Abstract
Due to the notable biological activity of isatin hydrazone compounds, we synthesized and structurally characterized three arylidene isatin hydrazone derivatives (3a-3c) utilizing the SBA-Pr-N-Is-Bu-SO3H nanocatalyst. A comparative investigation was performed through an integrated approach combining experimental assays and theoretical modeling. In vitro biological assays performed on normal umbilical vein endothelial cells (HUVECs) and breast cancer cell lines (MCF-7 and MDA-MB-231) revealed noteworthy anticancer activity, with all derivatives significantly reducing cancer cell viability in a concentration- and time-dependent manner. Furthermore, compound 3c exhibited the most potent antioxidant effect, achieving up to 80% inhibition at higher concentrations. Biomolecule (DNA and BSA) interaction studies were performed utilizing UV-Vis spectroscopic analysis and molecular docking simulations. The values of Kapp for the interaction of compounds 3a (1.01 × 104 M− 1), 3b (1.17 × 104 M− 1), and 3c (2.03 × 104 M− 1), along with docking simulations, indicate that the studied compounds are likely to bind to CT-DNA via a groove-binding mechanism. Lipophilicity assessments demonstrated that compounds 3b and 3c had significantly higher log P values than cisplatin, indicating enhanced lipid affinity and superior cellular membrane permeability potential. Comprehensive computational analyses, including DFT/TD-DFT, topology analysis, and ADME-Tox profiling, were employed to further validate the observed biological activity and pharmacokinetic potential. The convergence of computational insights and experimental findings provides robust validation for the potential of these compounds as promising anticancer agents.
Similar content being viewed by others
Introduction
Cancer remains a major global health concern, characterized by uncontrolled cell proliferation and metastasis, and ranks as the second leading cause of death worldwide after cardiovascular diseases1. Among cancers, breast cancer is the most frequently diagnosed type in women, accounting for 2.3 million new cases and 685,000 deaths in 20201, with projections estimating over 3.2 million annual cases by 20501. This growing burden is especially pronounced in developing countries, where delayed diagnosis and limited healthcare infrastructure hinder effective treatment.
Current therapies, especially chemotherapy, suffer from poor selectivity, severe systemic toxicity, and increasing drug resistance, all of which reduce therapeutic efficacy1. The clinical complexity of breast cancer arises from its heterogeneous subtypes, namely hormone receptor-positive, human epidermal growth factor receptor 2 (HER2)-positive2, and triple-negative3, each requiring tailored therapeutic approaches4. Overexpressed molecular targets like HER2 and epidermal growth factor receptor (EGFR) have emerged as key drivers of tumor progression, making them central to anticancer drug development5. Thus, novel agents that overcome resistance and enhance selectivity remain a pressing need.
Many anticancer agents act by disrupting DNA replication, inducing apoptosis in rapidly dividing cells. DNA-interacting drugs function through covalent or non-covalent mechanisms, with the latter offering enhanced specificity and reversibility while reducing off-target cytotoxicity6,7. Among non-covalent strategies, intercalation and groove binding are predominant. Intercalators such as doxorubicin distort DNA by inserting aromatic rings between base pairs and inhibiting topoisomerase II8,9, but their efficacy is often limited by dose-dependent toxicity.
Groove-binding molecules engage DNA via hydrogen bonding and hydrophobic interactions, fitting into the minor or major grooves with minimal helical distortion and high specificity8,10. Compounds such as distamycin analogs selectively recognize AT- or GC-rich sequences, with binding efficiency influenced by structural features like aromatic curvature and hydrogen-bonding side chains11,12. Recent studies highlight groove binders as promising tools for reducing chemoresistance and side effects. However, challenges related to pharmacokinetics and tumor heterogeneity persist13,14. To bridge these gaps, one promising avenue involves the exploration of heterocyclic scaffolds capable of achieving high binding selectivity, with isatin (1 H-indole-2,3-dione) emerging as a notable candidate.
Isatin has garnered attention due to its versatile pharmacophore features7,15. First synthesized in 1841, it is an endogenous compound derived from tryptophan metabolism16,17, with distinct sites, such as the benzene ring, amide nitrogen, and C3 carbonyl, amenable to targeted modifications17. Among its derivatives, isatin hydrazones, formed by condensing the C3 carbonyl group with aromatic hydrazines, have gained prominence in anticancer drug discovery18,19,20. These derivatives exploit the electron-rich aromatic moieties to enhance interactions with cancer-related enzymes, including tyrosine kinases, tubulin, and histone deacetylases, which regulate cancer cell proliferation and apoptosis21,22,23,24. Structural modifications, such as halogenation of the hydrazone-linked aryl moiety, can enhance binding affinity, pharmacokinetics, and target selectivity by fine-tuning steric and electronic features, thereby minimizing off-target interactions and improving drug–target engagement18,21,23,25. Clinically validated isatin-based drugs, such as sunitinib and nintedanib, exemplify the significant therapeutic potential inherent to the isatin scaffold5,26. Moreover, emerging evidence highlights hydrazone derivatives as promising candidates for overcoming drug resistance in cancer27.
Beyond oncology, isatin hydrazones exhibit neuroactive properties, cross the blood-brain barrier, and modulate monoamine oxidase activity, making them potential candidates for central nervous system therapies23,28. Importantly, the integration of aromatic moieties with the isatin core not only enables the design of multifunctional therapeutics but also provides opportunities to combine anticancer activity with favorable pharmacophoric features29,30. Despite progress in DNA-targeting therapies and isatin hydrazone development, challenges persist. Future efforts should focus on optimizing pharmacokinetics, enhancing tumor selectivity, and refining structural features. Computational design and advanced DNA-binding assays will be crucial for developing safer and more effective groove-specific anticancer agents.
In this study, we conducted a comprehensive comparative investigation of three structurally related isatin hydrazone derivatives, employing a synergistic combination of experimental and computational methodologies. The compounds were systematically evaluated for their in vitro anticancer and antioxidant activities, offering insights into their potential therapeutic relevance. To elucidate their mechanistic interactions at the molecular level, we assessed binding affinities with key biological targets, DNA, a primary locus for many chemotherapeutic agents, and bovine serum albumin (BSA), a critical transport protein in systemic circulation, using UV-Vis absorption spectroscopy and molecular docking simulations. In parallel with rigorous experimental investigations, a comprehensive array of cutting-edge computational approaches was deployed, including DFT calculations to unravel intricate electronic structures, quantum chemical topology analyses to decode bonding patterns and reactivity landscapes, sophisticated absorption-distribution-metabolism-excretion–toxicity (ADME-Tox) profiling to predict pharmacokinetic behavior, and structure-activity relationship (SAR) analysis to mechanistically rationalize and predict the potent bioactivities observed.
Experimental part
Materials and methods
All solvents were analytical grade, purchased from Merck, and used without further purification. Titrations were performed with doubly distilled water. CT-DNA (calf thymus DNA, Type-1, highly polymerized), bovine serum albumin (BSA, ~ 66,463 Da), and Tris-HCl buffer were procured from Sigma-Aldrich and used as received. Other chemicals, such as sodium hydroxide (NaOH), dimethyl sulfoxide (DMSO), sodium chloride (NaCl), 2-Octanol, ethanol (EtOH), and 2,2-diphenyl-1-picrylhydrazyl (DPPH) were sourced from Merck and employed without additional purification.
FT-IR spectra were obtained using a RAYLEIGH WQF-510 A spectrometer with KBr pellets, covering the spectral range of 600–4000 cm− 1 (Beijing Rayleigh Analytical Instrument Corporation, Beijing, China). 1H-NMR spectra were conducted using a VARIAN INOVA 500 MHz spectrometer utilizing DMSO-d6. The UV-Vis absorption spectra of the compounds, as well as their interactions with DNA and BSA, were recorded using an Analytic Jena Specord S600 spectrophotometer.
Synthesis of SBA-Pr-N-Is-Bu-SO3H nanocatalyst and Isatin hydrazone
SBA-Pr-N-Is-Bu-SO3H nanocatalyst and isatin hydrazone were prepared according to the previously reported articles31,32.
Synthesis of arylidene isatin hydrazones
First, the SBA-Pr-N-Is-Bu-SO3H nanocatalyst was heated at 120 ℃ for activation. Subsequently, 0.02 g of the nanocatalyst was added to a 10 mL flask containing a mixture of isatin hydrazone (1 mmol, 0.16 g) and various aldehydes (1 mmol each). The reaction mixture was refluxed in 5 mL of EtOH for 5 min. Upon reaction completion (monitored by TLC; eluent: ethyl acetate/n-hexane, 1:1 v/v), the resulting solid was dissolved in excess hot EtOH, and the nanocatalyst was separated via filtration. Finally, the arylidene isatin hydrazones (3a-c) were isolated.
Cell cytotoxicity assay
The cytotoxic effects of the compounds were tested against normal human umbilical vein endothelial cells (HUVEC) and breast cancer cell lines (MCF-7 and MDA-MB-231) utilizing the MTT colorimetric assay. Human breast cancer cell lines and normal cells were sourced from the Iranian Biological Resource Center (IBRC) in Tehran, Iran. HUVEC, MCF-7, and MDA-MB-231 cells were cultured in T25 flasks until they reached approximately 80% confluence. The cells were then seeded into 96-well plates and incubated for 24 h at 37 ℃ under 5% CO2 to ensure proper adhesion. Subsequently, the cells were treated with different concentrations of the compounds (0, 2.5, 5, 10, 25, 50, 100, 150, 250, and 500 µg/mL) for 24 and 48 h. Following treatment, 25 µL of MTT solution (Sigma) was added to each well, and the plates were incubated for an additional 3 h. The medium was then discarded, and the resulting formazan crystals were solubilized using 100 µL of DMSO. Finally, cell viability was evaluated by measuring the optical density (OD) at 490 nm with a microplate reader.
Antioxidant activity
The antioxidant potential of the compounds was assessed using the DPPH free radical scavenging assay, a widely recognized method for evaluating the antioxidant capability of compounds, extracts, and other bioactive agents33. The purple color of the DPPH solution disappears when an antioxidant agent is introduced into the reaction medium. The antioxidant donates an electron or a hydrogen atom to the DPPH radical, reducing it to a colorless or pale-yellow compound (DPPH-H). This reduction process leads to a decrease in absorbance at 517 nm. To prepare the DPPH radical solution, 4 mg of DPPH was dissolved in 100 mL of ethanol to obtain a 0.1 mMsolution. Various concentrations of the test compounds (10, 25, 50, 100, 250, 500, and 1000 µg/mL) were prepared and each concentration was mixed with an equal volume of the DPPH solution (1:1 ratio, v/v). The mixtures were vortexed for 10 s and then left to incubate in the dark at room temperature for 30 min. Afterward, the absorbance was measured at 517 nm with a spectrophotometer. Ascorbic acid was employed as the reference antioxidant control. The DPPH radical scavenging activity of the compounds was determined using the following Eq. (1):
DNA/BSA interaction
Sample preparations
Fresh stock solutions of BSA (3 mg/mL) and CT-DNA (1.5 mg/mL) were prepared in Tris-HCl buffer (pH 7.2) under gentle stirring at 4 °C to ensure homogeneity. Simultaneously, the isatin-hydrazone derivatives were dissolved in distilled water at a concentration of 10− 4 M. The purity of the DNA solution was verified via assessing the absorbance ratio at 260 nm and 280 nm (A260/A280), which was within the expected range of 1.8–1.9, indicating minimal protein contamination. Precise concentrations of the biomolecules were calculated using their respective molar extinction coefficients: ε260 = 6600 M− 1cm− 1 for DNA34 and ε280 = 44,300 M− 1cm− 1 for BSA35. All experimental mixtures were incubated at 298 K for spectral analysis.
UV–Vis titrations
UV–Vis spectroscopy was employed to monitor structural alterations in CT-DNA and BSA upon interaction with synthesized isatin-hydrazone derivatives at 298 K. For these experiments, two titrations were performed for each compound:
-
i.
CT-DNA Interaction Study: 50 µL of the CT-DNA stock solution was introduced into 1.45 mL of Tris-HCl buffer, yielding a total volume of 1.50 mL. The mixture was then gently agitated to ensure thorough homogenization and consistent distribution of components. Subsequent titrations were performed by incrementally adding varying concentrations of investigated compounds (3a-c). The concentration ranges applied were as follows: 0.00 to 9.64 µM for the 3a + CT-DNA system, 0.00 to 9.09 µM for the 3b + CT-DNA system, and 0.00 to 13.29 µM for the 3c + CT-DNA system.
-
ii.
BSA Interaction Study: 500 µL of BSA stock solution was mixed with 1.00 mL of Tris-HCl buffer, yielding a total volume of 1.50 mL. The mixture was gently agitated to ensure thorough homogenization. The final BSA samples were titrated via various concentrations of compounds (3a-c), which include 0.00 to 6.83 µM (for 3a + BSA), 0.00 to 10.18 µM (for 3b + BSA), and 0.00 to 8.53 µM (for 3c + BSA).
After each titration step, the resulting mixtures were maintained at a constant temperature of 298 K for a duration of two minutes to ensure equilibrium was reached. UV–Vis absorption spectra were subsequently recorded and analyzed to determine the apparent association constant (Kapp) for the biomolecule-compound interactions. The calculations were performed using the following Eq. (2)36:
In this analysis, A0 and A refer to the absorbance values of CT-DNA or BSA measured at wavelengths of 260 nm and 280 nm, respectively. Here, A0 denotes the absorbance in the absence of the studied compounds (3a-c), and A corresponds to the absorbance after each titration step involving the compounds. The variable [Ligand] corresponds to the concentration of the compound introduced at each stage. As the interaction progressed and the biomolecule binding sites became saturated, a stable UV-Vis absorbance profile was recorded, denoted as A∞. To evaluate the binding affinity of the compounds against the biomolecules, a double reciprocal plot was constructed via plotting 1/[Ligand] on the x-axis and 1/(A-A0) on the y-axis. From the slope and intercept of this linear plot (intercept divided by slope), the Kapp was determined. This parameter quantifies the interaction strength between the compounds and the biomolecules under the experimental conditions.
Furthermore, the thermodynamic stability of the compound-biomolecule complexes was assessed by calculating the standard Gibbs free energy change (ΔG°) utilizing the formula ΔG° = - RTlnKapp, where T is 298 K and R is the ideal gas constant (1.987 cal.mol− 1K− 1).
Lipophilicity evaluation
Lipophilicity (log P) of the synthesized isatin-hydrazone derivatives was determined by the shake-flask method37 using a 1:1 (v/v) mixture of 2-octanol and doubly distilled water (10 mL total). Each compound (0.001 g) was dissolved in the biphasic system, shaken overnight, and left to equilibrate for 1 h. The phases were separated, and compound concentrations in each were determined using UV-Vis spectroscopy. The partition coefficient was calculated as Eq. (3):
Here, [Ligand]Octanol Phase and [Ligand]Water Phase are related to the concentration of the isatin-hydrazone derivatives in octanol and water solvents, respectively.
Computational details
DFT perspectives
The geometries of compounds (3a-c) were optimized using DFT/B3LYP with the 6–311+g (d,p) basis set in Gaussian 09 W38 employing GaussView 6.0, within the CPCM model (water). Vibrational frequency analyses confirmed true minima. From the optimized geometries, HOMO–LUMO energies, MEP maps, thermodynamic parameters, DOS spectra, and quantum chemical descriptors were obtained. UV–Vis spectra were computed via TD-DFT in gas phase and with CPCM (water).
Topological analysis
Non-Covalent Interaction (NCI) analysis was performed with Multiwfn 3.839 and visualized using VMD package40. Reduced Density Gradient (RDG) isosurfaces were generated, along with Electron Localization Function (ELF) and Localized Orbital Locator (LOL) analyses.
ADME-Tox prediction
In the initial phases of drug development, the integration of in vitro techniques with advanced computational strategies has become a cornerstone for screening candidate molecules for optimal pharmacokinetic (ADME: absorption, distribution, metabolism, and excretion) and toxicological profiles41. Physicochemical, drug-likeness, and toxicity parameters of the compounds were predicted using OSIRIS DataWarrior (v06.04.01)42.
Molecular docking
To investigate the in silico potential biological activities of the compounds (3a-c) and their molecular interactions with Estrogen Receptor alpha (ERα), ERα–DNA complex, DNA, and BSA, molecular docking simulations were performed. These studies were carried out utilizing AutoDock 4.243 and AutoDock Tools 1.5.644. The output files of the compounds as ligands were obtained from DFT calculations. Receptor models were sourced from the RCSB Protein Data Bank: ERα (PDB ID: 3ERT), estrogen receptor DNA-binding domain bound to its response element (PDB ID: 1HCQ), DNA duplex (PDB ID: 1BNA), and BSA (PDB ID: 4OR0). Prior to docking, all macromolecules and ligands were prepared by assigning Kollman and Gasteiger partial charges, respectively. Grid maps defining the docking space were generated using AutoGrid, with parameters tailored to each target to encompass the ligand-binding or interaction site. For ERα (PDB ID: 3ERT), the grid box size was 70 × 90 × 70 Å3 with the center at X = 22.397, Y = − 3.396, Z = 21.988. For the ERα–DNA complex (PDB ID: 1HCQ), the grid box size was 110 × 100 × 110 Å3 with the center at X = 36.692, Y = 12.334, Z = 111.741. For DNA (PDB ID: 1BNA), the grid box size was 50 × 50 × 56 Å3 with the center at X = 14.779, Y = 14.954, Z = 14.819. For BSA (PDB ID: 4OR0), the grid box size was 70 × 60 × 60 Å3 with the center at X = 3.808, Y = 12.586, Z = 94.758. All grid maps were calculated with a standard grid spacing of 0.375 Å. Lamarckian Genetic Algorithm (LGA) was used for docking protocol, and docking visualization and interaction profiling were conducted using BIOVIA Discovery Studio.
Results and discussion
Isatin hydrazone (1) was reacted with different aldehydes (2) in the presence of SBA-Pr-N-Is-Bu-SO3H nanocatalyst to produce arylidene isatin hydrazones (3) in EtOH under reflux conditions for 5 min (Scheme 1).
Possible mechanism
First, the aldehyde is activated and protonated by the SBA-Pr-N-Is-Bu-SO3H nanocatalyst. The protonated aldehyde carbonyl (2) is then nucleophilically attacked by the amine group of isatin hydrazone (1), leading to the formation of intermediate A. Proton transfer is subsequently initiated in intermediate A, resulting in intermediate B, which is followed by water elimination to produce intermediate C. Finally, the target products (3a-c) are formed through deprotonation of intermediate C by the SBA-Pr-N-Is-Bu-SO3− (Scheme 2).
The time, yield, and melting point for the preparation of arylidene isatin hydrazones (3a-c) are demonstrated in Table 1.
Characterization
(Z)−3-(((E)−3-nitrobenzylidene)hydrazineylidene)indolin-2-one (3a), Orange powder, 3159 (N-H), 3084 (aromatic C-H), 1745 (C = O), 1618 (N-H), 1533 and 1348 (N-O), 1594, 1531, 1460 (aromatic C-C). 1HNMR (250 MHz, DMSO-D6): 6.9 (d, 2 H, Ar-H), 7.4 (s, 1H, Ar-H), 7.7 (m, 2 H, Ar-H), 8.4 (s, 2 H, -N = CH-Ar), 8.6 (s, 2 H, Ar-H), 10.9 (s, 1H, N-H) ppm. 13C NMR (62.5 MHz, DMSO-D6), 111.4, 116.5, 122.9, 123.9, 126.5, 129.1, 131.2, 134.4, 134.5, 135.4, 145.6, 148.7, 150.4, 157.3, 164.7 ppm. MS (m/e): 294 (M+, 10%), 266 (100%), 220(50%), 145 (15%), 118 (85%), 103 (45%), 91 (40%), 76 (55%)46.
(Z)−3-(((E)−2-methoxybenzylidene)hydrazineylidene)indolin-2-one (3b), Orange powder, 3150 (N-H), 3080 (aromatic C-H), 1728 (C = O), 1610 (N-H), 1593, 1544, and 1489 (aromatic C-C). 1HNMR (300 MHz, DMSO-D6): 11.05 (s, 1H, N-H), 8.81 (s, 1H, -N = CH-Ar-H), 8.10 (dd, 1H, Ar-H), 7.97 (d, 1H, Ar-H), 7.58 (td, 1H, Ar-H), 7.39 (td, 1H, Ar-H), 7.20 (d, 1H, Ar-H), 7.14 (t, 1H, Ar-H), 7.03 (t, 1H, Ar-H), 6.9 (d, 1H, Ar-H), 3.9 (s, 3 H, OMe) ppm. 13C NMR (62.5 MHz, DMSO-D6), C = 55.4 (OMe), 169, 141, 138, 117, 157, 119, 129, 123, 111, 131, 124, 127.7, 132.0, 121.1, 55.8, 157.3. MS (m/e): 279 (M+, 3%), 251 (100%), 208 (20.1%), 118 (32%), 91 (25%), 77 (19%)47.
((Z)−3-(((E)−4-fluorobenzylidene)hydrazineylidene)indolin-2-one (3c), Orange powder, 3165 (N-H), 3085 (aromatic C-H), 1726 (C = O), 1622 (N-H), 1596, 1546, and 1460 (aromatic C-C). 1HNMR (300 MHz, DMSO-D6): 10.65 (s, 1H, N-H), 8.63 (s, 1H, -N = CH-Ar-H), 8.06 (dd, 2 H, Ar-H), 7.90 (d, 1H, Ar-H), 7.40 (m, 3 H, Ar-H), 7.03 (t, 1H, Ar-H), 6.90 (d, 1H, Ar-H) ppm. 13C NMR (500 MHz, DMSO-D6), C = 166.05, 164.97, 159.94, 151.05, 145.53, 139.11, 134.25, 131.87, 130.57, 129.32, 127.50, 126.69, 122.82, 117.91, 116.85, 111.35, 110.43, 141, 138, 117, 157, 119, 129, 123, 111, 131, 124, 127.7, 132.0, 121.1, 55.8, 157.3. MS (m/e): 267 (M+, 7%), 239 (100%), 212 (9%), 118 (76%), 95 (71%)47,48.
The experimental absorption maximum wavelength (λmax) for the arylidene isatin-hydrazone (3a-c) was investigated. Two λmax were observed for 3a in 241, 320 nm, 3b in 245, 330 nm, and 3c in 245, 310 nm, which were attributed to the π\(\:\to\:\)π* and n\(\:\to\:\)σ* transitions, respectively. Also, it was observed that compounds (3a-c) have almost similar UV-Vis spectra.
Theoretical IR and UV-Vis spectra of the compounds (3a-c) were obtained at DFT/TD-DFT calculation at B3LYP/6–311+g (d,p) level with CPCM using water as the solvent. These spectra for the compounds along with the experimental spectra are illustrated in Figs. 1 and 2. A good agreement between the theoretical and experimental spectra was observed.
Cell proliferation suppression
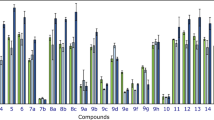
The in vitro cytotoxicity of compounds (3a-c) was evaluated against normal HUVEC cells and breast cancer cell lines (MCF-7 and MDA-MB-231) utilizing the MTT method at 24 and 48 h. The half-maximal inhibitory concentration (IC50) values for each of these compounds across normal and cancer cells are summarized in Table 2. The results revealed cytotoxic effects in normal cells at concentrations exceeding 200 µg/mL for 3a and 100 µg/mL for 3b and, with 50% cell death observed after 24 and 48 h of exposure. In contrast, compound 3c exhibited higher cytotoxicity profile within the 50–500 µg/mL concentration range, as depicted in Fig. 3 (a-c) and Table 2. Figure 3 (d-i) illustrates the anticancer effects of the compounds on breast cancer cells. The findings demonstrate that compounds (3a-c) effectively reduced cancer cell viability in a concentration- and time-dependent manner. Among them, compound 3c exhibited the most potent cytotoxicity (IC50 ~ 10 µg/mL) at all-time points, whereas compounds 3a and 3b required higher concentrations to achieve comparable effects. A progressive decline in cell viability was observed over time for all tested compounds. The presence of fluorine within the molecular structure of the compounds appears to enhance cellular membrane permeability, thereby increasing cytotoxic efficacy. The observed anticancer effects may be related to mechanisms such as the generation of reactive oxygen species (ROS) or disruption of key cellular pathways. Notably, despite the cytotoxicity of these compounds, their antitumor potential remains evident at concentrations lower than the toxic threshold. Furthermore, significant morphological changes were observed upon treatment. Under an inverted light microscope, spherical cell formation and shrinkage were clearly detectable across different concentrations, with some instances of cellular necrosis. The rounding of cells at higher concentrations indicates necrotic death, while shrinkage suggests apoptotic progression. Figure 4 presents the morphological effects of IC50 concentrations and higher, further supporting the cytotoxic and apoptotic potential of these compounds.
Cytotoxic effect of 3a (a), 3b (b), and 3c (c) against HUVEC cells after 24 and 48 h post-treatment. Cytotoxic effect of 3a (d), 3b (e), and 3c (f) against MCF-7 breast cancer cells after 24 and 48 h post-treatment. Cytotoxic effect of 3a (g), 3b (h), and 3c (i) against MDA-MB-231 breast cancer cells after 24 and 48 h post-treatment.
Changes in cancer cell morphology after treatment with compounds. The red arrows indicate the rounded and shrunken cells in each treatment. Control sample of MCF-7 cells (a). MCF-7 cells treated with 25 µg/mL of compound 3a (b). MCF-7 cells treated with 25 µg/mL of compound 3b (c). MCF-7 cells treated with 25 µg/mL of compound 3c (d). Control sample of MDA-MB-231 cells (e). MDA-MB-231 cells treated with 25 µg/mL of compound 3a (f). MDA-MB-231 cells treated with 25 µg/mL of compound 3b (g). MDA-MB-231 cells treated with 25 µg/mL of compound 3c (h).
Antioxidant activity
Figure 5 shows the percentage of free radical scavenging activity at various concentrations of compounds (3a-c) in comparison to the standard antioxidant control, ascorbic acid. As depicted in Fig. 5, the scavenging activity exhibited a concentration-dependent increase across all tested samples. The highest antioxidant activity was observed within the concentration range of 250 to 500 µg/mL for all compounds. However, lower concentrations also demonstrated free radical scavenging potential; for instance, at 50 µg/mL, the activity reached approximately 50% for all tested compounds. The standard control (blue bar) displayed the highest scavenging activity, nearing saturation at elevated concentrations, underscoring its superior antioxidant potential. Among the tested compounds, 3c exhibited the most pronounced antioxidant activity (80% in high concentration).
UV–Vis titrations
Given the promising anticancer activity exhibited by the synthesized isatin hydrazone derivatives (3a-c) against breast cancer cell lines, we extended our investigation to explore their molecular interactions with biomolecules using UV-Vis titration. DNA, being a primary target of numerous chemotherapeutic agents, and serum albumin, the principal carrier protein in systemic circulation, were selected to evaluate the compounds’ binding behavior and potential bioavailability.
DNA interaction
UV–Vis spectroscopy stands as a pivotal analytical method for elucidating the binding interactions between biologically active small molecules and macromolecules such as DNA or proteins49. These interactions frequently induce alterations in the compound’s electronic environment, which are reflected in its UV–Vis spectral profile. Variations in absorbance intensity, manifesting as either hyperchromism (an increase in absorbance) or hypochromism (a decrease in absorbance), along with wavelength shifts, such as bathochromic (red shift) or hypsochromic (blue shift) transitions50, serve as critical indicators of the nature and mode of binding. In particular, hypochromic effects accompanied by bathochromic shifts are typically indicative of an intercalative binding mechanism. This case occurs when the aromatic part of the ligand intercalates between the base pairs of DNA, resulting in strong π–π stacking interactions that perturb the DNA’s electronic structure51. Furthermore, a spectral profile showing hyperchromism and a hypsochromic shift suggests a non-intercalative binding pathway52. Such interactions may involve electrostatic attraction to the phosphate backbone or fitting within the major or minor grooves of the DNA helix, implying surface-level or groove binding rather than insertion between base pairs. UV–Vis absorption titration was conducted by maintaining a fixed volume of CT-DNA while varying the concentrations of compounds (3a-c), 0.00, 1.96, 2.59, 3.22, 3.84, 4.46, 5.06, 5.66, 6.25, 6.83, 7.40, 7.97, 8.53, 9.09, and 9.64 µM for 3a, 0.00, 4.46, 5.06, 5.66, 6.25, 6.83, 7.40, 7.97, 8.53, and 9.09 µM for 3b, and 0.00, 5.06, 5.66, 6.25, 6.83, 7.40, 7.97, 8.53, 9.09, 9.64, 10.18, 11.24, 12.28, and 13.29 µM for 3c. Figure 6 presents the UV-Vis absorption responses of CT-DNA upon gradual titration with varying concentrations of compounds (3a-c). Across all three systems, namely 3a + CT-DNA, 3b + CT-DNA, and 3c + CT-DNA, a marked increase in absorbance was observed with the incremental addition of the respective compounds, indicative of a hyperchromic effect. This phenomenon typically signals significant interaction between the DNA bases and the external molecules. Notably, in the spectra of the 3b + CT-DNA and 3c + CT-DNA systems, a significant hypsochromic shift (blue shift = 10 nm for 3b + CT-DNA and 12 nm for 3c + CT-DNA) in the absorption maxima was also evident. Such spectral behavior strongly suggests that the interaction mechanism does not involve intercalation between base pairs. Instead, it points toward a non-intercalative binding mode, most likely involving groove binding or electrostatic interactions along the DNA backbone. Since the studied compounds are neutral and lack an electrical charge, the likelihood of significant electrostatic attraction to the negatively charged phosphate backbone of DNA is minimal. Consequently, it is more plausible that the interaction occurs through groove binding mode, rather than through charge-based mechanisms. In this regard, molecular docking simulations serve as a valuable tool to detect the binding mode between the compounds and DNA.
By plotting 1/A-A0 against 1/[Ligand], the apparent binding constant (Kapp) was generated from the ratio of the intercept to the slope at 298 K, as illustrated in Fig. 6 (insets). The values of Kapp for the interaction of compounds 3a, 3b, and 3c with CT-DNA were calculated to be 1.01 × 104, 1.17 × 104, and 2.03 × 104 M− 1, respectively. It is important to highlight that the binding constants (Kb) for traditional intercalators like ethidium bromide and [Ru(phen)DPPZ] are typically in the range of 106 to 107 M− 153. Based on these values, it can be inferred that the isatin-hydrazone derivatives are likely to bind to CT-DNA via a groove-binding mechanism. The thermodynamic evaluation of the interactions between compounds 3a–c and CT-DNA revealed negative standard Gibbs free energy changes (ΔG°), calculated as −5.46 kcal/mol for 3a, −5.55 kcal/mol for 3b, and − 5.87 kcal/mol for 3c. These negative ΔG° values provide compelling evidence that the binding processes are energetically favorable and occur spontaneously under physiological conditions, indicating a strong intrinsic affinity of the compounds for the DNA substrate.
BSA interaction
Electronic absorption spectroscopy serves as a powerful analytical method for monitoring conformational alterations in proteins and assessing their interactions with small molecules, which is a key aspect in drug discovery and pharmacological research. In the present study, UV-visible spectroscopic titration was employed to investigate the binding affinity of isatin-hydrazone derivatives (3a–c) with BSA. UV–Vis absorption titration was conducted by maintaining a fixed volume of BSA while varying the concentrations of compounds (3a-c), 0.00, 1.96, 2.59, 3.22, 3.84, 4.46, 5.06, 5.66, 6.25, and 6.83 µM for 3a, 0.00, 3.22, 3.84, 4.46, 5.06, 5.66, 6.25, 6.83, 7.40, 7.97, 8.53, 9.09, 9.64, and 10.18 µM for 3b, and 0.00, 2.59, 3.22, 3.84, 4.46, 5.06, 5.66, 6.25, 6.83, 7.40, 7.97, and 8.53 µM for 3c. Figure 7 illustrates the absorption spectra of native BSA in comparison with the spectra obtained upon incremental addition of the isatin-hydrazone compounds. A pronounced hyperchromic effect was detected, characterized by a steady increase in absorbance intensity as the concentration of the compounds increased. This spectral behavior indicates the interaction between the derivatives and BSA. The observed enhancement in absorbance is attributed to microenvironmental changes surrounding the tryptophan residues within the BSA structure. Specifically, the interaction appears to perturb the polarity and hydrophobic balance in these regions, likely due to partial unfolding of the protein’s α-helical domains and an overall conformational rearrangement54. Such structural transitions strongly suggest the formation of a stable protein-ligand complex, demonstrating the effective binding potential of the isatin-hydrazone derivatives.
The Kapp values for compounds 3a, 3b, and 3c in their interaction with BSA were determined to be 6.61 × 103, 7.35 × 103, and 8.93 × 103 M− 1, respectively. These values offer important insights into the nature of the interaction between these isatin-hydrazone derivatives and BSA. Thermodynamic analysis of the binding interactions between BSA and the synthesized compounds 3a, 3b, and 3c revealed favorable free energy changes, with ΔG° values calculated at −5.21 kcal/mol, −5.27 kcal/mol, and − 5.39 kcal/mol, respectively. These negative ΔG° values indicate spontaneous binding processes, with compound 3c demonstrating the strongest affinity toward BSA among the three, as reflected by its more pronounced free energy reduction. When extending the investigation to CT-DNA, compound 3c demonstrated the highest binding affinity, with a Kapp of 2.03 × 104 M− 1, noticeably greater than those of compounds 2a and 2b, which showed values of 1.01 × 104 M− 1 and 1.17 × 104 M− 1, respectively. The Kapp values for the interaction between the compounds and DNA/BSA follow the order: 3c > 3b > 3a. This trend confirms the observed trend in their in vitro biological activity, suggesting a direct correlation between molecular binding strength and bioefficacy. Furthermore, the pronounced affinity of 3c toward CT-DNA suggests a strong molecular recognition capability, likely attributed to specific structural or electronic characteristics that enhance its interaction with the DNA helix. Conversely, while compound 3c maintained moderate affinity for BSA, its interaction was clearly more substantial than that observed for 3a and 3b, which bound more weakly. The observed variability in binding constants underscores the critical role of structural nuances within the ligand series (3a-c) in dictating their binding profiles. These findings not only advance our understanding of the selective interaction mechanisms of isatin-hydrazone derivatives but also emphasize their promise as molecular probes or therapeutic agents with tailored binding affinities toward specific biological macromolecules.
Lipophilicity evaluation
The partition coefficient between water and 2-octanol, commonly referred to as log P, is a pivotal pharmacokinetic indicator employed to assess the lipophilic character of chemical entities. This parameter serves as a proxy for a compound’s ability to dissolve in lipids, thereby providing insight into its potential to traverse the lipid-rich phospholipid bilayers of cellular membranes55. Compounds exhibiting elevated log P values typically demonstrate enhanced membrane permeability, enabling easier diffusion into adipose tissues and other lipid-dense biological environments. In the present study, the calculated log P values for the compounds 3a, 3b, and 3c were found to be 1.294, 2.297, and 2.225, respectively. These values indicate a moderate to high degree of lipophilicity. In contrast, cisplatin, a widely used chemotherapeutic agent, has a significantly lower log P value reported in the literature as − 2.28 ± 0.0756. The resulting lipophilicity hierarchy, 3b > 3c > 3a > cisplatin, highlights a marked difference in membrane permeability potential. Notably, compounds 3b and 3c, owing to their higher lipophilic profiles, are more likely to diffuse efficiently across cellular membranes compared to cisplatin.
DFT calculations
Structural parameters
Geometry optimization was performed for the compounds (3a-c) bearing a different substituent at varied positions on the aromatic ring (Fig. 8). The comparative analysis of their selected bond lengths and angles revealed that the electronic nature and position of the substituents exert a subtle yet noticeable influence on the molecular geometry. The selected bond lengths and angles for these compound are summarized in Table 3. The N1—C1 and C1—O1 bond lengths remain nearly constant across the series, suggesting minimal impact of substitution on this local environment. However, the C1—C8 bond is slightly shorter in 3b (1.523 Å) compared to 3a and 3c, possibly due to the electron-donating resonance effect of the ortho-OCH3 group, which may slightly enhance delocalization. A similar trend is observed in the C9—C10 bond, which is shortest in 3b (1.450 Å), indicating localized structural effects influenced by substituent proximity. Compound 3a uniquely shows characteristic nitro group bond lengths (C12—N4 at 1.473 Å, N4—O2 and N4—O3 both at 1.226 Å), while 3b displays typical ether linkage values (C11—O2 at 1.354 Å, O2—C16 at 1.433 Å), and 3c exhibits a C12—F1 bond length of 1.355 Å. The angular parameters show consistency, with only slight variations, e.g., the N1—C1—O1 angle slightly narrows in 3b (126.281°), likely due to steric effects from the ortho substituent. Overall, while the core scaffold remains structurally consistent, the electronic and steric properties of the substituents subtly modulate bond lengths and angles, revealing their influence on the molecular geometry.
Thermodynamic parameters
Statistical mechanics serves as a bridge between the microscopic molecular behavior and macroscopic thermodynamic properties. By analyzing energy states arising from molecular translational, rotational, vibrational, and electronic motions, it becomes possible to derive key thermodynamic quantities such as entropy and heat capacity, parameters that are crucial in understanding complex biological systems57. Among these parameters, entropy (S) is insightful, as it provides a measure of molecular disorder and aids in predicting the spontaneity and direction of biochemical interactions, including ligand-receptor binding events58. In this study, the thermodynamic profiles of compounds (3a-c) were explored through DFT calculations conducted in the gas phase utilizing B3LYP/6–311+g (d,p) level. The computed heat capacities at constant volume (Cv) for the respective compounds were 67.15, 67.73, and 61.46 cal/molK, indicating subtle variations in their ability to store thermal energy. Furthermore, the entropy values were found to be 140.62, 139.16, and 130.66 cal/molK, reflecting the degree of molecular complexity and freedom in each case. The calculated ground-state energies for 3a, 3b, and 3c were determined to be −1022.47, −932.46, and − 917.17 Hartree, respectively. Dipole moment values for 3a, 3b, and 3c were measured as 10.50, 6.32, and 5.01 Debye, respectively. The pronounced dipole moments observed in these molecules reveal a marked increase in their polar nature, stemming from robust internal dipole-dipole interactions59 that significantly influence their structural and chemical behavior.
MEP distribution and ESP arrow
To predict the reactive regions within the studied compounds, MEP maps were constructed. These visual representations serve as powerful analytical tools, illustrating the distribution of electrostatic potential across a molecule’s surface through a nuanced, color-coded format60. This technique provides an accessible yet detailed view of the electronic character of different molecular regions. In MEP diagrams, distinct colors convey varying degrees of electron density. Red areas reveal zones of high electron concentration, corresponding to regions with a significant partial negative charge, typically the most likely sites for electrophilic attack. Yellow regions, though still electron-rich, exhibit a slightly lower electron density than the red zones. Furthermore, blue signifies areas deficient in electron density, marked by a partial positive charge and often serving as likely sites for nucleophilic interactions. Green shades represent regions with neutral electrostatic potential61, suggesting minimal reactivity. MEP diagrams for compounds (3a-c) are illustrated in Fig. 9. MEP map of compound 3a reveals that the O1 atom associated with the isatin moiety, along with the O2 and O3 atoms from the nitro group, are predominantly located within regions of negative potential, represented by red to yellow zones on the MEP surface. These areas signify sites of high electron density, suggesting a strong propensity for electrophilic attack. Furthermore, the hydrogen atoms labeled H1, H8, H9, and H10 are situated in regions of positive potential (blue zones), marking them as likely centers for nucleophilic interaction due to their electron-deficient nature. MEP maps of compounds 3b and 3c reveal strikingly similar charge distribution profiles. In both molecules, the O1 atom emerges as a prominent site for potential electrophilic attack, whereas the H1 atom appears favorably positioned for nucleophilic interaction. The presence of pronounced red zones on the MEP surfaces, indicative of high electron density, demonstrates regions most conducive to hydrogen bonding. These electron-rich sites not only underscore the compounds’ chemical reactivity but also suggest optimal locations for cation sensing, thereby enhancing their potential utility in selective ion detection applications.
Figure 10 presents the electrostatic potential (ESP) maps derived from the Gaussian output files for the synthesized compounds. These maps reveal a pronounced concentration of negative electrostatic potential, represented by red-shaded regions, primarily surrounding electronegative atoms, most notably oxygen. This spatial distribution shows zones of heightened electron density, which make them favorable sites for potential electrophilic interactions. These findings align with the MEP distribution and predict the reactive behavior and chemical environment of these compounds.
HOMO and LUMO orbitals
The analysis of molecular orbitals, HOMO and LUMO, serves as a foundational approach in assessing the chemical behavior and reactivity patterns of molecular systems. A significant energy gap between the HOMO and LUMO levels typically reflects low molecular polarizability and reduced chemical reactivity, while also implying heightened kinetic stability62. Furthermore, the HOMO-LUMO energy gap is widely regarded as a fundamental parameter influencing a molecule’s reactive potential63. Figure 11 presents the molecular orbital distributions for compounds (3a-c). The HOMOs in these molecules demonstrate π-bonding characteristics, with electron density concentrated in regions predisposed to nucleophilic activity or electron donation. Furthermore, the LUMOs display π* anti-bonding traits, illustrating zones of electron deficiency that are primed to accept electrons, which plays a crucial role in electrophilic attacks or redox transformations. The estimated energy gaps for 3a, 3b, and 3c are 3.21, 3.26, and 3.34 eV, respectively. These values of energy gap suggest relatively lower energy requirements for electronic excitation or charge transfer between bonding and anti-bonding orbitals. This improved electronic mobility indicates a higher potential for reactivity, as electron transitions become more easily achievable.
Quantum parameters analysis
HOMO-LUMO theory was applied in order to obtain the chemical reactivity parameters of compounds (3a-c). By utilizing Koopmans approach64, ionization energy (I), electrophilicity index (ω), electronegativity (χ), chemical hardness (η), chemical softness (σ), maximum charge transfer capability index (∆Nmax), electron affinity (A), and chemical potential (µ), can be determined through HOMO and LUMO energy values. These parameters were determined utilizing the following Eqs. (4–11):
The quantum parameter values of the compounds are demonstrated in Table 4. The global electrophilicity index (ω) is a quantum parameter for assessing a molecule’s tendency to attract and accommodate additional electron density. The ω values exceeding 1.5 eV are indicative of potent electrophiles; those in the intermediate range of 0.9 to 1.4 eV reflect moderate reactivity, while values under 0.8 eV denote weak electrophilic nature65. The studied compounds display remarkably high ω values, ranging from 6.71 to 7.53 eV. These elevated values place them well within the domain of highly reactive electrophilic agents. The negative µ values observed for the isatin hydrazone derivatives signify their stability, indicating that these compounds are resistant to spontaneous decomposition into their elemental constituents under ambient conditions. One of the defining factors influencing a molecule’s reactivity and overall chemical behavior is its capacity for charge transfer. This characteristic is effectively captured by the ΔNmax parameter, which serves as a quantitative descriptor of a molecule’s electron-accepting potential. Quantum chemical analyses reveal that compound 3a demonstrates a markedly enhanced tendency to accept electrons compared to 3b and 3c. This pronounced difference is likely attributed to the strategic placement of the NO2 group at the meta position in 3a, a configuration known to exert a strong electron-withdrawing influence.
DOS spectra
Density of states (DOS) spectrum of the compounds (3a-c) was generated from the Gaussian output file using GaussSum software. This spectrum predicts the contributions to the HOMO and LUMO molecular orbitals. Employing DFT method, the DOS profiles were calculated and are illustrated in Fig. 12. In these spectra, the green curve corresponds to the occupied electronic states, denoting HOMO energy levels, while the red curve highlights the unoccupied or virtual states associated with LUMO energies. The evaluated values of EHOMO, ELUMO, and ΔE for 3a (−6.52, −3.31, and 3.21 eV), 3b (−6.31, −3.05, and 3.26 eV), and 3c (−6.44, −3.09, and 3.35) are closely aligned with those obtained from the HOMO and LUMO orbital analysis.
ADME-Tox prediction
In the initial stages of drug development, comprehensive early-stage screening of candidate molecules plays a pivotal role, primarily due to the substantial financial burden and logistical complexity associated with conducting in vivo experiments. To mitigate these challenges, researchers increasingly rely on computational approaches, which offer not only greater efficiency and scalability but also significantly reduced costs66. Among the widely adopted tools for assessing the drug-likeness of potential compounds is Lipinski’s Rule of Five (RO5). This rule suggests that a compound is more likely to be orally active if it meets the following criteria67: a log P value below 5, no more than 10 hydrogen bond acceptors, fewer than 5 hydrogen bond donors, and a molecular weight under 500 Daltons. A compound may still be considered viable if it deviates from only one of these parameters. In this study, in-silico ADME-Tox prediction has been employed to study the structural and pharmacokinetic behaviours of the isatin hydrazone derivatives (3a-c). Detailed results of compounds (3a-c) along with cisplatin (as a standard chemotherapeutic drug) are illustrated in Table 5. Analysis of the data reveals that the predicted logP values for the synthesized compounds are higher than that of cisplatin. This result aligns well with experimental observations, indicating an increased lipophilicity among these compounds compared to the reference drug. Furthermore, the isatin hydrazone derivatives (3a-c) satisfy all criteria outlined in RO5, suggesting favorable characteristics for oral administration and bioavailability. Moreover, these compounds exhibit superior drug-likeness and drug score indices relative to cisplatin, revealing their potential as promising candidates for therapeutic development. Importantly, predictive toxicity assessments show that derivatives (3a-c) are unlikely to elicit adverse effects such as irritation, tumorigenicity, mutagenicity, or reproductive toxicity.
Topological analysis
ELF and LOL
The ELF serves as a powerful analytical tool to interpret the spatial distribution of electron density within a molecule, particularly in distinguishing between localized electrons, such as those in bonding and lone pairs, and delocalized electrons commonly found in metallic or conjugated systems. The ELF is quantitatively expressed on a scale from 0.00 to 1.00, where values exceeding 0.50 are indicative of localized electronic regions68, while values below this threshold denote significant delocalization. Notably, regions surrounding hydrogen and carbon atoms frequently exhibit high ELF values, reflecting a strong tendency toward electron localization, both in bonding interactions and non-bonding electron pairs. Maximum ELF values, typically within the 0.80 to 1.00 Bohr range, correspond to zones of intense electron concentration and are often centered on electronegative elements such as carbon, nitrogen, oxygen, and fluorine. These findings align with expectations based on known chemical behavior and are visually captured in the topographical representation of Fig. 13 (a-f). Complementing the ELF, the LOL offers additional insight into molecular orbital character by mapping areas of localized versus delocalized electronic structure. The LOL contour map spans a range from 0.00 to 0.80 Bohr, with a color gradient where cooler tones (blue) denote weakly delocalized orbitals and warmer tones (red) highlight strongly localized regions. Interestingly, carbon and nitrogen atoms within the studied molecules are depicted in blue, while hydrogen atoms appear in red, suggesting that their electron densities surpass the upper boundary of the LOL mapping scale. This color representation underscores regions of heightened electronic density and reinforces the interpretive data derived from ELF analysis, as shown in Fig. 13 (g-i).
RDG-NCI
To investigate the noncovalent interactions (NCIs) in the studied compounds, Reduced Density Gradient (RDG) isosurface visualizations and scatter plots were analyzed for compounds (3a-c). The RDG isosurfaces, particularly the green regions that signify weak van der Waals interactions, revealed distinct interaction patterns among the compounds. RDG and RDG scatter patterns for all compounds are demonstrated in Fig. 14. In compound 3a, green isosurfaces were prominently observed between the O2 atom and H7, and between O3 and H8, suggesting the presence of weak intramolecular hydrogen bond-like interactions, likely facilitated by the electron-withdrawing effect of the nitro group in the meta position. Compound 3b exhibited green surfaces between hydrogen atoms H9 and H10, as well as between O2 and H6, indicative of intramolecular contacts and possible weak C–H···O interactions involving the ortho-methoxy substituent. In compound 3c, a faint green isosurface was observed between H7 and H8, indicating minimal noncovalent interactions. This observation aligns with the presence of a fluorine atom in the para position, which exerts a relatively weaker electronic and steric influence on intramolecular contacts compared to the other substituents. Across all three compounds, red RDG isosurfaces were consistently observed over the aromatic rings, characteristic of π–π stacking interactions or steric repulsion in those regions. Notably, the RDG scatter plot of compound 3c displayed a lower density of colored points compared to compounds 3a and 3b, suggesting a reduced number or strength of NCIs. This may be attributed to the limited electron-donating or -withdrawing capability of the para-substituted fluorine atom. Despite sharing a common molecular framework, the variations in substituent position and electronic nature markedly influence the distribution and intensity of noncovalent interactions. These findings underscore the sensitivity of NCIs to structural modifications, which could have implications for the reactivity, stability, and potential biological activity of these derivatives.
Molecular docking
Molecular docking is a powerful computational strategy employed to predict how small molecules, such as drugs or bioactive compounds, associate with biological targets like proteins or DNA. This technique provides significant results for the binding pose, binding affinity, and the specific molecular interactions. Furthermore, molecular docking serves as a vital bridge between theoretical modeling and empirical experimentation. In the present study, docking simulations were performed to evaluate the interaction potential of the selected compounds with DNA, BSA, and a specific anticancer target. The results are summarized below:
Anticancer docking
To investigate the potential anticancer mechanism of the synthesized arylidene isatin hydrazones (3a-c), docking studies were carried out with both the ligand-binding domain of Estrogen Receptor alpha (ERα, PDB ID: 3ERT) and the estrogen receptor DNA-binding domain bound to its response element (PDB ID: 1HCQ). This dual approach not only evaluates the compounds’ affinity for ERα but also examines their possible influence on receptor–DNA interactions, providing deeper insight into their mode of action.
All compounds displayed notable affinity for the ERα active site, with binding energies of −6.34, −6.49, and − 6.51 kcal/mol for 3a, 3b, and 3c, respectively. This ranking correlates well with cytotoxicity trends (3c > 3b > 3a). Compound 3a formed hydrogen bonds with TRP393 and GLU353, electrostatic attractions with GLU323, GLU353, and MET357, hydrophobic interactions with ILE326, LEU387, and PRO324, and multiple van der Waals contacts (PHE445, LYS449, ILE386, LEU354, HIS356, LEU327, PRO325, and PRO324). Compound 3b engaged in hydrogen bonding with PRO324 and GLU353, hydrophobic contacts with ILE326, MET357, LEU387, and PRO324, electrostatic attractions with GLU323 and GLU353, and van der Waals interactions involving PHE445, TRP393, LYS449, GLY390, ILE386, LEU354, HIS356, LEU327, and PRO325. Compound 3c exhibited hydrogen bonding with LYS449 and ILE386, hydrophobic interactions with PRO324 and ILE326, electrostatic attractions with GLU353 and LYS449, and van der Waals interactions involving TRP393, PRO325, GLU323, PHE445, GLY390, ILE389, LEU387, MET357, and HIS356. These results confirm that binding is stabilized through a synergistic network of hydrogen bonds, hydrophobic forces, electrostatic attractions, and van der Waals interactions. 3D and 2D interaction diagrams for these binding systems are demonstrated in Figs. 15 and 16, respectively.
To explore whether these ligands could influence ERα–DNA recognition, the compounds were docked to the receptor bound to its estrogen response element. All ligands occupied regions between the protein and DNA, engaging in interactions with both partners, indicative of a groove-binding mode with DNA. Binding energies were − 6.63, −6.67, and − 7.12 kcal/mol for 3a, 3b, and 3c, respectively, again supporting the trend 3c > 3b > 3a. 3D and 2D interaction diagrams for these binding systems are illustrated in Figs. 17 and 18, respectively. Compound 3a formed hydrogen bonds with GLN60 and MET42 residues and the DG29 base pair, hydrophobic contacts with GLN60 and LYS57, an electrostatic attraction with DT30, and van der Waals contacts with TYR41, ALA61, PRO44, CYS59, CYS43, SER58, and GLN60. Compound 3b established hydrogen bonds with the DG11 base pair and SER58, hydrophobic contacts with the DA10 base pair and LYS57, and van der Waals interactions with DT28, DT12, DG29, and LYS57, SER58, GLN60, CYS59, and MET42. Compound 3c displayed hydrophobic interactions with LYS57, halogen bonding with DG29, DA10, and DT28, and van der Waals contacts with DT30, GLN60, SER58, CYS59, ARG55, MET42, and ASN54. These observations suggest that, beyond receptor binding, the compounds, especially 3c, may modulate ERα’s interaction with its DNA target, potentially impacting transcriptional activity.
The complementary docking analyses reveal that the arylidene isatin hydrazones possess strong affinity for both ERα and its DNA-bound form, with 3c consistently showing the most favorable binding energies. The interactions within the ERα–DNA complex imply a capacity to bridge or perturb protein, DNA contacts, offering a plausible mechanism for transcriptional modulation in ERα-positive cancer cells. This integrated approach addresses both ligand-receptor affinity and potential influence on the receptor’s engagement with its genomic response element.
5
DNA interaction
In order to verify the experimental DNA interaction of the derivatives (3a-c), docking simulation was conducted against 1BNA duplex. Figure 19 indicates the binding interaction between isatin hydrazone derivatives and the DNA minor groove. 2D interaction map (Fig. 20) illustrates that compound 3a anchors to DNA via two hydrogen bonds formed with the cytosine base pairs DC9 and DC21. Additionally, it engages in a hydrophobic contact with DC21 and establishes eight van der Waals interactions involving several nucleotide residues, such as DA18, DT19, DT20, DT7, DA6, DA5, DT8, and DG22. Compounds 3b and 3c exhibit similar binding profiles, characterized by a combination of hydrogen bonding, hydrophobic forces, and multiple van der Waals interactions. These molecular interactions collectively contribute to the stabilization of the ligand-DNA complex. The ΔG° associated with the binding events were computed to be −6.51 kcal/mol for 3a, −6.65 kcal/mol for 3b, and − 7.17 kcal/mol for 3c. These values are consistent with the binding affinities determined from UV-Vis DNA titration studies, which confirm the order of interaction strength as 3c > 3b > 3a. This correlation not only validates the thermodynamic measurements but also supports the notion that compound 3c forms the most stable complex with DNA among the studied isatin hydrazone derivatives.
BSA interaction
The in silico BSA binding study of the compounds was performed to investigate the binding poses and the nature of the interaction between the isatin hydrazone derivatives and BSA. Figure 21 illustrates the molecular docking conformations of compounds (3a-c) in their interaction with BSA. The calculated binding energies for these compounds were − 5.75, −5.86, and − 6.44 kcal/mol, respectively, revealing a progressive enhancement in binding affinity from 3a to 3c. Notably, this computational trend mirrors the experimental UV-Vis titration data, further confirming the binding sequence of 3c > 3b > 3a. The 2D interaction profiles of these complexes are presented in Fig. 22. Analysis of these interaction maps indicates that all three compounds engage BSA through a synergistic network of non-covalent interactions, including hydrogen bonding, hydrophobic contacts, and van der Waals forces. Such multifaceted binding behavior underscores the structural compatibility of these compounds with the BSA binding domain and may contribute to their differential affinities.
RMSD and Ramachandran plot
To validate the docking protocol, redocking of the compounds into their respective binding sites was performed, and the Root-Mean Square Deviation (RMSD) values between the two docked poses were calculated. RMSD values were interpreted according to the following classification: good when RMSD ≤ 2.0 Å, acceptable when 2.0 Å < RMSD ≤ 3.0 Å, and bad when RMSD ≥ 3.0 Å69,70,71. All calculated RMSD values for the studied compounds (3a-c) fell below 2.0 Å (Table 6), indicating that the docking protocol reliably reproduced the observed binding orientations. This result confirms the robustness and reproducibility of the docking simulations.
The conformational quality and stability of the protein structures used in the docking studies were evaluated by Ramachandran (RC) plot analysis employing BIOVIA Discovery Studio Visualizer. The RC plot assesses the stereochemical quality of protein models by mapping the φ (phi) and ψ (psi) dihedral angles of amino acid residues, with regions classified as: most favorable (innermost blue contour), additionally allowed (outer pink contour), and disallowed (outside the contour). A higher percentage of residues in the most favorable regions indicates better structural quality and suitability for molecular docking studies72. For ERα (PDB ID: 3ERT), 95.52% of residues were located in the most favorable regions, 3.25% in the additionally allowed regions, and only 1.23% in the disallowed regions. The ERα–DNA complex (PDB ID: 1HCQ) showed 88.89% of residues in the most favorable regions, 7.64% in the additionally allowed regions, and 3.47% in the disallowed regions. For BSA (PDB ID: 4OR0), 96.90% of residues were in the most favorable regions, 2.75% in the additionally allowed regions, and just 0.35% in the disallowed regions. These results confirm that the used protein structures are of high stereochemical quality and structurally stable for docking simulations. The Ramachandran plots for each protein are presented in Fig. 23.
Structure-activity relationship (SAR) analysis
The comparative evaluation of the three synthesized arylidene isatin hydrazone derivatives, 3a, 3b, and 3c, highlights the impact of substituent type and position on their biological activities, including anticancer efficacy, DNA/BSA binding affinity, lipophilicity, and molecular docking performance. Compound 3a bears a meta-nitro (-NO2) group, a strong electron-withdrawing substituent. Despite its potential to increase oxidative stress and DNA damage via ROS generation, 3a showed the lowest biological activity across the tests. The meta position may limit conjugation with the π-system of the aromatic ring, thereby reducing interaction strength with biological targets such as DNA or proteins. Furthermore, 3a had the lowest lipophilicity (log P = 1.294), suggesting weaker membrane permeability, which aligns with its higher IC50 values for cancer cell lines. Compound 3b features an ortho-methoxy (-OCH3) group, which is an electron-donating substituent. This increases electron density on the aromatic ring and can enhance π–π stacking or hydrogen bonding interactions with biomolecules. The ortho position allows for possible intramolecular interactions that may stabilize the bioactive conformation. Compound 3b showed improved cytotoxicity and binding constants compared to 3a. Its higher lipophilicity (log P = 2.297) supports better cell membrane permeability and correlates with its superior anticancer effect. Compound 3c includes a para-fluoro (-F) substituent, which is moderately electron-withdrawing and has a strong influence on pharmacokinetic properties due to its small size and high electronegativity. Fluorine at the para position optimizes electronic distribution and molecular polarity, enhancing cell membrane permeability, binding interactions, and molecular docking affinity. This compound demonstrated the highest biological activity, with the lowest IC50 values across both MCF-7 and MDA-MB-231 cell lines, strongest DNA and BSA binding constants, and favorable docking affinities. The log P of 2.225 confirms its high lipophilicity, facilitating efficient cellular uptake.
Conclusions
Three arylidene isatin hydrazone derivatives (3a-c) were synthesized utilizing SBA-Pr-N-Is-Bu-SO3H nanocatalyst and characterized by spectroscopic techniques and theoretical methods. Anticancer activity against normal HUVEC cells and breast cancer cell lines (MCF-7 and MDA-MB-231) indicated that these compounds reduced cancer cell viability in a concentration- and time-dependent manner, with compound 3c showing the strongest cytotoxic effect. The enhanced activity may be attributed to fluorine’s role in enhancing cellular membrane permeability, thereby promoting intracellular uptake. Furthermore, the antioxidant assay demonstrated that 3c has the highest scavenging activity (80% at high concentration). The interaction of the synthesized compounds with DNA through groove binding was confirmed using UV-Vis titration and docking simulations. The Kapp values for the interaction between the compounds and DNA/BSA followed the order: 3c > 3b > 3a consistent with the observed trends in their in vitro biological activity. Detailed DFT calculations such as geometry optimization, MEP, ESP, HOMO-LUMO analysis, quantum reactivity parameters, and DOS spectra elucidated the compounds’ reactivity, active sites, stability, electron transfer, and electronic behaviors. ADME-Tox profiling, molecular docking, in vitro cytotoxicity assays, and DNA/BSA binding studies collectively affirmed the potential of these arylidene isatin hydrazone derivatives as promising agents for anticancer chemotherapy.
Data availability
Data will be made available on request to corresponding author.
References
Rasgania, J. et al. Synthesis of isatin-tagged thiadiazoles as anti-breast cancer leads: in-vitro and in-silico investigations. J. Mol. Struct. 1294, 136464 (2023).
Wu, V. S., Kanaya, N., Lo, C., Mortimer, J. & Chen, S. From bench to bedside: what do we know about hormone receptor-positive and human epidermal growth factor receptor 2-positive breast cancer? J. Steroid Biochem. Mol. Biol. 153, 45–53 (2015).
Yin, L., Duan, J. J., Bian, X. W. & Yu, S. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22, 61 (2020).
Zhu, Z., Jiang, L. & Ding, X. Advancing breast cancer heterogeneity analysis: insights from genomics, transcriptomics and proteomics at bulk and single-cell levels. Cancers 15, 4164 (2023).
Fayed, E. A., Eldin, R. R. E., Mehany, A. B., Bayoumi, A. H. & Ammar, Y. A. Isatin-Schiff’s base and chalcone hybrids as chemically apoptotic inducers and EGFR inhibitors; design, synthesis, anti-proliferative activities and in silico evaluation. J. Mol. Struct. 1234, 130159 (2021).
Kumar, N., Kaushal, R. & Awasthi, P. Non-covalent binding studies of transition metal complexes with DNA: A review. J. Mol. Struct. 1288, 135751 (2023).
Arshad, N. et al. Investigations on anticancer potentials by DNA binding and cytotoxicity studies for newly synthesized and characterized imidazolidine and thiazolidine-based Isatin derivatives. Molecules 27, 354 (2022).
Chowdhary, S. et al. Isatin-indoloquinoxaline click adducts with a potential to overcome platinum-based drug-resistance in ovarian cancer. Bioorg. Chem. 142, 106953 (2024).
Ali, A. Q. et al. Synthesis of platinum (II) complexes of Isatin thiosemicarbazones derivatives: in vitro anti-cancer and deoxyribose nucleic acid binding activities. Inorg. Chim. Acta. 416, 235–244 (2014).
Khan, H. Y., Ansari, M. F., Tabassum, S. & Arjmand, F. A review on the recent advances of interaction studies of anticancer metal-based drugs with therapeutic targets, DNA and RNAs. Drug Discov. Today. 104055 (2024).
Gresh, N., Ongaro, A., Demange, L., Zagotto, G. & Ribaudo, G. Sequence-selective recognition of the d (GGCGCC) 2 DNA palindrome by oligopeptide derivatives of mitoxantrone. enabling for simultaneous targeting of the two guanine bases upstream from the central intercalation site in both grooves and along both strands. ACS Omega. 9, 42309–42328 (2024).
Alniss, H. Y. et al. Structure-based drug design of DNA minor groove binders and evaluation of their antibacterial and anticancer properties. Eur. J. Med. Chem. 271, 116440 (2024).
Paul, A., Guo, P., Boykin, D. W. & Wilson, W. D. A new generation of minor-groove-binding—heterocyclic diamidines that recognize G· C base pairs in an AT sequence context. Molecules 24, 946 (2019).
Khan, G. S., Shah, A. & Barker, D. Chemistry of DNA minor groove binding agents. J. Photochem. Photobiol., B. 115, 105–118 (2012).
Kakkar, R. Isatin and its derivatives: a survey of recent syntheses, reactions, and applications. MedChemComm 10, 351–368 (2019).
Pokharel, Y. R. et al. Synthesis, characterization, and in vitro anticancer activity of a series of N (4) dimethyl isatin thiosemicarbazones and their copper (II) complexes. J. Mol. Struct. 1327, 141157 (2025).
Teng, Y. O. et al. Synthesis and anti-cancer activity evaluation of 5-(2-carboxyethenyl)-isatin derivatives. Eur. J. Med. Chem. 112, 145–156 (2016).
Munir, I. et al. Design, synthesis, in vitro, and in silico studies of novel isatin-hybrid hydrazones as potential triple-negative breast cancer agents. RSC Adv. 15, 948–965 (2025).
Ahmed, M. F. et al. Novel hydrazone‐isatin derivatives as potential EGFR inhibitors: synthesis and in vitro pharmacological profiling. Arch. Pharm. 356, 2300244 (2023).
Manoharan, A. et al. Assembling a cinnamyl pharmacophore in the C3-position of substituted isatins via microwave-assisted synthesis: development of a new class of monoamine oxidase-B inhibitors for the treatment of Parkinson’s disease. Molecules 28, 6167 (2023).
Al-Salem, H. S., Arifuzzaman, M., Issa, I. S. & Rahman, A. M. Isatin-hydrazones with multiple receptor tyrosine kinases (RTKs) inhibitory activity and in-silico binding mechanism. Appl. Sci. 11, 3746 (2021).
Vine, K., Matesic, L., Locke, J., Ranson, M. & Skropeta, D. Cytotoxic and anticancer activities of isatin and its derivatives: a comprehensive review from 2000–2008. Anti-Cancer Agents Med. Chem. (Formerly Curr. Med. Chemistry-Anti-Cancer Agents). 9, 397–414 (2009).
Al-Salem, H. S. et al. A series of isatin-hydrazones with cytotoxic activity and CDK2 kinase inhibitory activity: a potential type II ATP competitive inhibitor. Molecules 25, 4400 (2020).
Tumosienė, I., Jonuškienė, I., Kantminienė, K., Mickevičius, V. & Petrikaitė, V. Novel N-substituted amino acid hydrazone-isatin derivatives: Synthesis, antioxidant activity, and anticancer activity in 2D and 3D models in vitro. Int. J. Mol. Sci. 22, 7799 (2021).
Kumar, S. et al. Isatin-tethered halogen-containing acylhydrazone derivatives as monoamine oxidase inhibitor with neuroprotective effect. Sci. Rep. 14, 1264 (2024).
Yousef, M. A., Ali, A. M., El-Sayed, W. M., Qayed, W. S. & Farag, H. H. Aboul-Fadl, design and synthesis of novel isatin-based derivatives targeting cell cycle checkpoint pathways as potential anticancer agents. Bioorg. Chem. 105, 104366 (2020).
Chowdhary, S., Shalini, Arora, A. & Kumar, V. A mini review on isatin, an anticancer scaffold with potential activities against neglected tropical diseases (Ntds). Pharmaceuticals 15, 536 (2022).
Başaran, E. P., Köprü, S., Akkoç, S. & Türkmenoğlu, B. Investigation of newly synthesized fluorinated isatin-hydrazones by in vitro antiproliferative activity, molecular docking, adme analysis, and e-pharmacophore modeling. ACS Omega. 9, 26503–26518 (2024).
Ubaid, A. et al. Synthesis and structure–activity relationship (SAR) studies on new 4-aminoquinoline-hydrazones and isatin hybrids as promising antibacterial agents. Molecules 29, 5777 (2024).
Kumar, S., Nair, A. S., Abdelgawad, M. A. & Mathew, B. Exploration of the detailed structure–activity relationships of isatin and their isomers as monoamine oxidase inhibitors. ACS Omega. 7, 16244–16259 (2022).
Mohammadi Ziarani, G., Roshankar, S., Mohajer, F., Badiei, A. & Sillanpää, M. The synthesis of SBA-Pr-N-Is-Bu-SO3H as a new Hg2 + fluorescent sensor. Inorg. Chem. Commun. 146, 110100 (2022).
Panahande, Z., Mohammadi Ziarani, G., Feizi-Dehnayebi, M., Badiei, A. & Abu‐Dief, A. M. Synthesis and DFT calculation of Hg2+ fluorescence chemosensor based on a new hybrid organic–inorganic nanoporous material of SBA‐Pr‐Is‐Hy. Appl. Organomet. Chem. 39, e7818 (2025).
Kedare, S. B. & Singh, R. Genesis and development of DPPH method of antioxidant assay. J. Food Sci. Technol. 48, 412–422 (2011).
Reichmann, M., Rice, S., Thomas, C. & Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 76, 3047–3053 (1954).
Maity, A. et al. Preferential photochemical interaction of Ru (III) doped carbon nano dots with bovine serum albumin over human serum albumin. Int. J. Biol. Macromol. 137, 483–494 (2019).
Benesi, H. A. & Hildebrand, J. A spectrophotometric investigation of the interaction of iodine with aromatic hydrocarbons. J. Am. Chem. Soc. 71, 2703–2707 (1949).
Jagadeesan, S. et al. Water-soluble Co (III) complexes of substituted phenanthrolines with cell selective anticancer activity. Inorg. Chem. 52, 12535–12544 (2013).
M. Frisch, Gaussian 09, revision d. 01, Gaussian, inc. Wallingford CT, 201 (2009).
Lu, T. & Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 33, 580–592 (2012).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Sander, T., Freyss, J., Von Korff, M. & Rufener, C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 55, 460–473 (2015).
Zahirović, A. et al. Substituent effect in salicylaldehyde 2-furoic acid hydrazones: theoretical and experimental insights into DNA/BSA affinity modulation, antimicrobial and antioxidant activity. J. Mol. Struct. 1312, 138628 (2024).
Garrett, M. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Tools, A. 1.5. 6 (ADT)/MGL tools 1.5. 6. (The Scripps Research Institute, 2012) (2016).
Mohammadi Ziarani, G. M. et al. Spectroscopy investigations on chemosensor activities of isatin-hydrazones toward Ag+ and Hg2+ ions. J. Iran. Chem. Soc. 21, 2607–2622 (2024).
Gholamzadeh, P., Mohammadi Ziarani, G. & Badiei, A. Application of SBA-Pr-SO3H in the green synthesis of isatinhydrazone derivatives: Characterization, UV-Vis investigation and computational studies. J. Chil. Chem. Soc. 61, 2935–2939 (2016).
Khan, K. M. et al. Synthesis of bis-Schiff bases of isatins and their antiglycation activity. Bioorg. Med. Chem. 17, 7795–7801 (2009).
Maliyakkal, N. et al. Synthesis, biochemistry, and in silico investigations of isatin-based hydrazone derivatives as monoamine oxidase inhibitors. Appl. Biol. Chem. 67, 63 (2024).
Arif, R. et al. Facile synthesis of chalcone derivatives as antibacterial agents: Synthesis, DNA binding, molecular docking, DFT and antioxidant studies. J. Mol. Struct. 1208, 127905 (2020).
Sathyadevi, P. et al. Organometallic ruthenium (II) complexes: Synthesis, structure and influence of substitution at azomethine carbon towards DNA/BSA binding, radical scavenging and cytotoxicity. Eur. J. Med. Chem. 55, 420–431 (2012).
Barton, J. K., Danishefsky, A. & Goldberg, J. Tris (phenanthroline) ruthenium (II): stereoselectivity in binding to DNA. J. Am. Chem. Soc. 106, 2172–2176 (1984).
Ahamad, M. N. et al. A combined experimental and theoretical approach to investigate the structure, magnetic properties and DNA binding affinity of a homodinuclear Cu (ii) complex. New J. Chem. 43, 7511–7519 (2019).
Xu, Z. H., Chen, F. J., Xi, P. X., Liu, X. H. & Zeng, Z. Z. Synthesis, characterization, and DNA-binding properties of the cobalt (II) and nickel (II) complexes with salicylaldehyde 2-phenylquinoline-4-carboylhydrazone. J. Photochem. Photobiol., A. 196, 77–83 (2008).
Mohammadlou, F. et al. A new zinc (II) complex of 2-benzoimidazoledisulfide ligand: synthesis, X-ray crystallographic structure, investigation of CT-DNA and BSA interaction by spectroscopic techniques and molecular docking. J. Photochem. Photobiol. A. 443, 114830 (2023).
Rothwell, J. A., Day, A. J. & Morgan, M. R. Experimental determination of octanol – water partition coefficients of quercetin and related flavonoids. J. Agric. Food Chem. 53, 4355–4360 (2005).
Yilmaz, V. T. et al. Synthesis, structures and anticancer potentials of platinum (II) saccharinate complexes of tertiary phosphines with phenyl and cyclohexyl groups targeting mitochondria and DNA. Eur. J. Med. Chem. 155, 609–622 (2018).
Muthu, S. & Prabhakaran, A. Vibrational spectroscopic study and NBO analysis on tranexamic acid using DFT method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 129, 184–192 (2014).
Janani, S., Rajagopal, H., Muthu, S., Aayisha, S. & Raja, M. Molecular structure, spectroscopic (FT-IR, FT-Raman, NMR), HOMO-LUMO, chemical reactivity, AIM, ELF, LOL and molecular docking studies on 1-benzyl-4-(N-Boc-amino) piperidine. J. Mol. Struct. 1230, 129657 (2021).
Canakdag, M. et al. Comprehensive evaluation of purine analogues: cytotoxic and antioxidant activities, enzyme inhibition, DFT insights, and molecular docking analysis. J. Mol. Struct. 1323, 140798 (2025).
Kumar, R., Kamal, R., Kumar, V. & Parkash, J. Bifunctionalization of α, β-unsaturated diaryl ketones into α-aryl-β, β-ditosyloxy ketones: single crystal XRD, DFT, FMOs, molecular electrostatic potential, Hirshfeld surface analysis, and 3D-energy frameworks. J. Mol. Struct. 1250, 131754 (2022).
Dehghani, N., Mohammadi Ziarani, G., Feizi-Dehnayebi, M., Mirhosseyni, M. & Badiei, A. Synthesis and theoretical investigation of a new Hg2 + chemosensor based nanomagnetic particle (Fe3O4@ SiO2@ Pr–NCIM). Mater. Res. Bull. 183, 113200 (2025).
Feizi-Dehnayebi, M. et al. Probing the biological activity of Isatin derivatives against human lung cancer A549 cells: Cytotoxicity, CT-DNA/BSA binding, DFT/TD-DFT, topology, ADME-Tox, docking and dynamic simulations. J. Mol. Liq. 428, 127475 (2025).
Shakuntala, R., Keshavayya, J., Satyanarayan, N., Nippu, B. & Thippeswamy, B. Synthesis, spectroscopic, computational, and biological evaluation of pyrazole mono azo dyes. J. Mol. Struct. 1309, 138045 (2024).
Koopmans, T. Über Die Zuordnung Von Wellenfunktionen Und Eigenwerten Zu Den Einzelnen Elektronen Eines Atoms. Physica. 1, 104–113 (1934).
Shinde, R. A., Adole, V. A. & Jagdale, B. S. Antimicrobial and computational investigation of two 2, 3-dihydro‐1 H‐inden‐1‐one derived fluorinated chalcone motifs. Vietnam J. Chem. 59, 800–812 (2021).
Rajalakshmi, R. et al. In Silico studies: physicochemical properties, drug score, toxicity predictions and molecular Docking of organosulphur compounds against diabetes mellitus. J. Mol. Recognit. 34, e2925 (2021).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 (1997).
M.J. Pramila, D.A. Dhas, I.H. Joe, S. Balachandran, G. Vinitha, Structural insights,spectral, flourescence, Z-scan, CH… O/NH… O hydrogen bonding and AIM, RDG, ELF, LOL,FUKUI analysis, NLO activity of N-2 (Methoxy phenyl) acetamide. J. Mol. Struct. 1272, 134140 (2023).
Patil, P. et al. [MerDABCO-SO3H] Cl catalyzed synthesis, antimicrobial and antioxidant evaluation and molecular docking study of pyrazolopyranopyrimidines. J. Mol. Struct. 1242, 130672 (2021).
Nippu, B., Rahman, A., Kumaraswamy, H. & Satyanarayan, N. Design and synthesis of novel tetrazolo quinoline bridged isatin derivatives as potential anticancer leads against MIA PaCa-2 human pancreatic cancer cell line. J. Mol. Struct. 1263, 133103 (2022).
Zond, R. et al. Fe3O4@ SiO2@ CPTMS-PADETA nanocomposite-catalyzed one-pot three-component synthesis of spiroindoloquinazolines: a combined biological and computational profiling. J. Mol. Struct. 143368 (2025).
Nippu, B. et al. Design, synthesis and anticancer screening of Cu-catalyzed SnAr substituted pyridine bridged ring systems. J. Mol. Struct. 1277, 134829 (2023).
Acknowledgements
We are thankful for the support from the Research Council of Alzahra University. Senem Akkoc would like to thank Suleyman Demirel University Research Fund for financial support with project number TSG-2024-9516.
Author information
Authors and Affiliations
Contributions
Mehran Feizi-Dehnayebi: Project administration, Conceptualization, Writing - Original Draft, Writing - Review & Editing. Ghodsi Mohammadi Ziarani: Supervision-lead, Validation. Somayeh Reiisi: Formal analysis. Majid Farsadrooh: Writing - Review & Editing. Zahra Panahande: Data Curation. Hassan Mansouri-Torshizi: Investigation. Senem Akkoc: Resources, Methodology.
Corresponding authors
Ethics declarations
Consent of publication
All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feizi-Dehnayebi, M., Mohammadi Ziarani, G., Reiisi, S. et al. Biological evaluations and biomolecular interactions along with computational insights of arylidene isatin hydrazones synthesized using nanocatalyst. Sci Rep 15, 42705 (2025). https://doi.org/10.1038/s41598-025-26721-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26721-7