Abstract
Enterotoxigenic Escherichia coli (ETEC) of fimbriae types F4 and F18 causes post-weaning diarrhea (PWD) in pigs and susceptibility relates to host genotype. We examined if CHCF1/FUT1 genotypes were associated with ETEC-mediated PWD in seven Danish herds. Diarrhea was assessed in 1,919 pigs by evaluating perianal fecal staining in the first week after weaning. From each herd, twelve diarrheic and twelve non-diarrheic pigs were selected for genotyping, fecal dry matter (FDM) and microbiological testing. ETEC F4 diarrhea occurred only in CHCF1 heterozygous (RS) pigs. Furthermore, CHCF1 RS pigs had higher odds of diarrhea (Odds ratio (OR) 2.09; p = 0.026) and lower mean FDM (13.9%) than homozygous resistant (RR) pigs (18.6%; p = 0.0052). ETEC F4 was more frequently isolated from CHCF1 RS pigs compared to RR pigs (OR 8.88, p = 0.00005). Additionally, hemolytic E. coli shedding was higher in CHCF1 RS pigs (mean 47.4% vs. 24.1%; p = 0.00001). Too few FUT1 RR pigs were found for analyzing association between FUT1 genotypes and ETEC F18-mediated diarrhea. In conclusion, CHCF1 genotype was strongly associated with ETEC F4-mediated PWD under production settings. ETEC F4 resistance in pigs could help to reduce PWD occurrence and antibiotic use.
Similar content being viewed by others
Introduction
Post-weaning diarrhea (PWD) is a widespread disease problem in global pig production and treatment of PWD is a major cause of antibiotic consumption in pigs1. The condition is highly prevalent in the first weeks after weaning2. This is a challenging period where pigs are adapting to a solid diet, removal of sow milk and lactogenic immunity3, a novel environment, and a new social hierarchy4,5.
Enterotoxigenic Escherichia coli (ETEC) expressing F4 or F18 fimbriae is the main pathogen associated with PWD in newly weaned pigs6. Fimbriae enable ETEC to bind to specific receptors7,8 in the gut, facilitating colonization and infection. F4 fimbriae are currently described with three variants of antigens: ab/ac/ad9, whereas F18 fimbriae include two antigenic variants referred to as ab/ac10. Regarding E. coli F18, the F18ac variant is associated with PWD caused by ETEC, whereas the F18ab variant is associated with oedema disease10. Genetic resistance to ETEC F4 and/or F18 infection has been described in pigs11,12. In both cases, resistance is inherited in a Mendelian fashion as a recessive trait11,12.
A locus encoding for resistance to ETEC F4ab/ac was mapped to chromosome 13 by linkage analysis13. Since then, different research groups have progressed in narrowing in on the chromosomal region that contains the variant responsible for resistance14,15. Most recently, the responsible variant was hypothesized to be situated between the HEG1 and MUC13 genes, in an area without annotated genes16. In this region, the DNA-marker CHCF1 was highly reliable in predicting ETEC F4ab/ac adhesion phenotypes on enterocytes16. Our experimental infection trials confirmed that the CHCF1-marker was highly effective in predicting clinical susceptibility to ETEC F4ab/ac challenge17,18. However, as these were experimental infection trials with pigs from a single herd, it would be valuable to investigate if a similar strong link exists under common rearing conditions and across multiple herds.
Susceptibility to E. coli F18 adhesion is determined by a single nucleotide polymorphism (SNP) in the FUT1 gene on the porcine chromosome 619. An experimental infection study confirmed that one FUT1 allele was highly associated with clinical E. coli F18-mediated PWD20.
The main aim of this study was to investigate the association between CHCF1 genotype and ETEC F4-mediated diarrhea in newly weaned pigs reared under standard conditions in Danish commercial pig herds. We also assessed other disease-related outcomes, namely fecal dry matter content (FDM), shedding of hemolytic E. coli (% out of total bacterial growth) and presence of ETEC isolates. A secondary aim was to investigate the frequency of FUT1 genotypes and if possible, the association between FUT1 genotype and ETEC F18-mediated diarrhea in the same herds.
Materials and methods
Ethical statement
The study was an observational study, which was ethically assessed and approved by the Animal Ethics Institutional Board, University of Copenhagen (AEIRB approval number: 2022-11-PNH-018 A). All methods were carried out in accordance with Danish regulations involving animal research and methods are reported according to ARRIVE guidelines. Data was collected from May to December 2022. Investigators were blinded to genotypes at conduct and outcome assessment part of the study. Investigators were not blinded at the time of data analysis.
Study design
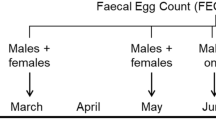
The study was a case-control study including 1,919 newly weaned pigs from seven Danish commercial weaner herds. Herd visits and sampling of pigs took place on the day when the farmers expected the occurrence of a PWD outbreak requiring treatment. The farmer was instructed to postpone the treatment until after the herd visit. In each herd, a weaner section housing newly weaned pigs was selected at random. Pigs were visually inspected for perianal fecal staining (PFS) and 12 pigs with and 12 without staining were selected by systematic random sampling. From each of these 24 pigs, an oral mucosa swab was obtained for genotyping of CHCF1 and FUT1 markers. Additionally, a fecal sample was obtained for determination of dry matter content and for bacterial cultivation targeting hemolytic E. coli, which was tested for the presence of fimbriae and toxin genes. The general study design is illustrated in Figure. 1.
Herd inclusion and description
All herds had a history of diarrhea outbreaks in the immediate post-weaning period. Five herds (herd 1,2,3,5) were identified by veterinarians, and three herds (4,6,7) by information on previous ETEC F4 recovery from PWD obtained from the Veterinary Laboratory, Kjellerup (Danish Agriculture and Food Council). The Veterinary Laboratory gave the owners of ETEC F4 positive herds the opportunity to participate in the study by providing them with our contact information, thus respecting confidential and private information and data. The use of medicinal zinc oxide was an exclusion criterion.
All herds belonged to the Danish SPF-System21 and were production herds producing pigs intended for slaughter. All herds reared crossbred pigs, Danish Landrace x Large White x Duroc (LY-D), with Danbred genetics. Weaning ages ranged from 3 to 5 weeks between herds and herd-specific information, such as weaning age and time of sampling appears in Table 1.
Inclusion of pens and pigs
Approximately 300 pigs per herd were examined for PFS. A sufficient number of pens to obtain this number were selected at random in each herd. Relief and sick pens were excluded from the study.
All the pigs in the included pens were examined individually for PFS. Perianal fecal staining was defined as moist fecal matter visible on the perianal skin. Pigs with perianal staining or observed diarrheic excretion were marked and the total number of both “+/PFS” and “-/PFS” pigs was determined. In each farm, pigs with and without PFS were selected at random among the 300 pigs initially examined to obtain a sample size of 12 pigs of each category.
Clinical examination and sampling procedure
The examination and sampling procedure (Fig. 1) of the 168 pigs (24 per herd) consisted of 4 steps: (1) Recording of individual body weight and sex; (2) Rectal swab-sampling using a sterile viscose swab (Sterile swabs with Amies gel and charcoal, SARSTEDT AG & Co. KG, Nümbrecht, Germany); (3) Collection of fecal samples for dry matter analysis; (4) Oral swab sampling for genotyping using two sterile cotton swabs per pig (Sterile dry swabs in impact resistant PP tubes, Avantor, Radnor, Pennsylvania, USA).
Laboratory analyses
Dry matter analysis
For a standardized and objective result, fecal dry matter analysis was performed for all fecal samples in accordance with Rydal et al.17. If the FDM percentage was ≥ 16.6%, feces classified as normal. If the percentage was < 16.6%, the sample was classified as diarrheic22.
To evaluate the diagnostic performance of PFS as a predictor for diarrhea, pigs that had been selected for fecal sampling were used for the analysis. Out of the total 168 newly weaned pigs selected for sampling, twelve pigs were excluded from the analysis as these pigs did not deliver a fecal sample at sample collection. The remaining 156 pigs were included in the analysis of FDM and diarrhea prevalence.
Cultivating fecal bacteria
Each swab was subjected to dilution streaking on a 5% calf blood agar plate followed by overnight incubation in ambient atmosphere at 37 °C. On each plate, the presence and relative proportion of hemolytic Enterobacterales-like colonies of the total colony count was registered. In addition, when present, one hemolytic colony per plate was collected for species identification by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry (Vitek MS RUO, BioMérieux; France) using Escherichia coli ATCC 8739 as reference strain and the software Saramis TM 3.5 (bioMérieux) for spectra interpretation. Confirmed E. coli isolates were subjected to multiplex PCR analysis for detection of genes encoding F4, F18 and the enterotoxins STa, STb and LT23.
DNA-marker tests
For each pig, a genomic DNA extraction from the oral swabs was performed for genetic determination of ETEC F4- and F18-resistance or susceptibility by using allele-markers CHCF1 and FUT1. For the genomic DNA preparation for PCR analysis, each cotton swab head was placed in Eppendorf tubes with caps filled with 100 µl of 0.9% NaCl. Additionally, 100 µl NaOH (25mM NaOH + 2mM EDTA) was added. To mix these solutions with the medium on the cotton swab, the Eppendorf tubes were vortexed for 5 s. All samples were heated at 100 °C for 20 min and were centrifuged for 30 s to get the mixed medium off the caps. Afterwards, 100 µl of Tris solution (40 mM Tris-HCl) was added to each Eppendorf tube, which was then vortexed again. All Eppendorf tubes were centrifuged for 30 s and 50–100 µl of each sample was transferred to new tubes. One µl from each of the crude DNA solutions was used for TaqMan genotyping of the CHCF116 and FUT119 loci according to the manufacturer’s instructions. Allele calling was performed on a Mx3000P qPCR System (Agilent, Santa Clara, California, USA).
Statistics
Data analysis and visualization were performed using R version 4.4.324. ETEC F4 diarrhea was the primary outcome of interest for comparison between CHCF1 genotypes. Secondary outcomes of interest were: FDM, diarrhea, hemolytic E. coli shedding, and ETEC F4 isolate presence. We could not analyze the association between FUT1 genotype and ETEC F18-mediated diarrhea due to too few FUT1 RR pigs.
The recordings of perianal fecal staining were used for calculation of prevalences of diarrhea within herds and mean diarrhea prevalence with standard deviation (SD) across herds.
The diagnostic performance of PFS as a predictor for diarrhea was assessed by using FDM as gold standard. FDM of < 16.6% of a sample was defined as diarrhea. Sensitivity, specificity, positive predictive value, negative predictive value, and accuracy of diarrhea prediction were calculated, with the formulas:
FDM and hemolytic E. coli shedding were analyzed using linear mixed models (using lme4 and broom.mixed package in R25,26 with CHCF1 genotype as the primary explanatory variable and herd as random effect. The model residuals were evaluated using QQ-plots for normal distribution.
Differences in the occurrence of diarrhea (FDM < 16.6%) between genotypes were analyzed with a generalized linear mixed model, where herd was included in the model as random effect.
A pig with ETEC-associated diarrhea was defined as FDM < 16.6% and simultaneous presence of ETEC at culture and ≥ 50% hemolytic Enterobacterales-like colonies on blood agar. Differences in occurrence of ETEC F4 diarrhea between CHCF1 genotypes, were analyzed using firth logistic regression27. In the model, CHCF1 genotype was included as explanatory variable. Pigs from herd 1 were excluded from the analysis, since ETEC F4 was not isolated in this herd.
The association between ETEC F4 isolation and CHCF1 genotype of the pig was analyzed using generalized linear mixed model, with CHCF1 genotype as explanatory variable and herd as random effect.
Results
Perianal fecal staining and diarrhea prevalence
Diarrhea prevalence varied between herds, with prevalences ranging from 4 to 16%, and the mean prevalence across herds was 10.1% (SD 4.9%) (Fig. 2). Perianal staining effectively predicted diarrhea with high positive predictive value, sensitivity, specificity, and accuracy (Table 2).
Diarrhea prevalence based on perianal fecal staining of pigs from each herd. The red dashed line shows the mean prevalence across herds. The number of pigs assessed for perianal fecal staining are listed for each herd above the bars. Herds were visited within the first week (ranging from 1–7 days) after weaning at the day where the respective herd would normally treat against diarrhea.
Distribution of CHCF1 genotypes, diarrhea determined by fecal dry matter, average shedding of hemolytic E. coli and ETEC F4 diarrhea occurrences
Table 3 summarizes distribution of CHCF1 genotypes, FDM, diarrhea/ETEC F4 diarrhea, shedding of hemolytic E. coli and recovery of ETEC F4 isolates within herds and overall.
FDM was determined in 156 pigs, where 67 (42.9%) pigs were determined to be non-diarrheic and 89 (57.1%) pigs to be diarrheic. The CHCF1 genotypes in the 156 pigs consisted of 84 CHCF1 RR pigs (53.8%) and 72 CHCF1 RS pigs (46.2%). A significantly higher proportion of CHCF1 RS pigs had diarrhea, compared with CHCF1 RR pigs (Table 3).
The amount of hemolytic E. coli shedding and the odds of ETEC F4 isolation were significantly higher for CHCF1 RS pigs (Table 3). All 15 cases of ETEC F4 diarrhea across herds were observed in CHCF1 RS pigs (Table 3). The ETEC virotype isolated from these cases was F4:STb: LT. The majority of cases of ETEC F4-mediated diarrhea (80%) were found in herds 6 and 7, while no cases were observed in herds 1, 2 and 5.
Distribution of FUT1 genotypes and ETEC F18 diarrhea occurrences
Seventy-two pigs (45.0%) were heterozygous susceptible (FUT1 RS), 85 (53.1%) were homozygous susceptible (FUT1 SS), and three (1.9%) were homozygous resistant (FUT1 RR), while oral swabs from eight pigs did not render a result for FUT1 genotype. None of the FUT1 RR pigs were diarrheic and they did not shed E. coli F4 or F18.
ETEC F18 diarrhea was confirmed in eight cases (4 pigs FUT1 SS and 4 pigs FUT1 RS), all originating from herd two.
Virulence profiles of E. coli strains isolated
Shedding of hemolytic E. coli was detected in a total of 105 pigs (out of the 168 pigs investigated). Virotypes of ETEC isolated in the trial are listed in Table 4. The virotype ETEC F4:STb: LT was the most frequently recovered ETEC virotype and it was isolated from 25 pigs from four herds. The second most frequent ETEC virotype was ETEC F18:STb: LT, which was isolated from 15 pigs from two herds. Hemolytic E. coli with F18 fimbriae but no enterotoxin genes were isolated from 33 pigs from three herds.
Discussion
The main aim of the study was to investigate the association between CHCF1 genotype and ETEC F4-mediated diarrhea under standard rearing conditions in Danish herds. We found complete association between CHCF1 genotype and ETEC F4-mediated diarrhea. This supports the conclusions of previous ETEC F4 challenge trials, where we found complete association between CHCF1 genotype and induced ETEC F4ab/ac diarrhea17,18. As for our secondary aim of investigating an association between FUT1 genotypes and ETEC F18-mediated diarrhea, we found a low prevalence of FUT1 RR pigs, which did not allow for a meaningful statistical comparison. Concerning the distribution of FUT1 genotypes, the vast overrepresentation of FUT1 susceptible genotypes, corresponds well with previous findings from another single herd17,18,28 and likely represents the Danish Danbred LY-D population well.
With this study, the significance of susceptibility in relation to ETEC F4-mediated diarrhea extends the observation in experimental challenge studies, where pigs originated from a single herd17,18. This is important for verifying the role of the CHCF1 genotype in Danish Danbred LY-D pigs. It is noteworthy that the CHCF1 marker was highly associated with ETEC F4 diarrhea under field conditions, where several viral and bacterial infections, as well as nutritional factors are involved and are much less controlled than what could be achieved in experimental infection trials. In fact, the associations were so strong that they exceeded what has been described for other DNA-markers for ETEC F4 susceptibility (MUC4 and MUC13) tested under experimental infection trial conditions29,30. In the current study, we did not test for other DNA-marker genotypes previously described to be associated with ETEC F4 susceptibility. One reason for this is that we expected all Danish pigs to be MUC4 homozygous, as there has been selection for MUC4 homozygosity in Denmark since 200331. This was confirmed in previous trials, where all tested pigs were MUC4 homozygous17,18. In the same studies, we also found that the MUC13 marker, previously found to be highly associated with ETEC F4 susceptibility32, was inferior to CHCF1 in predicting clinical ETEC F4ab susceptibility. This finding is aligned with in vitro experiments16, which suggests that CHCF1 is currently the most accurate marker for ETEC F4ab/ac susceptibility in pigs.
Only a single ETEC virotype, ETEC F4:STb:LT, was isolated from diarrhea samples, from which we also detected considerable hemolytic E. coli shedding (≥ 50% out of total bacterial growth). We previously used the same ETEC virotype as challenge strain in our experimental infection trials17,18. Furthermore, it is probably the most common ETEC virotype seen in connection with PWD in Europe33. We did not investigate fimbriae subtypes and therefore could not assess if recovered ETEC F4 isolates had F4ab/ac/ad subtypes. It should be noted that the CHCF1 marker is only related to susceptibility to ETEC F4ab/ac and not ETEC F4ad34. ETEC F4ac is recognized as the most prevalent variant recovered from pigs with PWD in the Western hemisphere31. However, the prevalence of ETEC F4ad might be higher in certain parts of Asia as indicated by a report from central China35.
In evaluating distribution of CHCF1 genotypes, it is important to recognize that the study might have a selection bias for herd inclusion. The first herds we investigated in the study were selected based on anamnesis of PWD, but not specifically ETEC F4-mediated PWD. Since very few positive cases of ETEC F4 were obtained, we decided to select herds based on a recent record of ETEC F4-mediated diarrhea (herd 4,6,7). Due to this selection criterium, we cannot infer that the overall 50–50% distribution of CHCF1 susceptible and resistant genotypes found in this study will represent all Danish herds with Danbred genetics. However, as we wanted to investigate association between CHCF1 genotype and ETEC F4 diarrhea in the field, it was important to find herds where pigs were exposed to ETEC F4 in the environment. Future studies that aim to describe the distribution of CHCF1 genotypes on a population level could be useful for better estimating the pros and cons of selection for ETEC F4ab/ac resistance.
In this study, we defined pigs with ETEC-mediated diarrhea (our primary outcome) based on a FDM < 16.6% with simultaneous isolation of ETEC and ≥ 50% hemolytic Enterobacterales-like colonies on blood agar. The use of FDM to define diarrhea enabled a more objective measure of diarrhea than scoring fecal consistency visually36 or relying on perianal fecal staining. The criterium of ≥ 50% hemolytic Enterobacterales-like colonies is in accordance with routine diagnostic recommendations37. This criterium may increase certainty in ETEC being the actual cause of diarrhea, when recovered from bacterial culture. Based on our current knowledge, we would also expect moderate to massive hemolytic E. coli growth in the presence of ETEC-mediated diarrhea38. However, it is possible that the 50% limit may not represent a valid cut-off in all cases. Finally, as we chose to investigate a single hemolytic E. coli isolate of the dominant colony morphology per pig, we might have overlooked other bacterial strains or species causing infection.
The diagnostic performance of perianal fecal staining as an indicator of diarrhea was high and performance fitted well with previous reports39. The agreement between perianal fecal staining and diarrhea is not perfect, since some diarrheic pigs may defecate without fecal staining. Furthermore, non-diarrheic pigs may get fecal staining from the pen from other pigs in the pen.
From a practical perspective, our results can be used to support genetic selection for ETEC F4 resistance, as the CHCF1 marker was very reliable for predicting ETEC F4 susceptibility under field conditions. Further genetic studies are needed to identify the molecular basis and genetic variants responsible for ETEC F4ab/ac resistance/susceptibility in pigs.
Conclusion
The CHCF1 genotype was strongly associated with ETEC F4-mediated diarrhea under field conditions and across multiple herds. We could not make any inference on the association between FUT1 resistant and susceptible genotypes and ETEC F18-mediated diarrhea as too few FUT1 resistant pigs were available. Our results suggest that the CHCF1-marker has potential use in identifying pigs that are clinically resistant to ETEC F4ab/ac. Therefore, the use of the CHCF1-marker could be a valuable tool in the control of ETEC F4ab/ac diarrhea in pig herds.
Data availability
The datasets analyzed for this study can be found in the zenodo repository, https://doi.org/10.5281/zenodo.15655499.
References
Lekagul, A., Tangcharoensathien, V. & Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 6, 7:100058. https://doi.org/10.1016/j.vas.2019.100058 (2019).
Rhouma, M., Fairbrother, J. M., Beaudry, F. & Letellier, A. Post weaning diarrhea in pigs: risk factors and non-colistin-based control strategies. Acta Vet. Scand. 59, 31. https://doi.org/10.1186/s13028-017-0299-7 (2017).
Deprez, P., Van den Hende, C., Muylle, E. & Oyaert, W. The influence of the administration of sow’s milk on the post-weaning excretion of hemolytic E. coli in the pig. Vet. Res. Commun. 10 (6), 469–478. https://doi.org/10.1007/BF02214010 (1986).
Coutellier, L. et al. Pig’s responses to repeated social regrouping and relocation during the growing-finishing period. Appl. Anim. Behav. Sci. 105, 102–114. https://doi.org/10.1016/J.APPLANIM.2006.05.007 (2007).
Jones, P. H., Roe, J. M. & Miller, B. G. Effects of stressors on immune parameters and on the faecal shedding of enterotoxigenic Escherichia coli in piglets following experimental inoculation. Res. Vet. Sci. 70 (1), 9–17. https://doi.org/10.1053/rvsc.2000.0436 (2001).
Frydendahl, K. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85, 169–182. https://doi.org/10.1016/S0378-1135(01)00504-1 (2002).
Van den Broeck, W., Cox, E., Oudega, B. & Goddeeris, B. M. The F4 fimbrial antigen of Escherichia coli and its receptors. Vet. Microbiol. 71, 223–244. https://doi.org/10.1016/s0378-1135(99)00174-1 (2000).
Coddens, A. et al. The age dependent expression of the F18 + E. Coli receptor on Porcine gut epithelial cells is positively correlated with the presence of histo-blood group antigens. Vet. Microbiol. 122, 332–341. https://doi.org/10.1016/J.VETMIC.2007.02.007 (2007).
Guinée, P. A. & Jansen, W. H. Behavior of Escherichia coli K antigens K88ab, K88ac, and K88ad in immunoelectrophoresis, double diffusion, and hemagglutination. Infect. Immun. 23 (3), 700–705. https://doi.org/10.1128/iai.23.3.700-705.1979 (1979).
Rippinger, P. et al. Designations F18ab and F18ac for the related fimbrial types F107, 2134P and 8813 of Escherichia coli isolated from Porcine postweaning diarrhoea and from oedema disease. Vet. Microbiol. 45 (4), 281–295. https://doi.org/10.1016/0378-1135(94)00141-i (1995).
Gibbons, R. A., Sellwood, R., Burrows, M. & Hunter, P. A. Inheritance of resistance to neonatal E. coli diarrhoea in the pig: examination of the genetic system. Theor. Appl. Genet. 51, 65–70. https://doi.org/10.1007/BF00299479 (1977).
Bertschinger, H. U., Stamm, M. & Vögeli, P. Inheritance of resistance to oedema disease in the pig: experiments with an Escherichia coli strain expressing fimbriae 107. Vet. Microbiol. 35, 79–89. https://doi.org/10.1016/0378-1135(93)90117-p (1993).
Edfors-Lilja, I. et al. The Porcine intestinal receptor for Escherichia coli K88ab, K88ac: regional localization on chromosome 13 and influence of IgG response to the K88 antigen. Anim. Genet. 26 (4), 237–242. https://doi.org/10.1111/j.1365-2052.1995.tb03250.x (1995).
Jørgensen, C. B. et al. Porcine polymorphisms and methods for detecting them. International patent number: WO/2004/048606. (2004).
Rampoldi, A. et al. The receptor locus for Escherichia coli F4ab/F4ac in the pig maps distal to the MUC4-LMLN region. Mamm. Genome. 22 (1–2), 122–129. https://doi.org/10.1007/s00335-010-9305-3 (2011).
Hu, D. et al. Effective genetic markers for identifying the Escherichia coli F4ac receptor status of pigs. Anim. Genet. 50 (2), 136–142. https://doi.org/10.1111/age.12770 (2019).
Rydal, M. P. et al. Pilot study on CHCF1 genotype in a pig challenge model for enterotoxigenic Escherichia coli F4ab/ac associated post-weaning diarrhea.. BMC Vet. Res. 18(1), 382 (2022).
Rydal, M. P., Jørgensen, C. B., Gambino, M., Poulsen, L. L. & Nielsen, J. P. Complete association between CHCF1 genotype and enterotoxigenic Escherichia coli F4ab-associated post-weaning diarrhea in a pig challenge trial. Vet. Microbiol. 282, 109771. https://doi.org/10.1016/j.vetmic.2023.109771 (2023).
Meijerink, E. et al. A DNA polymorphism influencing α(1,2)fucosyltransferase activity of the pig FUT1 enzyme determines susceptibility of small intestinal epithelium to Escherichia coli F18 adhesion. Immunogenetics 52, 129–136. https://doi.org/10.1007/s002510000263 (2000).
Frydendahl, K., Jensen, T. K., Andersen, J. S., Fredholm, M. & Evans, G. Association between the Porcine Escherichia coli F18 receptor genotype and phenotype and susceptibility to colonisation and postweaning diarrhoea caused by E. coli O138:F18. Vet. Microbiol 2 (1), 39–51. https://doi.org/10.1016/s0378-1135(02)00348-6 (2003).
Stege, H. et al. Prevalence of subclinical Salmonella enterica infection in Danish finishing pig herds. Prev. Vet. Med. 28 (44(3-4)), 175–188. https://doi.org/10.1016/s0167-5877(00)00103-3 (2000).
Eriksen, E. Ø., Sejersen, M. F. & Pedersen, K. S. The cotton swab method: an accurate and less invasive way to assess fecal consistency in weaned pigs. BMC Vet. Res. https://doi.org/10.1186/s12917-024-03888-1 (2024).
Zhang, W., Zhao, M., Ruesch, L., Omot, A. & Francis, D. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet. Microbiol. 123 (1–3), 145–152. https://doi.org/10.1016/j.vetmic.2007.02.018 (2007).
R Core Team. R: A Language and Environment for Statistical Computing (R.4.4.3) (The R foundation, 2025).
Bates, D., Maechler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67 (1), 1–48. https://doi.org/10.18637/jss.v067.i01 (2015).
Bolker, B. & Robinson, D. _broom.mixed: tidying methods for mixed models_. R package version 0.2.9.6, (2024). https://CRAN.R-project.org/package=broom.mixed
Heinze, G., Ploner, M., Jiricka, L. & Steiner, G. firth’s bias-reduced logistic regression. R package version 1, 33 (2023).
Rydal, M. P. et al. Post-weaning diarrhea in pigs from a single Danish production herd was not associated with the pre-weaning fecal microbiota composition and diversity. Front. Microbiol. 24, 14:1108197. https://doi.org/10.3389/fmicb.2023.1108197 (2023).
Jensen, G. M. et al. Experimental infection with Escherichia coli O149:F4ac in weaned piglets. Vet. Microbiol. 15(1–3), 243–249 (2006).
Middelkoop, A. et al. Effect of dietary tall oil fatty acids and hydrolysed yeast in SNP2-positive and SNP2-negative piglets challenged with F4 enterotoxigenic Escherichia coli. Sci. Rep. 24 (1), 2060. https://doi.org/10.1038/s41598-024-52586-3 (2024).
García, V. et al. F4- and F18-positive enterotoxigenic Escherichia coli isolates from diarrhea of postweaning pigs: genomic characterization. Appl. Environ. Microbiol. 86(23), e01913 (2020).
Ren, J. et al. Susceptibility towards enterotoxigenic Escherichia coli F4ac diarrhea is governed by the MUC13 gene in pigs. PLoS One. 7 (9), e44573. https://doi.org/10.1371/journal.pone.0044573 (2012).
Luppi, A. et al. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea in Europe. Porcine Health Manag. 1, 2:20. https://doi.org/10.1186/s40813-016-0039-9 (2016).
Rampoldi, A. et al. Inheritance of Porcine receptors for enterotoxigenic Escherichia coli with fimbriae F4ad and their relation to other F4 receptors. Animal 8 (6), 859–866. https://doi.org/10.1017/S1751731114000779 (2014).
Wang, J., Jiang, S. W., Chen, X. H., Liu, Z. L. & Peng, J. Prevalence of fimbrial antigen (K88 variants, K99 and 987P) of enterotoxigenic Escherichia coli from neonatal and post-weaning piglets with diarrhea in central China. Asian-Australasian J. Anim. Sci. 19, 1342–1346. https://doi.org/10.5713/ajas.2006.1342 (2006).
Pedersen, K. S. & Toft, N. Intra- and inter-observer agreement when using a descriptive classification scale for clinical assessment of faecal consistency in growing pigs.. Prev. Vet. Med. 98(4), 288–291 (2011).
Luppi, A. Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porcine Health Manag. 8, 3:16. https://doi.org/10.1186/s40813-017-0063-4 (2017).
Eriksen, E. Ø. et al. An observational field study of Porcine post-weaning diarrhea: clinical and Microbiological findings, and fecal pH-measurements as a potential diagnostic tool. Porcine Health Manag. 11 (1), 33. https://doi.org/10.1186/s40813-023-00325-x (2023).
Eriksen, E. Ø., Nielsen, J. P., Agerlin, M. V., Christensen, A. E. & Pedersen, K. S. Easy and reliable assessment of the prevalence of Porcine post-weaning diarrhoea. Prev. Vet. Med. 220, 106041. https://doi.org/10.1016/j.prevetmed.2023.106041 (2023).
Acknowledgements
The authors would very much like to thank Rasmus Jelle Shyler for helping on sampling days, and to laboratory technician Dan Friis Ryttov for performing the microbiological laboratory analyses.
Funding
This work was supported by the Innovation fund Denmark (IFD) [File number: 7076–00038B] and the Novo Nordisk Foundation Grant number: NNFSA210073688.
Author information
Authors and Affiliations
Contributions
KKL and MPR contributed equally and are shared first authors. JPN and ABA designed the study. ABA wrote the experimental protocol with input from MPR and JPN, while ABA and KKL performed the study and ABA compiled the dataset. MPR and KKL did the statistical analysis of the data and wrote the manuscript draft. MPR finalized the manuscript by editing the manuscript and writing the introduction, discussion and conclusion. CBJ was responsible for the genetic analyses and provided scientific insights into the discussion and interpretation of genotypical results. PD was responsible for the microbiological analyses and provided input in the assessment and interpretation of those results. All authors aided the revision of the manuscript and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rydal, M.P., Lyderik, K.K., Andersen, A.B. et al. Outbreaks of post-weaning diarrhea caused by ETEC F4 are strongly associated with CHCF1 genotype in Danish pigs. Sci Rep 15, 41748 (2025). https://doi.org/10.1038/s41598-025-26829-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26829-w