Abstract
Potato virus Y (PVY) is one of the most significant pathogens in the production of cultivated potato Solanum tuberosum worldwide. The most efficient way to combat viral diseases is the use of resistant varieties. The Rychc gene was discovered in the tuber-bearing wild potato relative Solanum chacoense. This gene is able to confer extreme resistance to PVY by rapidly halting virus multiplication in plants without the development of visible symptoms. Two molecular markers were developed to predict the presence of the Rychc gene and PVY resistance with 100% accuracy. In this study, the diversity of the Rychc gene and its relationship with resistance to PVY were investigated in genotypes from the Russian potato genbank. A collection of 60 genotypes originating from different populations was examined for PVY resistance and for the presence of Rychc using molecular markers. The observed phenotypes ranged from susceptibility to hypersensitive reaction and extreme resistance to PVY. Irrespective of the type of resistance, all the PVY-resistant plants carried the Rychc gene, indicating the prevalence of this resistance mechanism in S. chacoense. Unexpectedly, two S. chacoense individuals with the Rychc gene were found to be completely susceptible to PVY. Rychc sequences were evaluated via long-read targeted sequencing in all Rychc-positive genotypes. Despite the low overall sequence variation observed in Rychc gene, identical polymorphisms were found in Rychc alleles from susceptible S. chachoense genotypes, demonstrating the existence of nonfunctional Rychc allele unable to confer PVY resistance.
Similar content being viewed by others
Introduction
Potato is the most important non-grain crop worldwide. The use of vegetative reproduction by tubers makes potato especially vulnerable to various viral diseases, since many plant viruses are unable to enter true seeds but readily accumulate in tubers1. The viral load can multiply in successive vegetative generations, leading to severe yield losses2,3. Chemical control agents commonly used to combat various plant diseases are inefficient against viruses because of their intracellular lifestyle. As a result, modern potato cultivation largely depends on the use of virus-free tubers obtained from in vitro cultivated plants4. This practice is cumbersome and costly, and healthy plants can be readily infected again under field conditions2. Among potato viruses, arguably the most important is potato virus Y (PVY). PVY is distributed worldwide and causes yield losses of up to 80% in potato3,5. In addition to yield reduction, tuber quality is also compromised3. Under field conditions, PVY is primarily distributed by aphids in a non-persistent manner, which makes control of viral spread via insecticides challenging3,6, although some cultural practices, such as straw mulching or spraying with mineral oil, could have positive effects by impeding aphid infestation on potato plants4,7.
The most potent approach to combat viral diseases is the use of genetically resistant varieties8. In different potato species, two main types of resistance genes that act either through the hypersensitive response (HR) or confer extreme resistance (ER) have been identified9. In contrast to the hypersensitive response, which causes the development of necrotic lesions that restrict virus spread in plant tissues, extreme resistance is associated mainly with the formation of no or only minor necrotic lesions and presumably acts through halting of viral multiplication inside plant cells9. Multiple genes conferring either HR or ER resistance were discovered in different potato cultivars and wild relatives, named with “N” or “R” first letters according to the resistance type – Ny-1, Ny-2, Ny-Smira, Nychc, Nydms, Nytbr, Nyspl, Ny(o, n)sto, Ncspl, Nctbr, Nyadg for HR and Ryadg, Rysto(XI) and Rysto(XII), Ry-fsto, Ryhou, Rychc genes for ER5,8,10,11,12,13,14. Some novel sources of extreme resistance to PVY have been proposed recently15,16. While HR genes often confer strain-specific resistance, ER genes are generally effective against a broad spectrum of PVY strains. For example, the Rysto gene from S. stoloniferum Schlechtd. et Bché. is effective against all known PVY strains and even against another potyvirus, potato virus A8,17,18. Rysto protein is an immune receptor that recognizes the conserved region in potyviral coat proteins19. The Ryadg gene from S. tuberosum subsp. andigenа Hawkes confers ER to all strains of PVY13. The third most studied gene for extreme PVY resistance, Rychc from Solanum chacoense Bitter, also provides multistrain resistance to PVY20,21,22. Rychc was widely used in the selection of PVY-resistant potato8,23. Upon PVY infection, the phenotypic effect of the resistance conferred by Rychc gene in these cultivars varies from no symptoms to more pronounced necrotic areas developing around the site of inoculation20,21,24. After continuous efforts to create DNA markers and map Rychc, this gene was finally cloned from the 40−3 S. chacoense accession via map-based cloning and 40−3 BAC library analysis22. The transgenic expression of Rychc in two potato cultivars makes it resistant to PVY. Later, using hybrid genome assembly data, different Rychc allele variant was identified in the 184202-2 dihaploid potato line obtained after crossing of several Rychc-bearing potato cultivars25. Both identified Rychc alleles were highly similar, with few point mutations or short deletions/insertions between them, and were able to confer extreme resistance to PVY. While in the original study, the Rychc gene was suggested to have 5 exons and 4 introns22, further analysis of RNAseq data demonstrated that Rychc mRNA comprises only 4 exons25. Rychc encodes a Toll/interleukin-1 receptor (TIR)-nucleotide binding site (NBS)-leucine rich repeat (LRR)-type protein that is 911 aa in length. Curiously, the Rychc gene was shown to have two splicing variants, one for the 911 aa protein and another for the severely truncated 554 aa protein25. Using a known Rychc sequence, two PCR markers located directly within the gene sequence were developed. These two markers demonstrated complete linkage with both the presence of Rychc in the genome and PVY resistance22,25.
The investigation of plant R genes is hindered by their large number, cluster arrangement and repetitive nature of LRR domains26. Long-read sequencing technologies could be a valuable tool for studying R genes both separately and in their native genome arrangement. Recently, PacBio sequencing was used to evaluate the diversity of the Rysto gene in several Solanaceae species via targeted sequencing of Rysto amplicons27. In this study, a similar approach was used to discover the allelic diversity of the Rychc gene in a large collection of plants from the Russian potato genbank that had been previously classified as S. chacoense. These plants were characterized for PVY resistance, and the relationship between the presence of different Rychc allelic variants and resistance to PVY was investigated.
Materials and methods
Plant material
The study involved S. chacoense plants representing 11 accessions from the collection of the N.I. Vavilov Institute of Plant Genetic Resources (VIR). These accessions include S. chacoense material with a different origin and a different source of acquisition (Table 1). Expeditions throughout South America were performed by P. Zhukovskij and by L. Gorbatenko, who collected the accessions k-2732 and k-19769, respectively. Professor Jack Hawkes kindly provided accessions k-2861, k-7394 and k-22638 during his visit in VIR in 1990. Accession k-22638 was delivered as tubers and attributed by J. Hawkes as Solanum commersonii Dun., but later was recognized by a curator of Russian genbank as S. chacoense. Currently, its systematic position has not been completely resolved. The accessions k-21848, k-21849 and k-21854 were obtained from potato collection maintained in Ecuador (National Agricultural Research Institute, INIAP), and the accession k-22687 was obtained from the US Potato Genebank (USPG). Accessions of S. chacoense were obtained either as populations or as tubers and preserved as orthodox seeds in the VIR genebank. In 2017, the botanical seeds of 11 accessions S. chacoense from seed reproduction 1991–1992 were involved in a study of PVY resistance. Resistance to PVY was assessed among S. chacoense seedlings in the first year and their clonal plants in the second year by artificial inoculation in 2018–2019. All S. chacoense genotypes were subsequently maintained by the propagation of tubers annually in the VIR greenhouse. The second round of studies on PVY resistance was performed in 2021–2023. Owing to the unequal ability of the S. chacoense accession for clonal propagation, the number of plants that were saved for testing for molecular genetic analysis differed (Table 1). In total, 60 genotypes of S. chacoense were evaluated for resistance to PVY and the presence of DNA markers.
PVY infection
S. chacoense plants were tested in greenhouse by artificial inoculation. Nicotiana tabacum L. plants (‘Samsun’ variety) were used for PVY inoculum preparation. Three leaflets per one seedling of each S. chacoense accession were inoculated with sap from a mixture of tobacco plants displaying visible symptoms of ordinary and necrotic PVY strain infection. Ordinary strain infection displayed itself as mosaic and vein clearing, while necrotic strain infection – as vein necrosis on tobacco plants. The plant leaves identified for inoculation were powdered with carborundum powder, and the virus inoculum was then rubbed across the leaves. Symptoms of virus infection were evaluated after 10–14 days, and ELISA was carried out by taking leaf samples after 30 days. An ELISA double sandwich method28 was used to detect PVY in the leaves of all tested plants. The diagnostic kits used were produced in the A.G. Lorch Potato Research Institute (Russia) and Bioreba (Switzerland). S. chacoense plants showing no symptoms and no virus accumulation according to ELISA in the seedling test were retested next year as clonal plants. S. chacoense individuals were considered susceptible if the virus was detected by ELISA but were considered resistant if no virus was detected over two independent trials.
Screening the collection for the presence of DNA markers of the Ry chc gene
The isolation of plant genomic DNA was performed according to Doyle and Doyle29, with the incubation period extended to 2 h. Genomic DNA was diluted 100-fold prior to use. The PCR markers MG64-1722 and 1648F24/1648R2225 were used (Table S1, Figure S1). The total volume of the PCR mixture was 25 µl, which included 20 µl of water, 2.5 µl of buffer, 0.5 µl of each primer (10 µM stock solution), 0.5 µl of dNTPs (10 mM stock solution), 0.5 µl of Taq polymerase and 0.5 µl of template DNA. All PCR reagents were supplied by Evrogen (Russia). PCR was performed using a 96-well MiniAmp™ Plus Thermal Cycler (Thermo Fisher Scientific, USA). The PCR conditions for MG64-17 consisted of one cycle of 3 min at 94°С; 36 cycles of 30 s at 94°С, 30 s at 54°С, and 30 s at 72°С; and a final elongation of 10 min at 72°С, according to Li et al.22. The PCR conditions for 1648F24/1648R22 consisted of one cycle of 3 min at 94°С, followed by 35 cycles of 30 s at 94°С, 30 s at 55°С, and 1 min at 72°С, and a final elongation of 10 min at 72°С. PCR products were separated via electrophoresis on 1.5% agarose gels in 1× TAE buffer (40 mM Tirs, 20 mM acetic acid, and 1 mM EDTA) and visualized via ethidium bromide DNA gel stain (Biolabmix, Russia) and UV transillumination. All S. chacoense genotypes were evaluated with both markers at least twice.
Amplification of the full-length Ry chc gene
The primer pairs STRG1648f-F/STRG1648f-R and RyCDS-F/Ry5-R (Table S1) were used to amplify the full-length Rychc gene from genomic DNA. The total volume in the PCR assay was 20 µl, including 12.9 µl of water, 4 µl of buffer, 1 µl of each primer (10 µM stock solution), 0.4 µl of dNTPs (10mM stock solution), 0.2 µl of Phusion High-Fidelity DNA polymerase and 0.5 µl of template DNA. All PCR reagents were supplied by NEB (USA). The PCR conditions consisted of one cycle of 40 s at 98°С, followed by 32 cycles of 20 s at 98°С, 20 s at 63°С, and 2 min at 72°С, and a final elongation of 5 min at 72°С. For some difficult-to amplify samples, LongAmp Taq DNA polymerase (NEB, USA) was used instead of Phusion. All PCR products were separated via electrophoresis on 1.0% agarose gels in 1× TAE buffer and visualized through ethidium bromide DNA gel stain and UV transillumination.
Sequencing of the full-length Ry chc gene sequence on the MinION platform
The full-length Rychc gene sequences were isolated from the PCR mixture via the GeneJET Gel Extraction and DNA Cleanup Micro Kit (Thermo Fisher Scientific, USA). The quality of the isolated DNA was assessed by electrophoresis and spectrophotometry. The nanopore sequencing library was prepared using SQK-NBD114.96 (Oxford Nanopore Technology, Oxford, UK) and sequenced on FLO-MIN114 flow cell on a MinION sequencer. FAST5 data were base called and demultiplexing was performed via Guppy ver. 6.5.7 in high-accuracy mode with an NVIDIA GPU (RTX3060 12 GB).
Sequence analysis of the full-length Ry chc gene
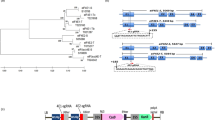
The adapter sequence was trimmed by guppy_barcoder. Quality control of the sequencing was performed by pycoQC (ver. 2.5.2). To obtain the complete sequence of the Rychc gene, the following programs were used: minimap2 (ver. 2.24-r1122) for raw data mapping, samtools (ver. 1.21) for data filtering and sorting, CLAIR3 (ver. v1.0.7) for SNP calling, WhatsHAP (ver. 2.2) for identifying the allelic state of the Rychc gene in samples, and amplicon_sorter.py to obtain the Rychc gene consensus sequence in its allelic state30. The protein sequences were obtained by annotating the gene exons according to data provided by Akai et al.25 (GenBank: LC726345.1) and then translating them according to the standard genetic code via the Geneious ver. 10.0.5 program. Domain annotation was performed by InterPro online software31. To make dendrograms, Rychc gene sequences or protein sequences were aligned using MUSCLE with default settings. Phylogenetic trees were generated using IQ-TREE 3.0.132, taking into account the optimal model selected by the program. All trees were visualized in iTOL33.
Results
Collection of S. chacoense genotypes evaluated for PVY resistance
Sixty S. chacoense individuals belonging to 11 accessions from the VIR (Table 1) were grown in the greenhouse to evaluate PVY resistance under artificial inoculation. Infection results from inoculation with the mixture of ordinary and necrotic PVY strains were symptoms in the aerial parts of the plants and data of ELISA double-antibody sandwich (DAS) test. A variety of phenotypic responses were observed in S. chacoense plants, ranging from no visible symptoms or small point (tiny) necroses to necrotic rings or spots on the upper side of the leaflets, necrosis along the veins of the underside of leaflets, mild mottle or rugosity, leaf distortion or collapse and dropping of intermediate leaves, which remained clinging to the stem. All S. chacoense individuals were divided into three groups according to phenotype evaluation and ELISA assessment. Twenty-nine plants presented no visible symptoms or tiny necrotic lesions (Fig. 1) and no PVY accumulation according to ELISA analysis were considered to be extremely resistant to PVY (Table 2). Twenty-two PVY-susceptible plants demonstrated diverse symptoms of viral infection and PVY accumulation in plant tissues. The remaining nine plants that developed stem necrosis without PVY accumulation were suggested to be resistant through a hypersensitive response mechanism (HR). Therefore, almost two-thirds of the investigated S. chacoense individuals were resistant to PVY. The ratio of resistant and susceptible individuals varied across the different S. chacoense accessions; 11 of the 14 plants in the 542 family (accession k-19769) were resistant, whereas none of 6 plants of 545 family (accession k-21849) and only 2 of the 9 plants in the 541 family (accession k-7394) were resistant, in accordance with previous study34.
Reaction of S.chacoense plants on PVY infection following mechanical inoculation. Extreme resistance – no systemic infection (A) with no lesions (not shown) or tiny necrosis on the inoculated leaf (B). Hypersensitive reaction – stem necrosis (C) with no systemic infection (D). Susceptibility – excessive stem necrosis (E) and systemic infection on leaves (F).
Evaluation of S. chacoense genotypes using molecular markers to Ry chc gene
According to the phenotypical evaluation, the S. chacoense genotypes were relatively clearly divided into PVY-susceptible and PVY-resistant ones, whereas the latter presented no PVY accumulation and either a lack of symptoms or a hypersensitive response to infection. Since most resistant plants do not show any symptoms of infection after artificial inoculation and Rychc-mediated extreme resistance is the only known mechanism of PVY resistance in S. chacoense characterized molecularly, S. chacoense collection was screened for the presence of the Rychc gene using two PCR markers, MG64-1722 and 1648F24/1648R2225. Both markers are located in the Rychc sequence, and a perfect correlation between the presence of the marker and PVY resistance was obtained in previous studies. Regardless of the type of resistance – hypersensitive response or extreme resistance – all PVY-resistant S. chacoense plants were uniformly positive for both MG64-17 and 1648F24/1648R22 (Table 2), which suggests that Rychc is present in these plants. The majority of the PVY-susceptible genotypes didn’t demonstrate the amplification of neither MG64-17 nor 1648F24/1648R22, indicating the absence of Rychc. Unexpectedly, 2 susceptible genotypes, W08 and W28 (both originate from accession k-22638), were positive for both markers, whereas in another susceptible genotype, W53 (accession k-7394), only MG64-17 but not 1648F24/1648R22 was amplified (Table 2; Fig. 2). To check the specificity of the PCR, the marker amplicons were purified and Sanger sequenced. In W53, the MG64-17 sequence was similar but not identical to the previously published Rychc sequence from 40−3 S. chacoense22 and was clearly the result of misamplification of distinct R gene. In contrast, for both the MG64-17 and 1648F24/1648R22 primer pairs, the amplicon sequences from W08 and W28 plants were identical to the sequence of Rychc from the 40−3 S. chacoense genotype (Figure S2). Therefore, Rychc or at least its fragment is present in two PVY-susceptible genotypes from the investigated collection, W08 and W28. These genotypes belong to the k-22,638 accession with a contradictional taxonomy (see Materials and methods).
Amplification and SMRT sequencing of the full-length Ry chc genes
Despite the presence of the Rychc gene, W08 and W28 plants were clearly susceptible to PVY, demonstrating virus accumulation in plant tissues, systemic necrosis at the whole-plant level and growth inhibition. Apparent non-functionality of Rychc in some plants could be due to modifications of gene sequences, such as point mutations, deletions or recombinations. It is known that R genes are especially prone to diverse modifications and rearrangements, which make important contributions to the rapid evolution of these genes35,36,37. Targeted SMRT sequencing could be used to study full-length gene sequences from multiple plants using different barcodes. To amplify the full-length Rychc sequences from Rychc-bearing S. chacoense plants, the previously developed primer pair STRG1648f-F/STRG1648f-R was used25. These primers flanked the whole Rychc ORF, approximately 90 bp before the start codon and 900 bp after the stop codon. After optimization of the PCR conditions, the PCR fragment of the expected size was amplified from the most marker-positive S. chacoense genotypes (Table S2). However, for few marker-positive plants, no band or only a very weak band was obtained, possibly due to the difficulty of long fragment amplification from plant DNA samples contaminated with different PCR-inhibiting components. More surprisingly, in some S. chacoense genotypes that were negative for Rychc markers, PCR products with sizes similar to the Rychc amplicon length were amplified (Table S2). To improve the robustness and specificity of PCR, alternative primers were developed to amplify Rychc, with the forward primer at the beginning of the first exon and the reverse primer in the 3’-UTR (Figure S3). Amplification with this primer pair, named RyCDS-F/Ry5-R, was also suboptimal, with no bands in some Rychc-bearing S. chacoense plants, and amplification of the product of the correct size in some marker-negative PVY-susceptible plants (Table S2). However, using both the STRG1648f-F/STRG1648f-R and RyCDSF/Ry5-R primer pairs, for every marker-positive S. chacoense plant including two PVY-susceptible genotypes W08 and W28, amplicon of the correct size was obtained with at least one primer pair. We also tried some more primer pairs for full-length Rychc amplification, but with poor efficiency and specificity (data not shown).
All PCR products of the correct size from either marker-positive or marker-negative plants were purified, individually barcoded and sequenced in parallel via MinION. In total, 88 PCR products from either one or two primer pairs were sequenced for 50 plants — all 38 PVY-resistant and 12 PVY-susceptible plants — including W08, W28 and ten marker-negative S. chacoense genotypes. Sequencing data were sorted to individual S. chacoense genotypes using the barcodes and analysed. Sequences very similar to those of previously published Rychc genes were obtained from all marker-positive S. chacoense plants, either extremely resistant or hypersusceptible to the PVY, and also from PVY-susceptible genotypes W08 and W28. In contrast, no Rychc-like sequences were found in any of the 10 marker-negative S. chacoense plants. All sequences obtained from these PVY-susceptible genotypes were somewhat similar to Rychc and represented another R genes (data not shown), which were apparently misamplified due to the sequence similarity with Rychc. Such superfluous R genes were also identified in sequenced amplicons from some Rychc-bearing plants, suggesting co-amplification of the Rychc gene and some homologous R genes from these plants. However, the implemented approach was suitable to obtain Rychc gene sequences from all marker-positive S. chacoense plants and to clearly distinguish them from any co-amplified genes at the stage of data analysis.
Diversity of Ry chc sequences in S. chacoense genotypes
Five alleles of Rychc were found in this study (Fig. 3). Most genotypes from different S. chacoense accessions carry allele 1, which is equal to Rychc from 40−3 S. chacoense22. Allele 2 has the single SNP relative to allele 1 (Supplementary File 1). It was discovered in fewer genotypes limited to accessions k-19759 and k-19769 (Table 1; Fig. 3). Allele 3 which was abundant and distributed between several accessions has several SNPs relative to allele 1 and small polymorphic region in the first intron with two adjacent deletions of 4 bp and 7 bp and three nucleotide substitutions (Supplementary File 1, Figure S4). Rychc alleles 4 and 5 were detected in a small number of plants from the single accession k-22638. Rychc alleles from all S. chacoense genotypes showed marked similarity. Besides the region in the first intron, no deletions or insertions were identified in the whole amplified Rychc sequences, and only 19 SNPs were discovered between all identified genes in the whole Rychc ORF (Supplementary File 1). Curiously, exons were enriched in SNPs relative to introns. Apart from the aforementioned polymorphic region in the first intron, as few as three SNPs were found in all three Rychc introns with a total length of 701 bp, with two SNPs in the first intron and one in the third intron. In contrast, in four Rychc exons with an overall length of 2730 bp, 16 SNPs were found. Among these 16 SNPs in exones, 9 were nonsynonymous, suggesting mild positive selection operating on the Rychc sequence (dN/dS = 1,286).
Five Rychc alleles encoded three protein variants, with three most widespread alleles 1, 2 and 3 encoded for the same protein. The predicted protein sequences of the different Rychc variants were compared with each other and with previously published ones (Table 3, Supplementary File 2). There are 14 polymorphic amino acid residues between alleles from 40−3 S. chacoense accession22 and 184202-2 dihaploid potato line25. Only four new polymorphic positions were identified in Rychc protein variants from this study. 18 polymorphic positions were discovered in all currently investigated full-length Rychc protein variants (Table 3). All protein sequences from this study were more similar to 40−3 Rychc than to 184202-2 Rychc (Table 3). Among the 3 identified protein variants, one was prevalent, whereas other two were minor. The most common variant was found in 37 out of 40 studied S. chacoense plants and in 37 of 38 PVY-resistant plants, both extremely resistant and hypersusceptible to PVY. It was equal to Rychc previously identified in 40−3 S. chacoense accession (Table 3)22. The remaining 2 protein variants were identified for the first time. The next most frequent Rychc variant was found only in three S. chacoense plants, two of which, W08 and W28, were susceptible to PVY, and one, W25, mounted hypersensitive response on PVY infection. This variant has seven SAAPs (single amino acid polymorphisms) relative to the common Rychc, five of which are shared with 184202-2 Rychc, while two, R502L and Q518L, are unique (Table 3). Both W08 and W28 plants were homozygous for this allele, whereas W25 was heterozygous. Since both S. chacoense plants with this Rychc allele present in the homozygous state were susceptible to PVY, whereas all the other Rychc-bearing genotypes, and they only, were resistant, we concluded that this Rychc protein variant was nonfunctional against PVY infection. The remaining Rychc protein variant was found in two heterozygous W24 and W25 plants (Table 3), both of which were resistant to PVY. It differed from the most common Rychc variant with 6 SAAPs, of which 4 were shared with 184202-2 Rychc, while R159K and S624R polymorphisms were found only in this allele. While in W24 genotype this allele was in combination with common Rychc variant typical for resistant plants, in W25 it coexisted with apparently non-functional Rychc allele. W25 plant was resistant to PVY through HR, similarly to several plants homozygous for common Rychc allele (Table 1), which suggests that Rychc variant identified in genotypes W24 and W25 was functional against PVY infection despite of several polymorphisms relative to common Rychc. Collectively these results indicate that among two new Rychc protein variants identified in this study, one was functional against PVY infection while the other was nonfunctional. While multiple SAAPs were observed in 184202-2 Rychc protein and in several alleles in this study, only two polymorphisms R502L and Q518L found in the single Rychc allele apparently have drastic negative influence on Rychc functionality. According to the annotation of the domain organization of Rychc, the R502 and Q518 residues were located between the NB-ARC domain and the LRR domains of this protein (Figure S5). Since PCR markers MG64-17 and 1648F24/1648R22 locate in the end of the last exon and in 5’-UTR far outside the gene region bearing polymorphisms (Figures S1,S5), both markers cannot provide any discrimination between functional and nonfunctional Rychc alleles.
All S. chacoense genotypes bearing Rychc protein variants other than the most common, W08, W24, W25, and W28, belong to the same S. chacoense family 548, which was raised from the seeds of accession k-22638 (Fig. 4; Table 1). This family also comprises the W12 genotype, which has a “common” Rychc allele, and the W10 genotype, which lacks Rychc. Hence, all the biodiversity of the Rychc protein sequences identified in this study was limited to individuals of a single S. chacoense accession. K-22638 accession originated from tubers that were kindly provided to VIR collection by Prof. J. Hawkes, and this accession was attributed by him as S. commersonii Dun. This accession was subsequently reclassified as S. chacoense on the basis of its distinctive features, such as the petiolulate acute to acuminate leaflets, the terminal leaflet hardly larger than the laterals, and the uniformly white corolla. Certain forms of S. chacoense were erroneously identified as S. commersonii and have been reported in previous studies38. Variation of S. chacoense and S. commersonii complicates their taxonomy and several features common for both species have been reported in natural habitat. Both S. chacoense and S. commersonii are spread through several provinces of Argentina and Uruguay, and their representatives were included within diploid microspecies described by Bukasov and Hawkes39,40. The rare events of hybridization between S. chacoense and S. commersonii observed under natural and experimental conditions39,40 indicate possible gene flow between these wild potato relatives. Taking this into account, the k-22638 accession seems to be an outlying S. chacoense variant genetically distinct from more typical plants of this species and close to S. commersonii, which could be a reason for the presence of nontypical Rychc alleles in this accession. To examine the possible presence of Rychc in S. commersonii, published genomes of this species (GenBank numbers GCA_001239805.1; GCA_018258275.1; GCA_029007595.1; GCA_029582365.1; GCA_029582665.1) were searched for the presence of Rychc-like genes. Gene very similar to Rychc was found in none of them. In contrast, in the published S. chacoense genome M6_v5.0 (SpudDB) the Rychc gene was found (Table S3). It is also cannot be excluded that the k-22638 accession is related to another wild potato species S. malmeanum Bitter. A recent report by Nicolao et al.41 brings new light to this wild relative. Former taxonomic classification of S. malmeanum was rather complicated, due to geographical distribution patterns that partially overlap with S. commersonii and S. chacoense and several shared morphological traits41. Morphological variability of S. malmeanum, S. commersonii and S chacoense and non-consistent criteria used by different botanists could have leaded a mistake in classifying accessions of those sympatric species. Currently, there are no high-quality genome assemblies available for S. malmeanum to check for the presence of Rychc gene.
Phylogenetic tree showing the distribution of different Rychc protein variants between various families of S. chacoense plants (different colors for different accessions). Proteins from the 40−3 S. chacoense and 184202-2 dihaploid potato line are also included. All families, except for the k-22638 family which comprise several protein variants, have a single Rychc variant. A0A3Q7I9U8 (UniProt database) was used as outgroup.
Discussion
In this study, phenotypic and genotypic evaluation of S. chacoense plants from multiple accessions demonstrated that the PVY resistance trait is widespread in S. chacoense and that this resistance is due to the presence of the Rychc gene. The abundance of PVY-resistant plants in this species is in accordance with previously published data (42 and references therein). In addition to Rychc, the existence of some genes conferring HR to PVY was previously proposed in S. chacoense43. In this study, the presence of Rychc in all the PVY-resistant plants and the susceptibility of all the Rychc-negative plants strongly suggest that Rychc is the only factor associated with PVY resistance in the investigated S. chacoense collection. Since this gene was identified in 40 out of 60 S. chacoense plants of diverse origins, Rychc-mediated resistance could be the widespread mechanism of PVY resistance in this potato species. It was shown earlier that the most common mode of Rychc action is extreme resistance with a lack of visible necrosis, although in some cases, a hypersensitive response also occurs20,22,25. Both types of defense responses to PVY infection in Rychc-bearing S. chacoense plants were observed in this study. Extreme resistance was the most common, since less than one-fourth of the plants with resistance to PVY (9 of 38) developed significant necrotic lesions on leaves or the stem necrosis, indicating the hypersensitive response. A comparison of Rychc sequences from plants displaying either ER- or HR-type resistance to PVY revealed that Rychc genes were identical between them. Differences in the observed phenotype could therefore be related to the local variabilities in plant growing conditions, unequal infection or some inter-genotype variations influencing the mounting of defence responses. The dependence of R gene-based resistance on environmental conditions is a well-studied phenomenon44,45. Since the genes associated with extreme resistance belong to the same NB-LRR gene family as classical R genes conferring HR and seem to operate through at least partially similar mechanism5,45,46,47, unfavourable conditions could also hinder the mounting of the ER, leading to delayed action, which cannot rapidly block virus spread. This results in the generation of visible necrosis instead of mounting resistance at the single-cell level48.
In contrast to individuals with a hypersensitive response to PVY that demonstrated large necrosis but no PVY accumulation in plant tissues, two Rychc-bearing genotypes, W08 and W28, displayed systemic PVY infection. We believe that this is the first time that plants with the Rychc gene were found to be susceptible to PVY. An earlier study indicated that some PVY multiplication could be observed in Rychc-bearing potato plants; however, this multiplication was detected only under especially unfavourable conditions, such as permanent high temperature, and systemic infection was rarely detected by ELISA49. Both the PVY-susceptible W08 and W28 genotypes had the same Rychc variant in the homozygous state. Apparently, two adjacent non-conserved amino acid changes, R502L and Q518L, which are unique to this allele, perturbed the functionality of Rychc protein to a sufficient extent to make it ineffective against PVY. It was previously shown that even a single amino acid change could have a drastic influence on the activity or specificity of NB-LRR proteins44,50,51. The reason for the existence of the apparently nonfunctional Rychc allelic variant is elusive. Among hundreds of NB-LRR genes present in the genomes of higher plants, many nonfunctional variants exist, which accumulate mutations that disturb their functionality52. This process of R gene attenuation is at least partially promoted by the pleiotropic effects inherent to some R genes, which makes the selection of such non-functional variants favourable in pathogen-free conditions53,54,55. Alternatively, the identified apparently inactive Rychc variant could have altered specificity and play a role in resistance to viruses other than PVY or to some specific PVY strains. The specificity of pathogen recognition by NB-LRR proteins is mainly defined by the LRR domain, which is the least conserved part of this class of proteins56. According to domain prediction, the R502L and Q518L polymorphisms from the susceptible Rychc allele are located adjacent to the LRR domain but outside of it. Rychc sequences were especially conserved in the second part of this gene (Supplementary file 1), and C-terminal LRR domain was almost identical between different Rychc variants (Table 3, Supplementary File 2). It is uncertain could the R502L and Q518L polymorphisms have any effect on pathogen recognition specificity, or they rather modify Rychc functionality in a different way. Curiously, the single amino acid change in the region between NB-ARC and LRR was sufficient to make the Arabidopsis R gene SNC1 constitutively active without interaction with pathogens57.
S. chacoense is the most vigorous and adaptable of all the wild potato species in South America. Its natural distribution range is very extensive, from sea level to high mountains in Argentina, Paraguay, Uruguay, Brazil and Bolivia. There is morphological evidence for gene introgression from certain mountain species, such as Solanum microdontum Bitt., Solanum tarijense Hawkes, Solanum spegazzinii Bitt., and Solanum kurtzianum Bitt. et Wittm. ex Engl., that have resulted in a wider variety of S. chacoense habitats. Variation of S. chacoense and S. commersonii complicates their taxonomy and several features common for both species have been reported in natural habitat39. Despite the study of multiple S. chacoense plants obtained from diverse sources, which led to the identification of the unanticipated “susceptible” Rychc allele, the overall diversity of Rychc was low at both the gene and protein levels. Besides the single polymorphic region in the first intron, few SNPs were found in the whole Rychc ORF, especially in the non-coding regions. Only five Rychc alleles were found, with the most common allele was equal to the one previously discovered in 40−3 S. chacoense22. Moreover, three major Rychc alleles encoded the same protein. Two other minor Rychc alleles were discovered in only 4 out of 40 genotypes, all of which belong to the same accession k-22638. The pronounced allelic diversity of Rychc gene in plants of k-22638 accession apparently reflects its overall genetic distance from the other studied accessions, since k-22638 either represents an intermediate form between species or even belongs to species S. commersonii or S. malmeanum rather than to S. chacoense. Two minor Rychc protein variants had 6 or 7 polymorphisms relative to the common Rychc allele, whereas the previously identified 184202-2 Rychc had 14 polymorphisms (Table 3). In accordance with Akai et al.25, an 184202-2 dihaploid line was obtained after crossing of several potato cultivars harboring the Rychc gene from the ‘Konafubuki’ cultivar which, in turn, received it from a doubled S. chacoense plant24. Among all Rychc protein variants identified to date, Rychc from 184202-2 is the most distinct from the common Rychc variant. Even in S. chacoense plants of the accession k-22638, which seems to be rather unique and close to S. commersonii or S. malmeanum, Rychc variants were less different from the common Rychc than 184202-2 Rychc was (Table 3). This could suggest that the ancestral doubled line used in the development of ‘Konafubuki’ was rather unusual among S. chacoense species. Alternatively, 184202-2 Rychc sequences readily accumulated multiple point mutations after transfer from ancestral doubled S. chacoense line to potato cultivars. Rapid accumulation of mutations could be related to any pleiotropic effects, such as autoimmunity, that are frequently observed for plant R genes and largely depend on the genetic background55,58. Several polymorphisms were shared between 184202-2 Rychc and alleles from the k-22638 accession (Table 3), suggesting their common origin. The low overall diversity of Rychc sequences and the prevalence of the most common Rychc allele, which is effective against multiple PVY strains in different genetic backgrounds, favour the use of S. chacoense in potato breeding programs as a source of PVY resistance. Additional studies are needed to find out the taxonomic determination of the k-22638 accession from Russian potato germplasm collection and to check the possible involvement of Rychc genes in resistance to PVY in species close to S. chacoense, namely S. malmeanum or S. commersonii.
One of the most serious drawbacks of the use of molecular markers in plant breeding is the lack of full linkage between the marker and trait of interest because the marker locus and trait locus are typically located proximally but are not the same. However, for the Rychc gene, two recently introduced PCR markers are located within its sequence22,25, ensuring full linkage between the marker and the gene. This was confirmed in this study because all marker-positive S. chacoense plants were also Rychc positive, except for the single W53 genotype, in which one marker provided misleading results due to the misamplification of different R gene. However, the use of markers was insufficient to reliably predict PVY resistance in S. chacoense plants because of the existence of an apparently non-functional Rychc allele. To address this concern, modern sequencing technologies provide relatively simple and easy approach for studying full-length genes from multiple biological samples simultaneously. Molecular markers could be used to initially identify genes of interest in individual plants before attempting to amplify the full-length gene sequences. If closely linked markers were not developed but the expected gene sequence is available, several primer pairs could be used to amplify short gene fragments, as was recently done to identify the Rysto gene in multiple accessions of different Solanum species27. After this initial evaluation, full-length amplification of target genes, such as Rysto or Rychc using several primer pairs is applicable to isolate the gene of interest from different plants (27 and this study). Both primer pairs used in this study lead to some false-positive Rychc amplification from plants that do not contain this gene and to co-amplification of some additional R genes together with Rychc. However, such misamplified genes could be identified and omitted at the stage of data analysis. Although PacBio is frequently used for targeted long-fragment sequencing, the use of more assessable MinION allows similar results to be achieved59. Since both technologies allow the sequencing of full-length genes as single molecules without fragmentation on small pieces, there are no hurdles related to the sorting of short DNA pieces between highly homologous genes upon data analysis.
Conclusions
In this study, the allelic diversity of Rychc gene was examined in relation to resistance to PVY in a diverse collection of potato plants related to S. chacoense and, in case of k-22638 accession, possibly to other potato species. This investigation revealed that resistance to PVY is widely distributed in this species and is associated with the Rychc gene. Overall allelic variation of Rychc was low and in terms of coding sequence was restricted to the single S. chacoense accession, which is closely related to another wild potato species S. commersonii or S. malmeanum. In a few plants, a Rychc allele incapable to provide PVY resistance was discovered. While previously developed molecular markers cannot discriminate this allele, amplicon targeted sequencing was applied to investigate it. This approach allows for easy and robust identification of Rychc sequences in multiple plant genotypes.
Our results emphasize the importance of the relevance, accuracy and completeness of information in plant gene bank management. Challenges related to differences between potato classification systems have been noticed in the Global Strategy for the Conservation of Potato, which was developed through a collaborative effort of potato collection curators from 32 genebanks and their partners in potato research60. The international community has recognized the evaluation and characterization of potato accessions as a priority measure towards a harmonization of potato taxonomy. A comprehensive study of k-22638 accessions remains to be conducted in future research. Plant morphology and genetic variability of S. chacoense and related wild species preserved at the Russian potato genebank require a more thorough characterization. The sequence data obtained in our study linked to passport information are essential for gaining a deeper understanding of the diversity of potato genes involved in virus resistance.
Data availability
Gene and protein sequences are available under GenBank numbers PV815852, PV815853, PV815854, PV815855 and in Supplementary Files 1 and 2.
References
Khurana, S. M. Potato viruses and their management. In Diseases of fruits and vegetables, volume II: diagnosis and management (ed. Naqvi, S. A. M. H.) 389–440Springer, (2004).
Palukaitis, P. Resistance to viruses of potato and their vectors. Plant. Pathol. J. 28, 248–258. https://doi.org/10.5423/PPJ.RW.06.2012.0075 (2012).
Lacomme, C. & Jacquot, E. General characteristics of potato virus Y (PVY) and its impact on potato production: an overview. In Potato Virus Y: biodiversity, pathogenicity, Epidemiology and Management (ed (eds Lacomme, C. et al.) 1–19 (Springer, (2017).
Dupuis, B. et al. Potato virus Y: control, management and seed certification programmes. In Potato Virus Y: biodiversity, pathogenicity, Epidemiology and Management (ed (ed Lacomme, C.) 177–206 (Springer, (2017).
Quenouille, J., Vassilakos, N. & Moury, B. Potato virus Y: a major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol. Plant. Pathol. 14, 439–452. https://doi.org/10.1111/mpp.12024 (2013).
Radcliffe, E. B. & Ragsdale, D. W. Aphid-transmitted potato viruses: the importance of Understanding vector biology. Am. J. Potato Res. 79, 353–386. https://doi.org/10.1007/BF02870173 (2002).
Kirchner, S. M. et al. Comparison of straw mulch, insecticides, mineral oil, and Birch extract for control of transmission of potato virus Y in seed potato crops. Potato Res. 57, 59–75. https://doi.org/10.1007/s11540-014-9254-4 (2014).
Valkonen, J. P., Gebhardt, C., Zimnoch-Guzowska, E. & Watanabe, K. N. Resistance to potato virus Y in potato. In Potato Virus Y: biodiversity, pathogenicity, Epidemiology and Management (ed (eds Lacomme, C. et al.) 207–241 (Springer, (2017).
Valkonen, J. P. Elucidation of virus-host interactions to enhance resistance breeding for control of virus diseases in potato. Breed. Sci. 65, 69–76. https://doi.org/10.1270/jsbbs.65.69 (2015).
Valkonen, J. P. T., Slack, S. A., Plaisted, R. L. & Watanabe, K. N. Extreme resistance is epistatic to hypersensitive resistance to potato virus YO in a solanum tuberosum subsp. andigena-derived potato genotype. Plant. Dis. 78, 1177–1180. https://doi.org/10.1094/PD-78-1177 (1994).
Solomon-Blackburn, R. M. & Bradshaw, J. E. Resistance to potato virus Y in a Multitrait potato breeding scheme without direct selection in each generation. Potato Res. 50, 87–95. https://doi.org/10.1007/s11540-007-9031-8 (2007).
van Eck, H. J. et al. Graphical genotyping as a method to map Ny(o,n)sto and Gpa5 using a reference panel of tetraploid potato cultivars. Theor. Appl. Genet. 130, 515–528. https://doi.org/10.1007/s00122-016-2831-y (2017).
Herrera, M. D. R. et al. Molecular and genetic characterization of the Ryadg locus on chromosome XI from Andigena potatoes conferring extreme resistance to potato virus Y. Theor. Appl. Genet. 131, 1925–; (1938). https://doi.org/10.1007/s00122-018-3123-5 (2018).
Mangal, V. et al. Updates on molecular markers linked to potato virus Y (PVY) resistance genes in potato. Potato J. 48, 173–180 (2021).
Torrance, L. et al. Natural resistance to potato virus Y in solanum tuberosum group Phureja. Theor. Appl. Genet. 133, 967–980. https://doi.org/10.1007/s00122-019-03521-y (2020).
Elison, G. L., Park, J., Novy, R. G. & Whitworth, J. L. A potential new source of extreme resistance to potato virus Y in the potato variety bistra. Am. J. Potato Res. 101, 248–256. https://doi.org/10.1007/s12230-024-09954-6 (2024).
Barker, H. Inheritance of resistance to potato viruses Y and A in progeny obtained from potato cultivars containing gene Ry: evidence for a new gene for extreme resistance to PVA. Theor. Appl. Genet. 93, 710–716. https://doi.org/10.1007/BF00224066 (1996).
Grech-Baran, M. et al. Extreme resistance to potato virus Y in potato carrying the Rysto gene is mediated by a TIR-NLR immune receptor. Plant. Biotechnol. J. 18, 655–667. https://doi.org/10.1111/pbi.13230 (2020).
Grech-Baran, M. et al. The Rysto immune receptor recognises a broadly conserved feature of potyviral coat proteins. New. Phytol. 235, 1179–1195. https://doi.org/10.1111/nph.18183 (2022).
Sato, M., Nishikawa, K., Komura, K. & Hosaka, K. Potato virus Y resistance gene, Rychc, mapped to the distal end of potato chromosome 9. Euphytica 149, 367–372. https://doi.org/10.1007/s10681-006-9090-y (2006).
Mori, K. et al. Germplasm release: Saikai 35, a male and female fertile breeding line carrying solanum phureja-derived cytoplasm and potato cyst nematode resistance (H1) and potato virus Y resistance (Rychc) genes. Am. J. Potato Res. 89, 63–72. https://doi.org/10.1007/s12230-011-9221-4 (2012).
Li, G. et al. Rychc confers extreme resistance to potato virus Y in potato. Cells 11, 2577. https://doi.org/10.3390/cells11162577 (2022).
Elison, G. L., Novy, R. G. & Whitworth, J. L. Russet potato breeding clones with extreme resistance to potato virus Y conferred by Rychc as well as resistance to late blight and cold-induced sweetening. Am. J. Potato Res. 98, 411–419. https://doi.org/10.1007/s12230-021-09852-1 (2021).
Hosaka, K., Hosaka, Y., Mori, M., Maida, T. & Matsunaga, H. Detection of a simplex RAPD marker linked to resistance to potato virus Y in a tetraploid potato. Am. J. Potato Res. 78, 191–196. https://doi.org/10.1007/BF02883544 (2001).
Akai, K. et al. De Novo genome assembly of the partial homozygous dihaploid potato identified PVY resistance gene (Rychc) derived from solanum chacoense. Breed. Sci. 73, 168–179. https://doi.org/10.1270/jsbbs.22078 (2023).
Andolfo, G., Dohm, J. C. & Himmelbauer, H. Prediction of NB-LRR resistance genes based on full-length sequence homology. Plant. J. 110, 1592–1602. https://doi.org/10.1111/tpj.15756 (2022).
Paluchowska, P. et al. Diversity of the Rysto gene conferring resistance to potato virus Y in wild relatives of potato. BMC Plant. Biol. 24, 375. https://doi.org/10.1186/s12870-024-05089-2 (2024).
Clark, M. F. & Adams, A. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 34, 475–483. https://doi.org/10.1099/0022-1317-34-3-475 (1977).
Doyle, J. J. & Doyle, J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19, 11–15 (1987).
Vierstraete, A. R. & Braeckman, A. R. Amplicon_sorter: a tool for reference-free amplicon sorting based on sequence similarity and for Building consensus sequences. Ecol. Evol. 12, e8603. https://doi.org/10.1002/ece3.8603 (2022).
Blum, M. et al. InterPro: the protein sequence classification resource in 2025. Nucleic Acids Res. 53 https://doi.org/10.1093/nar/gkae1082 (2025).
Wong, T. et al. IQ-TREE 3: Phylogenomic Inference Software using Complex Evolutionary Models. Preprint at (2025). https://doi.org/10.32942/X2P62N
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Rogozina, E. V. et al. Genetic diversity of solanum pinnatisectum Dun. And solanum chacoense Bitt. By resistance to potato virus Y And results of DNA analysis. Russ J. Genet. 55, 1330–1337. https://doi.org/10.1134/S1022795419110139 (2019).
Meyers, B. C., Kaushik, S. & Nandety, R. S. Evolving disease resistance genes. Curr. Opin. Plant. Biol. 8, 129–134. https://doi.org/10.1016/j.pbi.2005.01.002 (2005).
Wessler, S. R. & Carrington, J. C. Genome studies and molecular genetics: the consequences of gene and genome duplication in plants. Curr. Opin. Plant. Biol. 8, 119–121. https://doi.org/10.1016/j.pbi.2005.01.015 (2005).
Durrant, W. E., Wang, S. & Dong, X. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. U S A. 104, 4223–4227. https://doi.org/10.1073/pnas.0609357104 (2007).
Hawkes, J. G. The Potato: evolution, Biodiversity and Genetic Resources (Belhaven, 1990).
Hawkes, J. G. & Hjerting, J. P. The Potatoes of Argentina, Brazil, Paraguay and Uruguay: a Biosystematic Study (Clarendon, 1969).
Hawkes, J. G. & Hjerting, J. P. The Potatoes of Bolivia: their Breeding Value and Evolutionary Relationships (Oxford Science, 1989).
Nicolao, R., Gaiero, P., Castro, C. M. & Heiden, G. Solanum malmeanum, a promising wild relative for potato breeding. Front. Plant. Sci. 13, 1046702. https://doi.org/10.3389/fpls.2022.1046702 (2023).
Cai, X. K., Spooner, D. M. & Jansky, S. H. A test of taxonomic and biogeographic predictivity: resistance to potato virus Y in wild relatives of the cultivated potato. Phytopathology 101, 1074–1080. https://doi.org/10.1094/PHYTO-02-11-0060 (2011).
Cockerham, G. Genetical studies on resistance to potato viruses X and Y. Heredity 25, 309–348. https://doi.org/10.1038/hdy.1970.35 (1970).
Zhu, Y., Qian, W. & Hua, J. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 6, e1000844. https://doi.org/10.1371/journal.ppat.1000844 (2010).
Balint-Kurti, P. The plant hypersensitive response: concepts, control and consequences. Mol. Plant. Pathol. 20, 1163–1178. https://doi.org/10.1111/mpp.12821 (2019).
Künstler, A., Bacsó, R., Gullner, G., Hafez, Y. M. & Király, L. Staying alive–is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant. Pathol. 93, 75–84. https://doi.org/10.1016/j.pmpp.2016.01.003 (2016).
Ross, B. T., Zidack, N. K. & Flenniken, M. L. Extreme resistance to viruses in potato and soybean. Front. Plant. Sci. 12, 658981. https://doi.org/10.3389/fpls.2021.658981 (2021).
Hong, X. et al. Searching for plant NLR immune receptors conferring resistance to potyviruses. Crop J. 12, 28–44. https://doi.org/10.1016/j.cj.2023.11.010 (2024).
Ohki, T., Sano, M., Asano, K., Nakayama, T. & Maoka, T. Effect of temperature on resistance to potato virus Y in potato cultivars carrying the resistance gene Rychc. Plant. Pathol. 67, 1629–1635. https://doi.org/10.1111/ppa.12862 (2018).
T Bryan, G. et al. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant. Cell. 12, 2033–2045. https://doi.org/10.1105/tpc.12.11.2033 (2000).
Zhang, J. et al. Single amino acid change alters specificity of the multi-allelic wheat stem rust resistance locus SR9. Nat. Commun. 14 https://doi.org/10.1038/s41467-023-42747-9 (2023).
Lozano, R. et al. Genome-wide identification and mapping of NBS-encoding resistance genes in solanum tuberosum group Phureja. PLoS One. 7, e34775. https://doi.org/10.1371/journal.pone.0034775 (2012).
Tian, D., Traw, M. B., Chen, J. Q., Kreitman, M. & Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis Thaliana. Nature 423, 74–77. https://doi.org/10.1038/nature01588 (2003).
Karasov, T. L., Chae, E., Herman, J. J. & Bergelson, J. Mechanisms to mitigate the trade-off between growth and defense. Plant. Cell. 29, 666–680. https://doi.org/10.1105/tpc.16.00931 (2017).
Barragan, A. C. & Weigel, D. Plant NLR diversity: the known unknowns of pan-NLRomes. Plant. Cell. 33, 814–831. https://doi.org/10.1093/plcell/koaa002 (2021).
Takken, F. L. & Goverse, A. How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant. Biol. 15, 375–384. https://doi.org/10.1016/j.pbi.2012.05.001 (2012).
Zhang, Y., Goritschnig, S., Dong, X. & Li, X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant. Cell. 15, 2636–2646. https://doi.org/10.1105/tpc.015842 (2003).
Wan, W. L. et al. Genetics of autoimmunity in plants: an evolutionary genetics perspective. New. Phytol. 229, 1215–1233. https://doi.org/10.1111/nph.16947 (2021).
Becerra-Wong, M. et al. Exploring S-RNase diversity in the Andean black Cherry (Prunus serotina) using minion sequencing: a cost-effective approach with increased genotyping resolution. Euphytica 219, 100. https://doi.org/10.1007/s10681-023-03227-9 (2023).
Nagel, M. et al. Global strategy for the conservation of Potato. Global crop diversity trust. Bonn Ger. https://doi.org/10.5447/ipk/2022/29 (2022).
Funding
This work was supported by the Russian Science Foundation Grant №21-76-10050, https://rscf.ru/project/21-76-10050/, and by Ministry of Science and Higher Education of the Russian Federation (theme No. 122042700045-3).
Author information
Authors and Affiliations
Contributions
NZ and ER conceived and designed the experiment. ER and AG participated in sample preparation. AA and NZ performed molecular studies. AA analysed the sequencing data. NZ and ER wrote the manuscript. AG and AA corrected the manuscript. All authors read the manuscript and approved it for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aleksandr, A., Alena, G., Nikolay, Z. et al. Role of Rychc gene diversity in the resistance of wild potato relative Solanum chacoense to potato virus Y. Sci Rep 15, 42055 (2025). https://doi.org/10.1038/s41598-025-25963-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-25963-9