maintaining microenvironmental homeostasis and the regenerative capacity of the subcutaneous fascia19,20.
Cell Communication Analysis of CCs in the Subcutaneous Fascia. A. Overview of intercellular signaling networks within the subcutaneous fascia, generated by ligand-receptor scoring matrix of subcutaneous fascia. B. Expression profiles of ligands secreted by CCs and their corresponding receptor proteins in target cells. C. Heatmap illustrating the intensity and specificity of intercellular communication signals originating from CCs. D. Sankey diagram depicting the flow of signaling interactions between CCs and other cell types focused on homotypic CC communication. E. Expression levels of Rho pathway-related genes regulated by Abl1, highlighting their enrichment in CCs. F. Heatmap showing the overall expression of Abl1-regulated target genes across cell populations in the subcutaneous fascia, demonstrating their pronounced upregulation in CCs.
CCs exhibited significant enrichment of contact-dependent signaling pathways, particularly the Efna5-Epha3 and Efnb1-Ephb4 axes. Further investigation into the downstream transcriptional regulator Abl1, a key effector of the Efn-Eph signaling axis, revealed that Abl1 target genes, such as Dock4, a Rho pathway gene, were markedly upregulated in CCs compared to other cell populations (Fig. 2F, Supplementary Table 2). These genes regulate cell-ECM adhesion and the MAPK pathway (Supplementary Table 3). Additionally, multiple Rho pathway-related genes, including Epha3 and members of the Dock protein family, were significantly upregulated in CCs (Fig. 2E, F). These Abl1-regulated target genes have been shown to modulate cytoskeletal reorganization and mechanoresponsiveness through Rho pathway activation, underscoring the possible involvement of CCs in coordinating cellular dynamics within the fascial microenvironment.
Substantial validation of CCs from the subcutaneous fascia by Immunofluorescence
To validate the corporeal presence of CCs identified in scRNA-seq analysis, we performed CD34-immunofluorescent staining in vivo and in vitro. In vivo immunofluorescence (IF) reaction uncovered longitudinal CCs in the subcutaneous fascia (Fig. 3A). They also circled blood vessels on paraffin sections. To elucidate the three-dimensional (3D) morphology from the linear CCs on 2D sections, we further employed Z-stack confocal microscopy to image CD34-immunostained fascia tissue. It was demonstrated that the linear CCs observed in 2D sections were, in fact, sheet-like structures (Fig. 3B, B1, and Video 1).
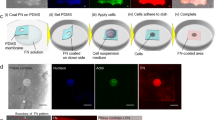
Immunofluorescence identification of CD34 + Cells in vivo and in vitro. A. In situ Immunofluorescence staining of CD34 + cells on paraffin section showing their present within the fascial tissue. B. Z-stack confocal microscopy imaging from image A, revealing the 3D membranous architecture of CD34 + cells in the fascia (Video 1: Dynamic visualization of CD34 + cells 3D membranous structures). B1. Magnified view of image B, highlighting sheet-like CD34 + membranous cells. C. Schematic representation of magnetic-activated cell sorting (MACS) for isolating and primary culture of CD34 + cells from rat subcutaneous fascia. D. CD34 + immunofluorescence staining of primary cultured CMCs, demonstrating their flat, membranous morphology. D1. High-magnification of image D, showing the tunneling nanotubes (triangular arrow) between CMCs (pentagram) were observed. Scale bars (A, D, D1) = 20 μm.
CCs were isolated from rat subcutaneous fascia using magnetic-activated cell sorting (MACS) and primary cultured in vitro (Fig. 3C). Immunofluorescence staining of cultured CCs showed an extremely thin or pellicular morphology (Fig. 3D, D1), consistent with the flat structures (Fig. 3B, B1) in vivo. They exhibited thin and broad membranous shapes with some tunneling nanotubes (TNTs)21 between adjacent CCs (distinguished from isolated filopodia22) and, here, we designated them as the “Cd34 + membranous cells” (CMCs). Together, those findings proved the existence of CMCs with thin and broad externals as a distinct stromal population in the fascia.
In addition, double IF data showed that CMC co-expressed CD34 with F-actin, TRPV4, CX43, Tsg101, Cdh1 or Itgb1 respectively (Supplementary Figure), which responded to the cytoskeleton of microfilaments, mechanoreceptors, gap junctions, exosomes, adherens junctions and focal adhesions separately.
Characteristics of CMCs in vitro under scanning electron microscope (SEM)
To detect the complete appearance and stereological features of the CMC, primary cultured CMCs from the fascia were carefully analyzed in vitro under SEM. CMCs were so thin that they had the pellicular or imprinted appearances (Fig. 4A, A1) which were CD34 positive by immune colloidal gold identification (Fig. 4B, B1). The CMC with a slightly protuberant nucleus had a membrane area of 8,000–15,000µm2 (Fig. 4A). Bulging cytoskeletons extended beneath its cytoplasmic membrane (Fig. 4C, C1). With fewer microvilli, the CMC was generally smooth on its membrane surface (Fig. 4A-F).
Natures of CMCs in vitro under SEM. A. Low-magnification image revealing the stamp-like appearances of primary cultured CMCs (pentagram). A1. A pellicular CMC with small flat nucleus (Nu) from image A, demonstrating its broad area. B. CD34 + immunogold labeled CMC. B1. High-resolution of image B, indicating CD34 + gold particles (yellow arrow) on CMC membrane. C. A primary cultured CMC closely stuck to the matrix. C1. High-resolution of photograph C, showing the prominent microfilament bundles (magenta arrow) extending beneath the cytoplasmic membrane of a CMC. D. Widespread focal adhesions (white arrow) between the CMC and ECM. D1. High-resolution of photograph D, indicative of focal adhesions (white arrow) anchoring to the ECM. E. TNT (green triangular arrow) and membrane bridging (white double arrow) between CMCs, suggesting special intercellular communications. E1. High-magnification of photograph E. F. Developed adhesive junctions between CMCs. F1. High-magnification of photograph F, indicating the adhesive junctions (blue arrow). G. Matrix fiber-like mesh, and membrane bridges (white double arrow) and TNT (green triangular arrow) between CMCs. G1. High-magnification of photograph G, showing the matrix fiber-like mesh (white triangle) beneath CMC membrane. Scale bar (A) = 100 μm, 40 μm in (A1); Scale bars (B, D1) = 400 nm, 200 nm in (B1); Scale bars (C, C1, E) = 5 μm; Scale bars (D, F, G) = 2 μm; Scale bars (F1, E1, G1) = 1 μm.
There were widespread focal adhesions between the CMC and matrix (Fig. 4D, D1). TNTs, special “membrane bridges” (Fig. 4E, E1), and abundant adhesion junctions (Fig. 4F) rose between CMCs each other. Marginal filamentous processes from the CMCs were often observed, which frequently segmented or foamed (Fig. 5), resulting in the generation of many small extracellular vesicles. All those ultra-structures provide evidence of mechanosensitivity and cell communication23,24.
Marginal processes of primary cultured CMCs and their segmentation and foaming. A. Marginal processes (white arrow) from the CMC (pentagram). A1. High-magnification of photograph A, indicating marginal filamentous processes and their segmenting and foaming (green arrow). B. Segmentation and foaming of some detached marginal processes (white arrow). C. EVs (red arrow) generated from the segmentation and foaming of detached marginal processes. Scale bar (A) = 10 μm; Scale bars (A1, B) = 2 μm; Scale bar (C) = 400 nm.
In addition, the matrix fiber-like mesh was secreted beneath the CMCs (Fig. 4G, G1).
In situ investigation of CMCs and their spatial relationships with surroundings in the fascia under SEM
To explore the features of in situ CMCs and their spatial relationships with surroundings, we implemented SEM observation of the fascia (Fig. 6A, A1). In fact, because of the frequent block and covering of other structures, or their flaky nature, the CMC was often seen as fragmental sheet-like structures rather than a whole cell (Fig. 6B-I).
In Vivo CMCs and their spatial relationships with surroundings in the fascia under SEM. A. Global overview of rat skin and its subcutaneous fascia of abdominal midline, showcasing the stratified tissue organization. A1. Magnification of A, showing multilayer structures of the fascia. Epidermis (Ep); dermis (De); subcutaneous fascia (Sf); panniculus carnosus (Pc). B. Visualization of CMCs alternated with flat collagen bundles (Fcb) in the fascia. B1. High-magnification of photograph B, indicating the tensile “hammock” CMC (pentagram) pulled by a mechanical scaffold with the fibers (white arrow) in the fascia. C. Multilayer CMCs (pentagram) in the fascia. flat collagen bundles (Fcb). C1. Magnification of C, showing smooth CMC membrane surface (pentagram) and collagen fiber bundles (Cb) beneath CMC. D. CMC (pentagram) direct contact with neighboring lymphocyte (Lc). E. The translucent CMC (pentagram) on a stiff collagen bundle (Cb) that formed irregular tissue micro-tunnels (TMT) in the fascia. F. CMCs (pentagram) incompletely encapsulated a hair follicle (Hf). G. EVs (short white arrow, purple pseudo-color) on CMC membrane (pentagram) in the fascia. fiber mesh (black arrow) in the fascia. H. Folding marginal processes (white double arrow) from the CMC (pentagram). I. Collagen fibers (white dashed line) and collagen fiber bundles (Cb) on surface of the CMC (pentagram). Scale bar (A) = 500 μm; 200 μm in (A1); Scale bar (B) = 20 μm; 5 μm in (B1); Scale bars (C, F, H, I) = 4 μm; Scale bars (D, C1, E, G) = 2 μm.
Multi-laminar CMCs were staggered with the collagen-bundles in the fascia (Fig. 6B, B1, C). Like the “hammock” appearances, pellicular CMCs were pulled by extracellular fibers in different directions, obtaining a special tension state (Fig. 6B, B1, C, G). Lymphocytes (Fig. 6D) and extracellular vesicles (Fig. 6G) were nearby or in contact with the CMCs, implying that they could move on broad membrane of CMC guided by CMC-derived signals (Fig. 2A, B). Extremely thin, even translucent CMCs (Fig. 6E) covered stiff collagen bundles which can develop uneven tissue micro-tunnels filled with tissue fluid in the fascia25. They always incompletely covered the hair follicle (Fig. 6F).
Without microvilli, the CMC membrane surface was always smooth (Fig. 6B1, C1, D, E), while its marginal processes rose. The marginal processes were always fold back on the smooth surface in vivo (Fig. 6H). Collagen fibers and their bundles (Fig. 6I) extended on the surface of CMC membrane, implying the assembling of extracellular fibers.
In situ ultrastructural natures of CMCs in the fascia under transmission electron microscope (TEM)
According to cell stereology, a 3D laminar structure corresponds to a 2D line (Fig. 7A). As a result, uninterrupted long “linear” structures (Fig. 7B) under TEM coincided with the broad laminal CMCs (Fig. 6C) under SEM, which was further confirmed by CD34 + immunocolloidal gold reaction (Fig. 7C, C1).
TEM Photograph of CMC and its associated ultrastructures in the fascia in vivo. A. Stereological illustration of 3D laminae corresponding to that of 2D lines. B. Long linear CMCs (black arrow) alternated with collagen bundles (Cb) in the fascia under TEM. C. Immunocolloidal gold-labeled CD34 + linear CMC (black arrow) in situ under TEM. C1. Magnification of photograph C, indicating CD34 + 12-nm-diameter gold particles (yellow arrow). D. Ultrathin CMC (black arrow) closely covered the collagen bundles (Cb). D1. Magnification of photograph D, strengthening the CMC (black arrow) could be thinner than a single collagen fibril (white dashed line). E. The thin CMC (black arrow) circling a blood vessel (Bv) in the fascia. Mast cell (Mc); smooth muscle cell (Smc). F. Thin CMCs (black arrow) circling a nerve, including myelinated nerve fibers (Mnf) and unmyelinated nerve fibers (Unf) in the fascia. G. Several layer thin CMCs (black arrow) circling a stem cell niche (pentagram) of the hair papilla. (Hp) attached to a hair follicle (Hf). H. A thin CMC (black arrow) circling a collagen bundle (Cb). I. Segmentation and foaming (magenta arrow) of marginal processes (black triangular arrow) from the CMC in fascia. J. Abundant extracellular vesicles (blue arrow) around the CMC extension (black arrow). K. A winding CMC (black arrow) among the lymphocyte (Lc), macrophage (Mp), adipocyte (Ac) and blood vessel (Bv) and its pericyte (Pc). L. Contact (black dashed oval) between the plasma cell (P) and CMC (black arrow). Scale bars (B, F, H, K) = 2 μm; Scale bar (C) = 100 nm, 50 nm in (C1); Scale bars (D, I, J, L) = 1 μm; 500 nm in (D1); Scale bar (E) = 4 μm; Scale bar (G) = 8 μm.
Under TEM, long linear CMCs with small flat nuclei were distributed often alternating with the collagen bundles in the fascia (Fig. 7B). With extremely thin cytoplasm, their extensions were often 20–30 nm in thickness (Fig. 7D, D1), even thinner than a single collagen fibril, which differed from other flat cells including fibroblasts. CMCs often surrounded the blood vessel (Fig. 7E, K), nerve (Fig. 7F), stem cell niche of the hair papilla (Fig. 7G), and collagen bundles (Fig. 7H), suggesting the integrations of the CMC with other systems including the nerve and circulation system. Corresponding to the SEM data (Fig. 5), segmentation and foaming of marginal processes from the CMC often took place (Fig. 7I). Extracellular vesicles like the EVs were distributed around the CMCs (Fig. 7J). Lymphocytes (Fig. 7K), macrophages (Fig. 7K), mast cells (Fig. 7E) and plasma cells (Fig. 7L) stood next to the CMC and its extensions, indicating the cross-talking between immune elements and the CMC.
With higher magnification, such developed organelles as rough endoplasmic reticular (Fig. 8A), mitochondria (Fig. 8A, C), caveolae (Fig. 8B, C), and microfilament cytoskeletons (Fig. 8C) were detected in the CMCs and their prolongations. There were abundant adherens junctions (Fig. 8B) and gap junctions (Fig. 8D, D1) between the CMCs. Typical focal adhesions (Fig. 8D, D2, E) frequently arose between them and the matrix in the fascia. All those elements under TEM associated to cell communication.
Detailed ultrastructures of the CMC. A. Ultra-photograph of rough endoplasmic reticulum (double arrow) and mitochondria (Mi) of the CMC in the fascia. B. Adherens junctions (black dashed oval) between CMCs in the fascia. Caveolae (short black arrow). C. Well-developed microfilaments (pentagram) within the CMC. Caveolae (short black arrow); mitochondria (Mi). D. Gap junction between CMCs and focal adhesion between the CMC and extracellular matrix. D1 and D2. Large magnifications from photograph D, highlighting the Gap junction (red dashed oval) and focal adhesion (black triangular arrow) respectively. E. Developed focal adhesion (black triangular arrow) between the CMC and extracellular matrix. Scale bars (A, D, E) = 400 nm; Scale bars (B, C, D1, D2) = 200 nm.
Discussion
Identifying the CMC in fascia had posed significant challenges in this study. (1) in situ CMCs were frequently blocked from view or enshrouded partially by other structures in the fascia; (2) pellicular CMC was so broad or long that it was hardly accommodated in a single field, resulted in most in vivo fragments rather than a whole CMC shape under SEM and TEM; (3) compared with common solid tissues, the fascia texture containing CMCs is soft and viscoelastic, eventuated in the difficulty of sample treatment26. (4) CMCs were easily confused with other flat cells like fibroblasts in the fascia, which needs a professional distinguishment. Concerning those questions, we successfully isolated CMCs from the fascia and primarily cultured them in vitro, which could show the complete CMC shapes without any covering effects. Along with the scRNA-seq, IF, 3D Z-stack confocal imaging, immunogold-labeling, TEM, and especially the stereoscopical SEM, we technically validated CMCs from different ways in vivo and in vitro, which contributed to identify them from other flat cells27. Fascia samples were properly treated following the protocols in this paper, assuring the repeatability of results.
The identification of CD34 + membranous cells in subcutaneous fascia highlights a significant gap in our understanding of fascial cellular composition, as previous studies have largely overlooked this substantial cell population (~ 20% of total cells in subcutaneous fascia)13. This oversight likely reflects a fundamental difference in research approaches: while previous studies have primarily focused on functional aspects such as wound healing10, stem cell therapy11, and angiogenesis12, our study directly addresses intrinsic cellular identity through comprehensive morphological characterization combined with single-cell transcriptomics. This methodological shift from function-oriented to identity-oriented research has revealed previously unrecognized cell populations that may have been overlooked due to the lack of systematic cellular characterization in prior fascial research.
It is suggested that previously unknown laminar CMC features under SEM mostly corresponded to those under TEM. They were as follows: (1) CMCs were intimately interwoven with extracellular fiber architectures, getting tensional state within the fascia. With widespread integrin-related focal adhesions, cadherin-related adherens junctions, and high expression of TRPs and PIEZOs on the broad membrane, CMCs were the novel mechanosensors, which could perceive and transduce the physical signals along the fascia. (2) Frequent segmentation and foaming of the marginal processes from CMCs resulted in many EVs. The latter, along with developed microfilament cytoskeleton, endoplasmic reticular, mitochondria, caveolae, CX43 gap junctions and TNTs21 indicated vibrant CMC-associated communications in the fascia. (3) With broad membrane surface, CMCs advantaged the contact with nerves, blood vessels, immunocytes and interstitial fluid, which can integrate different regulatory elements like nerves, vascula and immunocytes into an entirety within the fascia. (4) As the laminar boundary, CMCs incompletely covered or encapsulated some special structures, thereby sculpting distinct microenvironments within the body.
Accompanied with the bioinformation data in this study, those in vivo and in vitro morphological findings, substantiated that CMCs possessed the potential to form a mechano-biological signaling axis28. The CMCs expressed high levels of mechanosensitive ion channels (e.g., TRPs and PIEZOs), converting physical stimuli into intracellular calcium signals29,30 that activate Rho pathways (e.g. upregulation of Dock4)31,32,33, thereby regulating cytoskeletal reorganization34,35 and ECM remodeling (e.g. expression of elastin, Eln)36. Unlike the common mechanosensors (e.g. fibroblasts37, the expansive membrane architecture of CMCs with developed focal adhesions allowed them to integrate mechanical signals across larger areas more efficiently. Through Cx43 + gap junctions and TNTs, they could transmit signals to neighboring cells, forming a “distributed sensing network"38 within the fascia. Moreover, CMCs achieved multi-scale signal integration via specialized structures: frequent segment and vesiculation of the marginal processes, producing EVs that could carry signaling molecules such as WNT and BMP, potentially participating in long-range regulation39,40. Through the Eph receptor-ligand axis (e.g., EphA3- EFNA5) and EVs, CMCs could widely interact with neural, vascular, and immune cells, integrating multi-system signals41,42.
The discovery of CMCs holds potential implications. Theoretically, the role as “mechano-signal transducers” could explain how fascia integrates mechanical signals across the body43, providing a cellular basis for its functions in sports medicine (e.g., injury repair)4, chronic diseases (e.g., fibrosis)44, and traditional Chinese medicine (e.g., “meridian” conduction mechanisms)45. Practically, CMC-derived EVs and secreted factors (e.g., BMP/FGF) might serve as targets for tissue engineering scaffolds or regenerative medicine39, while their mechanosensitivity abnormalities could be linked to fascial pain syndromes, offering new intervention strategies46.
Materials and methods
Animal and sample preparation
All animal assays were meticulously managed by the Animal Laboratory Centre of Nanjing Agricultural University, adhering to the designated Standard Operating Procedures (SOP) for animal feeding within a Specific Pathogen-Free (SPF) barrier. For this study, 30 SPF Sprague Dawley (SD) rats were utilized as biological replicates. Euthanasia was performed by isoflurane inhalation followed by cervical dislocation. Subcutaneous fascia tissue samples were carefully dissociated from the ventral midline with a width of 2 cm and a total length of 7 cm. The fascia tissue was assiduously detached from adipose and skin attachments, ensuring the purification of the samples for further analysis.
Tissue dissociation and cell purification for scRNA-seq
The process of tissue dissociation and cell purification for scRNA-seq was meticulously executed. The fascia tissues were placed in a sterile culture dish containing 10 ml of 1x Dulbecco’s Phosphate-Buffered Saline (DPBS; Thermo Fisher, Cat. no. 14190144) on ice to eliminate the residual blood and adipose, followed by mincing on ice.
We employed a specially formulated dissociation solution, composed of the collagenase type 4 (Sigma-Aldrich, Cat. no. C5138), elastase (Sigma-Aldrich, Cat. no. e1250), and 10 ug/mL DNase I (Thermo Fisher, Cat. no. en0523) dissolved in PBS with 5% Fetal Bovine Serum (FBS; Thermo Fisher, Cat. no. SV30087.02), to digest the fascia. The fascia tissues were dissociated at 37℃ with a shaking speed of 50 rpm for approximately 40 min. We collected the dissociated cells at 20-minute intervals to maximize cell yield and viability.
Cell suspensions were filtered by the sequence of nylon cell strainer from 100 μm to 40 μm, and residual red blood cells were eliminated using 1X Red Blood Cell Lysis Solution (Thermo Fisher, Cat. no. 00–4333-57). The dissociated cells were then washed with 1x DPBS containing 2% FBS. Cell viability was assessed using 0.4% Trypan blue staining and the Countess® II Automated Cell Counter (Thermo Fisher), all samples of suspension cells were filtered by Dead Cell Removal Kit (Miltenyi Biotec, Cat. No. 130-090-101), and only samples with > 95% living cells were used for next steps.
Suspension cells were prepared for 10X library construction and 3’ sequencing, water-in-oil solution was prepared by the Chromium™ Single Cell Controller (10X Genomics), The remaining cells were transferred into a 12-well plate for culture, with the suspension from each well being transferred into a new well after 2 h. A time gradient for cell attachment was established up to 4 h, with cell slides placed in each well for Immunocytochemistry sample preparation. This rigorous methodology ensures the integrity and viability of the cells for subsequent analyses.
10x library preparation and sequencing
Beads, each carrying a unique molecular identifier (UMI) and cell barcodes, were loaded to near saturation, ensuring each cell was paired with a bead in a Gel Beads-in-emulsion (GEM). Polyadenylated RNA molecules hybridized to the beads after exposure to cell lysis buffer. The beads were then collected into a single tube for reverse transcription.
During cDNA synthesis, each cDNA molecule was tagged at the 5’ end (corresponding to the 3’ end of a messenger RNA transcript) with a UMI and a cell label indicating its cell of origin. The 10x beads were then subjected to second-strand cDNA synthesis, adaptor ligation, and universal amplification. Sequencing libraries were prepared using randomly interrupted whole-transcriptome amplification products to enrich the 3’ end of the transcripts linked with the cell barcode and UMI.
All subsequent procedures, including library construction, were performed according to the standard manufacturer’s protocol (Chromium SingleCell 3ʹ v3.1). The sequencing libraries were quantified using a High Sensitivity DNA Chip (Agilent) on a Bioanalyzer 2100 and the Qubit High Sensitivity DNA Assay (Thermo Fisher Scientific). The libraries were sequenced on a NovaSeq6000 (Illumina) using 2 × 150 chemistry.
The scRNA-seq data analysis pipeline
Commenced with the raw sequencing FASTA data, which was processed using the Cellranger 7.2.0 tool to generate expression matrices. These matrices were subsequently imported and integrated for analysis with the Seurat 4.4.0 package within the R-4.3.0 platform. Quality control was rigorously performed by calculating outliers based on counts, features, and mitochondrial gene ratios, resulting in a dataset comprising 13,224 high-quality cellular samples. The downstream analysis in Seurat adhered to the official data processing guideline, augmented by the inclusion of the Harmony algorithm for batch effect correction. This enabled the derivation of UMAP dimensionality reduction, followed by manual annotation of cell types, which were stored as RDS objects for subsequent analyses.
In this study, gene set enrichment analysis (GSEA) was conducted in accordance with the guidelines of the ClusterProfiler47 package, while cell communication analysis was executed using the CellCall18 package as per its recommended protocol.
Primary CMCs extraction and magnetic-activated cell sorting
Complete subcutaneous fascial tissue spanning from the xiphoid cartilage to the pubic symphysis was collected as 2-cm wide symmetrical strips centered along the abdominal midline. After scraping and mincing the subcutaneous fascia, tissues were digested at 37 °C with an enzymatic solution containing collagenase type 4 (Solarbio, C8160), Dnase (Solarbio, D8071), hyaluronidase (Solarbio, H8030). and elastase (Solarbio, E8210). Sequential filtration through 100-µm and 40-µm meshes was performed, followed by red blood cell lysis solution (Biosharp, BL503A) treatment.
All consumables and magnetic beads for MACS enrichment were sourced from RWD (CSAK-01). Single cells from rat subcutaneous fascia were resuspended in separation buffer (Thermo Fisher, 00–4222-57) and incubated with Cd31-Biotin conjugated antibody (Invitrogen, 13–0311-81) for 15 min. After washing off unbound antibodies, the cells were incubated with streptavidin-coated magnetic nanoparticles for another 15 min. The cells were then resuspended in 500 µl of separation buffer and added to a cell sorting column placed in a magnetic separator. The flow-through was collected as the unlabeled negative fraction. The negative fraction was collected and incubated again with Cd34-Biotin conjugated antibody (Invitrogen, 13–0341-81) and magnetic beads, followed by sorting as described above. The column was washed with separation buffer, and the retained magnetically labeled positive cells were the target CMCs. Suspension cells were cultured in DMEM/F12 medium (Gibco, Cat. No. 11330032) supplemented with 10% FBS and 1% penicillin-streptomycin (Solarbio, Cat. No. P7630). The cells were incubated under 5% carbon dioxide, 95% humidity, and a temperature of 37℃ in a cell incubator (Thermo Fisher).
Morphological fixation
Samples underwent fixation in a solution containing 4% paraformaldehyde for IF and 2.5% glutaraldehyde in PBS for Electron Microscopy, ensuring the preservation of cellular architecture.
Immunofluorescence (IF)
The hydrated paraffin-embedded tissue sections underwent antigen fixation through incubation in 0.01 M sodium citrate at 98 °C for 5 min. Post-incubation, sections were treated with primary antibodies, followed by fluorescently labeled secondary antibodies and DAPI staining (Beyotime, C1005). Rhodamine-labeled phalloidin (1:100, Abclonal, RM02835) in 1% BSA and 0.1% Triton X-100 was used to stain F-actin. Fluorescent images were acquired using a confocal microscope (Carl Zeiss, LSM900).
Antibodies Employed in This Investigation: Anti-CD34 (1:200, Abclonal, A13929); Anti-CD34 (1:100, Santa Cruz Biotechnology, sc-74499); Anti-PDGFRα (1:200, Santa Cruz Biotechnology, sc-398206); Anti-Connexin43 (Cx43, 1:100, Santa Cruz Biotechnology, sc-59949); Anti-E-cadherin (Cdh1, 1:100, Santa Cruz Biotechnology, sc-8426); Anti-integrin β1 (Itgb1, 1:100, Abclonal, A23497); TSG101 (1:100, Huabio, ET1701-59) (typical marker for extracellular vesicles48; Anti-transient receptor potential cation channel subfamily V member 4 (TRPV4, 1:100, Affinity, DF8624); CoraLite488-conjugated Goat Anti-Rabbit IgG (1:200, Proteintech, SA00013-2); CoraLite594-conjugated Goat Anti-Mouse IgG (1:200, Proteintech, SA00013-3).
Transmission electron microscopy (TEM)
Fascia samples were initially fixed in a 2.5% glutaraldehyde solution for 48 h at 4 °C, followed by post-fixation in 1% osmium tetroxide for 2 h under the same temperature conditions. The specimens underwent dehydration through a graded acetone series (30 to 100%), each for 10 min. Subsequently, the tissues were embedded in epoxy resin. Ultrathin sections, approximately 50 nm in thickness, were mounted on copper grids and subjected to staining with uranyl acetate and lead citrate. The prepared sections were then observed using a Hitachi HT7800 transmission electron microscope operating at an acceleration voltage of 80 kV.
Scanning electron microscopy (SEM)
Tissue blocks measuring 2 × 2 × 1 mm³ were fixed in 2.5% buffered glutaraldehyde for 12 h at 4 °C. Following fixation, the samples were rinsed three times with PBS, each wash lasting 15 min. Subsequently, the samples underwent gradient alcohol dehydration and were subjected to tert-butanol replacement three times. After freeze-drying to ensure structural integrity, the samples were mounted onto the specimen stage. A 10 nm gold film was then applied to the samples using an ion-sputtering device to enhance conductivity. Finally, the samples were examined using a SEM (Regulus 8100, Hitachi, Japan).
Immunogold cell slides for SEM
Cells were fixed with 0.5% glutaraldehyde and 3% PFA in PBS for 1 h at 4 °C, followed by blocking with 3% BSA for 30 min. Subsequently, the cells were incubated with an anti-CD34 primary antibody for 3 h at 37 °C. After washing with 1% BSA, the cell samples were incubated with a secondary antibody (goat anti-rabbit IgG conjugated to 35-nm colloidal gold particles, 1:50 dilution, Solarbio, K1034G-G35) for 1 h. The subsequent steps followed standard scanning SEM protocols. Finally, the samples were imaged using a Hitachi Regulus 8100 scanning electron microscope.
Immunogold sample preparation for TEM
Following tissue sample preparation to meet the TEM analysis requirements, the specimens were fixed with 0.5% glutaraldehyde and 3% PFA in PBS. Subsequently, ultrathin sections were prepared as described for TEM above, without the osmium tetroxide post-fixation step. The sections were then subjected to immunolabeling with an anti-CD34 primary antibody (Abclonal, A13929) and a secondary antibody conjugated to 12 nm colloidal gold (Abcam, ab105298). After post-fixation and staining, the samples were examined using TEM.
Ethics approval
The experimental procedure was approved by the Animal Welfare and Ethics Committee of Nanjing Agricultural University (approval No20220427087). All experiments were performed in accordance with relevant guidelines and regulations. The study was reported in accordance with ARRIVE guidelines.
Data availability
The scRNA-seq data sets have been deposited online in the NCBI GEO datasets under the accession code No. GSE261489.([https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE261489](https:/www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE261489)).
References
Bordoni, B. et al. Fascial nomenclature: update 2024. Cureus 16, e53995. https://doi.org/10.7759/cureus.53995 (2024).
Stecco, C. et al. Towards a comprehensive definition of the human fascial system. J. Anat. https://doi.org/10.1111/joa.14212 (2025).
Wilke, J., Schleip, R., Yucesoy, C. A. & Banzer, W. Not merely a protective packing organ? A review of fascia and its force transmission capacity. J. Appl. Physiol. Bethesda Md. 1985 124, 234–244. https://doi.org/10.1152/japplphysiol.00565.2017 (2018).
Benjamin, M. The fascia of the limbs and back–a review. J. Anat. 214, 1–18. https://doi.org/10.1111/j.1469-7580.2008.01011.x (2009).
Kumka, M. & Bonar, J. Fascia: a morphological description and classification system based on a literature review. J. Can. Chiropr. Assoc. 56, 179–191 (2012).
Adstrum, S., Hedley, G., Schleip, R., Stecco, C. & Yucesoy, C. A. Defining the fascial system. J. Bodyw. Mov. Ther. 21, 173–177. https://doi.org/10.1016/j.jbmt.2016.11.003 (2017).
Ghosh, S. K. Human cadaveric dissection: a historical account from ancient Greece to the modern era. Anat. Cell. Biol. 48, 153–169. https://doi.org/10.5115/acb.2015.48.3.153 (2015).
Stecco, C., Macchi, V., Porzionato, A., Duparc, F. & De Caro, R. The fascia: the forgotten structure. Ital. J. Anat. Embryol. 116, 127–138 (2011).
Ajimsha, M. S., Shenoy, P. D. & Gampawar, N. Role of fascial connectivity in musculoskeletal dysfunctions: A narrative review. J. Bodyw. Mov. Ther. 24, 423–431. https://doi.org/10.1016/j.jbmt.2020.07.020 (2020).
Correa-Gallegos, D. et al. CD201(+) fascia progenitors choreograph injury repair. Nature 623, 792–802. https://doi.org/10.1038/s41586-023-06725-x (2023).
Chen, G. et al. Fascia-derived stem cells enhance fat graft retention by promoting vascularization through the HMOX1-HIF-1alpha pathway. Stem Cell Res. Ther. 16, 92. https://doi.org/10.1186/s13287-025-04204-w (2025).
Shi, Z., Yao, C., Shui, Y., Li, S. & Yan, H. Research progress on the mechanism of angiogenesis in wound repair and regeneration. Front. Physiol. 14, 1284981. https://doi.org/10.3389/fphys.2023.1284981 (2023).
He, J. et al. Single-cell analysis reveals distinct functional heterogeneity of CD34(+) cells in anagen wound and diabetic wound. Biochem. Biophys. Res. Commun. 639, 9–19. https://doi.org/10.1016/j.bbrc.2022.11.080 (2023).
Pellegrini, M. S. F., Popescu, L. M. & Telocytes Biomol. Concepts 2, 481–489 https://doi.org/10.1515/BMC.2011.039 (2011).
Jean-Beltran, P. M., Mathias, R. A. & Cristea, I. M. A portrait of the human organelle proteome in space and time during cytomegalovirus infection. Cell Syst 3, 361-373.e366. https://doi.org/10.1016/j.cels.2016.08.012 (2016).
Zanini, F. et al. Northstar enables automatic classification of known and novel cell types from tumor samples. Sci. Rep. 10, 15251. https://doi.org/10.1038/s41598-020-71805-1 (2020).
Surek, C. C. Facial anatomy for filler injection: the superficial musculoaponeurotic system (SMAS) is not just for facelifting. Clin. Plast. Surg. 46, 603–612. https://doi.org/10.1016/j.cps.2019.06.007 (2019).
Zhang, Y. et al. CellCall: integrating paired ligand-receptor and transcription factor activities for cell-cell communication. Nucleic Acids Res. 49, 8520–8534. https://doi.org/10.1093/nar/gkab638 (2021).
Zhu, X. J. et al. BMP-FGF signaling axis mediates Wnt-Induced epidermal stratification in developing mammalian skin. PLoS Genet. 10, e1004687. https://doi.org/10.1371/journal.pgen.1004687 (2014).
Gumede, D. B., Abrahamse, H. & Houreld, N. N. Targeting Wnt/β-catenin signaling and its interplay with TGF-β and Notch signaling pathways for the treatment of chronic wounds. Cell. Communication Signal. 22. https://doi.org/10.1186/s12964-024-01623-9 (2024).
Ljubojevic, N., Henderson, J. M. & Zurzolo, C. The ways of actin: why tunneling nanotubes are unique cell protrusions. Trends Cell. Biol. 31, 130–142. https://doi.org/10.1016/j.tcb.2020.11.008 (2021).
Sartori-Rupp, A. et al. Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat. Commun. 10, 342. https://doi.org/10.1038/s41467-018-08178-7 (2019).
Sun, Z. Q., Guo, S. S. & Fässler, R. Integrin-mediated mechanotransduction. J. Cell. Biol. 215, 445–456. https://doi.org/10.1083/jcb.201609037 (2016).
Rustom, A., Saffrich, R., Markovic, I., Walther, P. & Gerdes, H. H. Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010. https://doi.org/10.1126/science.1093133 (2004).
Bai, X. et al. Tissue Micro-channels formed by collagen fibers and their internal components: cellular evidence of proposed meridian conduits in vertebrate skin. Microsc Microanal. 26, 1069–1075. https://doi.org/10.1017/s1431927620024381 (2020).
Berardo, A., Bonaldi, L., Stecco, C. & Fontanella, C. G. Biomechanical properties of the human superficial fascia: Site-specific variability and anisotropy of abdominal and thoracic regions. J. Mech. Behav. Biomed. Mater. 157. https://doi.org/10.1016/j.jmbbm.2024.106637 (2024).
Zhang, C., Ye, M., Bush, P. & Hu, B. H. Heterogeneity in macrophages along the cochlear spiral in mice: insights from SEM and functional analyses. Front. Cell. Neurosci. 17, 1222074. https://doi.org/10.3389/fncel.2023.1222074 (2023).
Cheng, D., Wang, J., Yao, M. & Cox, C. D. Joining forces: crosstalk between mechanosensitive PIEZO1 ion channels and integrin-mediated focal adhesions. Biochem. Soc. Trans. 51, 1897–1906 (2023).
Wang, L. et al. Structure and mechanogating of the mammalian tactile channel PIEZO2. Nature 573, 225–229. https://doi.org/10.1038/s41586-019-1505-8 (2019).
Naba, A. Mechanisms of assembly and remodelling of the extracellular matrix. Nat. Rev. Mol. Cell Biol. 25, 865–885. https://doi.org/10.1038/s41580-024-00767-3 (2024).
Alonso-Matilla, R., Provenzano, P. P. & Odde, D. J. Physical principles and mechanisms of cell migration. npj Biol. Phys. Mech. 2. https://doi.org/10.1038/s44341-024-00008-w (2025).
Mosaddeghzadeh, N. & Ahmadian, M. R. The RHO family gtpases: mechanisms of regulation and signaling. Cells 10. https://doi.org/10.3390/cells10071831 (2021).
Hantschel, O. Structure, regulation, signaling, and targeting of Abl kinases in cancer. Genes Cancer. 3, 436–446. https://doi.org/10.1177/1947601912458584 (2012).
Yamaguchi, H., Kasa, M., Amano, M., Kaibuchi, K. & Hakoshima, T. Molecular mechanism for the regulation of rho-kinase by dimerization and its Inhibition by fasudil. Structure 14, 589–600. https://doi.org/10.1016/j.str.2005.11.024 (2006).
Haga, R. B. & Ridley, A. J. Rho gtpases: regulation and roles in cancer cell biology. Small GTPases. 7, 207–221. https://doi.org/10.1080/21541248.2016.1232583 (2016).
Malik, R., Lelkes, P. I. & Cukierman, E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 33, 230–236. https://doi.org/10.1016/j.tibtech.2015.01.004 (2015).
Liebman, C., McColloch, A., Rabiei, M., Bowling, A. & Cho, M. in Current Topics in Membranes Vol. 86 (eds Levitan, I. & Trache, A.) 143–184 (Academic Press, 2020).
Ribeiro-Rodrigues, T. M., Martins-Marques, T., Morel, S., Kwak, B. R. & Girao, H. Role of connexin 43 in different forms of intercellular communication - gap junctions, extracellular vesicles and tunnelling nanotubes. J. Cell Sci. 130, 3619–3630. https://doi.org/10.1242/jcs.200667 (2017).
Yoo, D. et al. Functionalized extracellular vesicles of mesenchymal stem cells for regenerative medicine. J. Nanobiotechnol. 23, 219. https://doi.org/10.1186/s12951-025-03300-6 (2025).
Kestecher, B. M. et al. Reduced Circulating CD63(+) extracellular vesicle levels associate with atherosclerosis in hypercholesterolaemic mice and humans. Cardiovasc. Diabetol. 23, 368. https://doi.org/10.1186/s12933-024-02459-w (2024).
Zhu, Z. et al. EFNA5 suppresses cell proliferation and tumor metastasis in hepatoma via epithelial-to-mesenchymal transition. Discover Oncol. 15, 572. https://doi.org/10.1007/s12672-024-01454-7 (2024).
Pasquale, E. B. Eph receptor signaling complexes in the plasma membrane. Trends Biochem. Sci. 49, 1079–1096. https://doi.org/10.1016/j.tibs.2024.10.002 (2024).
Fede, C. et al. Evidence of a new hidden neural network into deep fasciae. Sci. Rep. 11, 12623. https://doi.org/10.1038/s41598-021-92194-z (2021).
Zuo, Z., Huang, P., Jiang, Y., Zhang, Y. & Zhu, M. Acupuncture attenuates renal interstitial fibrosis via the TGFbeta/Smad pathway. Mol. Med. Rep. 20, 2267–2275. https://doi.org/10.3892/mmr.2019.10470 (2019).
Bai, X. et al. The cellular mechanism of acupuncture for ulcerative colitis based on the communication of telocytes. Microsc. Microanal. 29, 1190–1204. https://doi.org/10.1093/micmic/ozad028 (2023).
Singhmar, P. et al. The fibroblast-derived protein PI16 controls neuropathic pain. Proc. Natl. Acad. Sci. U.S.A. 117, 5463–5471. https://doi.org/10.1073/pnas.1913444117 (2020).
Wu, T. et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Camb). 2, 100141. https://doi.org/10.1016/j.xinn.2021.100141 (2021).
Théry, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 7, 1535750. https://doi.org/10.1080/20013078.2018.1535750 (2018).
Acknowledgements
We provide our high regards to Professor William V. Holt (University of Sheffield, UK) for his help in improving the quality and English language of the manuscript and in terms of scientific meaning.
Funding
The authors received no funding for this work.
Author information
Authors and Affiliations
Contributions
H.H.X., M.L. and C.Q.S. conceived and designed research; H.H.X., M.L., L.L., Y.M., F.L.L. and Y.T. performed experiments and analyzed data; H.H.X. and M.L. interpreted results of experiments, prepared figures, and drafted manuscript; M.L., Z.Z.W., Y.J.M., D.M. and C.Q.S. edited and revised manuscript. All authors approved final version of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Material 3
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, H., Mei, L., Zhang, Z. et al. Laminar CD34+ membranous cells with mechanosensitive properties in subcutaneous fascia of the abdominal midline. Sci Rep 15, 39032 (2025). https://doi.org/10.1038/s41598-025-25651-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-25651-8