Abstract
The complete mitochondrial genome of the Peruvian Paso Horse was assembled using PacBio HiFi long reads, resulting in a high-quality circular genome of 16,617 bp comprising 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and a control region. Nucleotide composition and gene structure were consistent with other equine mitogenomes. Codon usage analysis revealed a bias toward CUA (Leu), AUA and AUC (Ile), suggesting translational optimization. Thirty-five heteroplasmic variants were identified, predominantly located in RNA genes (12 S rRNA and tRNA-Phe), with allele frequencies between 0.10 and 0.60 and no substitutions in protein-coding genes, consistent with purifying selection. Repeat analysis detected a single 192 bp tandem repeat and two short tandem repeat (STR) motifs (CATAA and TCT) within the control region, supporting their functional role in mitochondrial replication. Comparative mitogenomic alignment with 14 representative breeds showed high collinearity and structural conservation, with localized variability in the control region (~ 16,000–16,600 bp) and minor divergence at positions ~ 1,500 and ~ 10,000 bp. Phylogenetic analysis positioned the Peruvian Paso Horse within a European breed clade, closely related to Westphalian, Maremmano, and individuals from Germany and Serbia with potential connections to Asian lineages. These results provide valuable insights for equine mitogenomics, conservation, and evolutionary studies.
Similar content being viewed by others
Introduction
Since horses were first brought to America by Spanish settlers, many local breeds have taken root in the New World. These breeds are the result of the mix of different horses that came from Europe. Over 400 years of natural selection, Creole horses in various parts of South America have adapted to their local environments1. Breeders have selectively enhanced traits such as conformation, stamina, strength, and gait in these local populations. The Peruvian Paso Horse (PPH) (Equus caballus) is a breed native to Peru2 recognized as part of country´s cultural heritage3. The PPH is specially known for its distinctive gait, the paso llano4. Previous studies have investigated aspects of the breed´s reproduction5,6 and heritability of performance traits7,8,9. However, research on the Peruvian Paso Horse remain limited.
Genetic characterization and understanding genome structure are essential tools for breed conservation, as well as for informing reproductive strategies and management practices10. Mitochondrial genomes in animals are typically 14–20 kb circular DNA molecules. Due to their higher mutation rate compared to nuclear DNA and a mix of conserved and variable regions, mtDNA is widely used for inferring phylogenetic relationships at various taxonomic levels11. To date, numerous mitochondrial genomes of domestic horses were published12,13. mtDNA is routinely used at both the population and species levels in studies of phylogeography14,15, phylogenetics16,17,18,19,20 and paleogenomics21. However, mitochondrial genome includes specific complex regions, particularly repetitive sequences and segmental duplications, sometimes located within the control region (CR), which have traditionally posed difficulties in resolution. In theory, when repeat sequences exceed the length of sequencing reads, assembly is restricted by the limits of these repetitive elements22,23.
Traditionally, most mitochondrial genome has been assembled using Sanger sequencing or second-generation technologies such as sequencing by synthesis (e.g., Illumina sequencers, San Diego, CA, USA) and semiconductor sequencing (e.g., Ion Torrent systems from Thermo Fisher Scientific). These approaches are limited by their relatively short read lengths, which pose challenges when assembling repetitive or structurally complex regions24. In contrast, third-generation sequencing (TGS) technologies such as PacBio and Oxford Nanopore can generate long reads (> 10–20 kbp), enabling complete coverage of mitochondrial genomes in a single read. These methods have only recently been applied to assemble high-quality mitogenomes in mammals, especially for resolving complex regions25,26.
In this study, we present, the first complete mitochondrial genome of the Peruvian Paso Horse, sequenced using PacBio HiFi long-read technology. This represents the first published mitogenome of this breed obtained through TGS. Our results highlight the potential of long-read sequencing to resolve challenging mitochondrial regions, such as the D-loop and other repetitive elements. This study contributes to a deeper understanding of the maternal lineage of the PPH and provides valuable insights for phylogenetic analyses, breed conservation, and future genomic research.
Results
Genome organization
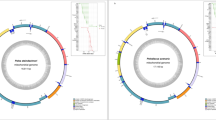
The mitochondrial genome was recovered from whole-genome PacBio HiFi reads using MitoHiFi. The final circularized assembly had an average depth of approximately 310.3×, ensuring a highly accurate reconstruction and reliable variant detection. The assembled mtDNA has a length of 16,617 bp. This genome comprises 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes and one control region (D-loop) (Table 1; Fig. 1). The heavy (H) strand harbored the majority of genes, including 12 protein-coding genes and 14 tRNA, whereas the light (L) strand encoded ND6 and eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro). The elemental composition of this genome was distributed as follows: 24.44% Adenine (A), 25.13% Thymine (T), 25.62% Cytosine (C), and 24.81% Guanine (G). The complete mitochondrial genome sequence has been deposited in the GenBank database under accession number PQ663616. The corresponding BioProject and BioSample identifiers are PRJNA1149496 and SAMN43249583 respectively.
Mitochondrial genome map of the PPH, with a total length of 16,617 bp. Different genomic elements are represented using various colors according to the legend. Genes encoded on the heavy strand are positioned outside the circle, while those on the light strand are placed inside. The innermost gray ring represents the (G + C) content, where darker areas indicate higher (G + C) percentages.
Protein coding genes (PCGs) and codon usage bias (CUB)
Ten of the PCGs (ND1, COX1, COX2, ATP8, ATP6, COX3, ND4, ND4L, ND5, ND6, and CYTB) use ATG as the start codon, while two genes (ND2 and ND3) initiate with ATA. Regarding stop codons, four genes (ND1, ND2, ATP8 and ND3) have the complete TAG stop codon, while COX1, COX2, ATP6, ND4L, ND5 and ND6 terminate with TAA and CYTB with AGA. Additionally, ND4 exhibit an incomplete TA(A) stop codon, whereas COX3 has a truncated T(AA) stop codon. PCGs also exhibit an AT bias, with AT content ranging from 54.6% in CYTB to 64.2% in ATP8. Additionally, the length of the PCGs varies significantly, from 204 bp in ATP8 to 1815 bp in NAD5 (Table 2). The amino acid (AA) codon usages were assessed by calculating relative synonymous codon usage (RSCU) values in 13 PCGs. A total of 3645 codons were encoded by 13 PCGs, and the most frequently used codons were CUA (7.81%), AUC (5.6%), and AUA (5.13%) (Fig. 2A, Supplementary Table S1). Analysis of codon usage bias based on 100 codons per thousand (CDpT) values revealed that the highest frequencies were observed for codons corresponding to isoleucine (Ile), leucine (Leu4), and threonine (Thr), indicating a strong codon preference for these amino acids. In particular, the codon CUA (Leu) exceeded 100 codons per thousand, followed by AUA and AUC (Ile), and UCC (Ser), as shown in Fig. 2B”.
.
rRNA, tRNA, and non-coding intergenic regions
The two rRNA genes (12 S and 16 S) had a combined length of 2,556 bp and were flanked by tRNAPhe and tRNALeu2 (Table 1; Fig. 1). A total of 22 tRNA genes were identified, spanning 1,500 bp, with lengths ranging from 56 bp (tRNAAsn) to 75 bp (tRNALeu2) (Table 1). The heavy (H) strand encoded 14 tRNA genes, while the light (L) strand encoded eight. All tRNA genes exhibited a typical cloverleaf secondary structure, except for tRNASer (Fig. 3). Non-coding intergenic regions included the origin of replication, intergenic spacers, and the control region. The origin of replication was 38 bp long and located between tRNAAsn and tRNACys. Additionally, the control region (D-loop) measured 1152 bp and was flanked by tRNAPro and tRNAPhe (Table 1; Fig. 1).
Heteroplasmy, repetitive sequences and comparative mitogenomic analysis
A total of 35 heteroplasmic positions were detected, most of which were located in mitochondrial RNA genes: 85.7% in 12 S rRNA and 11.4% in tRNA-Phe, with only a single variant in the control region. No heteroplasmy was observed in protein-coding genes (Supplementary table S2). All variants exhibited allele frequencies between 0.10 and 0.60 (AF = 0.171 ± 0.094) with a mean coverage of 50.6×. Tandem repeat analysis identified one major repeat located at positions 16,126–16,317, corresponding to the final ~ 500 bp of the mitogenome and mapping to the control region, as commonly observed in horses and other mammals (Supplementary Table S3). In addition, two short tandem repeats (STRs) were detected: a pentanucleotide motif (CATAA) with three copies, and a trinucleotide motif (TCT) with four copies (Supplementary Table S4).The alignment of the PPH mitochondrial genome with 14 other representative horses breeds revealed a conserved genomic structure, with no major rearrangements among sequences. The PPH showed high collinearity with all breeds; however, notable variability was detected in the control region (~ 16,000–16,616 bp), particularly when compared to Mongolian and Chinese ancient horses. Minor divergence was also observed around positions ~ 1,500 bp and ~ 10,000 bp in some breeds, such as Marwari and Thoroughbred. These differences may reflect breed-specific variants, especially in non-coding intergenic or hypervariable regions (Supplementary Figure. S1).
Phylogenetic analysis
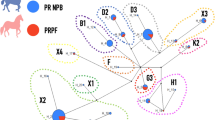
The mitochondrial genome of the PPH clustered within haplogroup B, according to the classification by Achilli et al. (2012). Our phylogenetic analysis, which included 681 mitochondrial genomes, among them the same reference individuals used by Achilli et al. (2012), reproduced the original clustering patterns. This assignment was supported by the Maximum Likelihood phylogenetic tree topology, where the PPH sequence grouped with reference genomes of haplogroup B with high bootstrap support. Additionally, phylogenetic analysis identified 14 major groups (Supplementary Table S6). Group 2 (with > 70% support) shows that the PPH is mostly related to south and central European representatives. Breeds included in the well-supported group of PPH are Maremmano, Westphalian, Holsteiner, Shagya - Arab and two individuals from Germany and Serbia. Moreover, major group 2 also comprises individuals from the Thoroughbred breed originating in Asia and Oceania, along with Kinsky horses and other representatives from Central Europe (Fig. 4). On the other hand, no clear pattern was observed linking horse evolutionary relationships to either geographic origin.
Maximum Likelihood phylogenetic tree of 681 horses inferred from mitochondrial genomic sequences. Bootstrap support values are indicated as black bullets and shown only for branches with > 70% support. The black numbers positioned above certain branches (e.g., 1 to 14) indicate major phylogenetic groups or clades identified in the analysis. Highlighted in the red box is a zoomed-in portion of the tree showing the position of several specific breeds, including the Peruvian Paso Horse (marked with an asterisk), alongside Westphalian, Maremmano, Holsteiner, and Shagya-Arab horses. Images of these breeds are shown on the left for visual reference: Westphalian (Photograph: Association of Westphalian Horse Breeders, https://westfalenpferde.de/en/), Maremmano (Source: Associazione Nazionale Allevatori del Cavallo Maremmano, Italy. http://www.anamcavallomaremmano.com), Peruvian Paso Horse (Source: UNTRM) Holstein (Source: Holsteiner Verband, Germany (https://www.holsteiner-verband.de and Shagya-Arab (Source: Bábolna Nemzeti Ménesbirtok és Tangazdaság Zrt., Hungría, https://babolnamenes.hu/lovak/termekkategoria/5181-gazal-xxii/).
Discussions
The Peruvian Paso Horse is a breed of significant cultural and genetic value, renowned for its distinctive gait and well-defined lineage. The assembly and annotation of the PPH mitochondrial genome using PacBio Long-Read technology yielded a high-quality circular contig of 16,617 bp, whose organization aligns with the classical structure reported in other equine mitogenome studies27. This genetic arrangement, with the majority of genes located on the heavy (H) strand and a smaller number on the light (L) strand, reflects the conserved pattern in Equus caballus mitogenomes and suggests stability in genomic organization throughout evolution28,29.The nearly balanced nucleotide composition—24.44% Adenine, 25.13% Thymine, 25.62% Cytosine, and 24.81% Guanine—indicates a stable profile similar to that observed in other studies of equine mitochondria, which may be associated with a low mutation rate and efficient maintenance of mitochondrial genome integrity30.
The analysis of PCGs and CUB in the mitochondrial genome of the PPH reveals implications about evolution and translational efficiency in equine mitochondria. Codon usage bias is influenced by multiple factors such as mutation pressure, natural selection, GC content, nucleotide skewness, gene expression levels, protein secondary structure, biochemical properties, transcription, and open reading frame length31. However, the main drivers of CUB are natural selection and mutation pressure acting on the background nucleotide composition32. Consequently, certain synonymous codons are favored over others, which in turn can promote adaptive evolution33. The start codon ATG and stop codon TAA were the most abundant in the PPH mitocondrial genome, like in previous studies34,35,36. ND4 exhibits an incomplete stop codon (TA[A]), and COX3 shows a truncated codon (T[AA]). This variability in start and stop codons is consistent with other vertebrates13,37 and equines mitogenomes may be related to post-transcriptional mechanisms that complete incomplete codons during mRNA maturation37,38. These incomplete stop codons are completed through polyadenylation at the 3’ end of the mRNA, thereby converting them into functional stop codons39,40. The analysis of CUB, based on RSCU values, revealed that the most frequent being CUA (7.81%), AUC (5.6%), and AUA (5.13%). In particular, the high frequency of the CUA codon for leucine, along with the elevated utilization of codons for isoleucine (AUC and AUA), suggests that there is an optimization in codon usage that could reflect the relative abundance of the corresponding tRNAs and an adaptation to maximize the efficiency of mitochondrial protein synthesis. This pattern of CUB is fundamental for understanding the evolution of the translational machinery in mitochondria41,42.
The ribosomal RNA genes (12 S and 16 S) in the PPH mitochondrial are flanked by tRNAPhe and tRNALeu2. This organization is consistent with the mitochondrial genomes of other mammals, where rRNA genes are highly conserved and play a crucial role in mitochondrial protein synthesis as integral components of the small (12 S) and large (16 S) ribosomal subunits43. This conserved structure is essential for maintaining translational efficiency in mitochondria, as previously reported in mammalian species such as Rattus norvegicus44. Most tRNAs exhibit a typical cloverleaf secondary structure, except for tRNASer, which lacks the D-arm, a common feature in mammalian mitogenomes45. This variation in secondary structure may influence the efficiency of aminoacylation and translation46.
The non-coding intergenic regions include the origin of replication, intergenic spacers, and the control region (D-loop). Overall, the organization and characteristics of the non-coding intergenic regions in the PPH mitochondrial genome exhibit a high degree of conservation with other mammalian mitogenomes, indicating strong evolutionary constraints to maintain essential functions in gene expression and mitochondrial DNA replication44. This structural conservation highlights the importance of mitochondrial genome stability in equine evolution and suggests potential applications in phylogenetics and breed conservation.
The detection of 35 heteroplasmic positions in the mitochondrial genome of the PPH, predominantly located in mitochondrial RNA genes, diverges from patterns reported in other equid studies. For instance, Xu & Arnason (1994) found extensive heteroplasmy in the control region of Equus caballus, with variable repeat numbers of the motif GTGCACCT45. In Chinese indigenous horses detected heteroplasmy in the coding region for Cyt b and D-loop by PCR-RFLP, but at low frequencies and with primer sampling bias47. In contrast, our data show no heteroplasmy in protein-coding regions, suggesting stronger purifying selection acting on functional domains48. The tandem repeat analysis revealed a single element located within the last ~ 500 bp of the mitochondrial genome, consistent with the so-called “ETAS-like repeats” described in the control region of horses and other mammals. These repetitive sequences have been implicated in mitochondrial replication regulation and exhibit high intra- and interspecific variability, highlighting their evolutionary and functional relevance24,39. In the comparative analysis, a high degree of structural conservation was confirmed between the PPH mitogenome and those of other horse breeds, with no evidence of major rearrangements. However, increased variability was detected, particularly in the control region and around positions 1,500 and 10,000 bp. These differences are consistent with previous mitogenomic studies, in which hypervariable regions of the mitochondrial genome have proven crucial for distinguishing breeds and ancient lineages49,50. The application of long-read sequencing technologies, such as PacBio HiFi, enables accurate detection of heteroplasmic variants and resolution of complex repetitive elements in mitochondrial genomes. These approaches overcome the limitations of short-read data, allowing comprehensive characterization of mitogenomic variability at both the nucleotide and structural levels.
The maximun likelihood phylogenetic analysis identified 14 major groups that provide valuable insights into the maternal lineage of the PPH and its evolutionary relationships with other equine breeds. Of the 681 mitochondrial genomes analyzed in our study, 82 correspond to the reference sequences used by Achilli et al.13. When classified under our grouping system, these sequences were distributed across Groups 1 to 12, maintaining clustering patterns consistent with those reported by Achilli. Groups 5 and 8 to 12 correspond exclusively to haplogroups E and G, respectively, while Groups 1 and 2 encompass the full range of major lineages defined in their study (A–D, J–R), reflecting broader mitochondrial diversity. This correspondence supports the validity of our grouping approach and demonstrates its ability to capture both the diversity and phylogenetic structure previously reported, while also revealing finer resolution within certain haplogroups. Traditionally, it has been assumed, based on historical documents, that the Peruvian Paso Horse descends from horses introduced to Peru by the Spanish in the 16th century, specifically from Andalusian and Barb breeds51. The breed likely envolved under unique ecological conditions and management practices in Peru, giving rise to the distinct modern phenotype52. In our phylogenomic tree, the PPH was placed within major group 2, clustering closely with individuals from Southern European breed Maremmano and five Central European breeds: Westphalian, German, Serbian, Holsteiner, and Shagya-Arab. These breeds are generally characterized by their use in sport and utility purposes and share history of selective breeding in Central Europe. For example, the Maremmano is an Italian breed traditionally used for cavalry and agricultural work53, while the Westphalian and Holsteiner horses are known for their performance in dressage and show jumping54. These findings suggest a shared maternal ancestry and support previous hypotheses regarding the complex origin of the PPH. They also highlight its genetic affinity with European lineages, particulary of Mediterranean origin, as reported for Italian breeds53. Additionally, group 2 also included individuals from Asia and other Central Europe representatives. This pattern is consistent with previous phylogenetic studies that identified genetic signals from ancient Asian horse lineages in modern breeds17,18. The presence of such Asian mitochondrial haplotypes may be attributed to historical horse trade routes or introgression events that occurred before or during early colonial expansion19. Interestingly, the accessions reported for the Andalusian breed, (JN398430 and JN398443.1) showed closer genetic relationships to individuals from Asian (Chinese and Mongolian), Central European (Polish, Maremmano, and Hungarian Coldblood), Middle Eastern (Iranian), and Northern Europe breeds (Exmoor Pony) rather than to the PPH. This suggests a more complex pattern of ancestry and gene flow than previously assumed. The presence of European equine mitochondrial haplotypes in South American breeds has been reported in previous studies14,15, further supporting this genetic connection with PPH. Similar clustering patterns have been observed using both D-loop sequences and complete mitochondrial genomes14,19,46 reinforcing the phylogenetic structure described here. Finally, no clear pattern was observed linking mitochondrial lineage distribution with current breed classification or geographic origin. The application of genomic tools has profoundly reshaped our understanding of domestication, ancestry, and the diversification of domesticated species, spanning both crops and animals. High-throughput sequencing technologies and advanced phylogenomic methods have clarified the complex origins of staple crops54,55,56. Our mitogenomic analysis of the PPH contributes to a broader understanding of breed origin, lineage diversification, and conservation priorities. The integration of long-read sequencing and mitochondrial phylogenetics provides a high-resolution view into the maternal ancestry of this culturally iconic breed, echoing the advances achieved in crop evolution studies. The complete mitochondrial genome generated here represents a valuable genomic resource for future research in equine genetics, the conservation of native breeds, and mitochondrial evolutionary studies.
Materials and methods
Sample collected and genomic DNA sequencing
A representative individual of the PPH breed named “Amunet” (Register Number YN-19315 in the National Association of Peruvian Paso Horse Breeders, born in 2013) residing at the Experimental Station (Luya, Amazonas, Peru) of UNTRM, was selected for sampling. Blood was collected using a vacutainer containing EDTA as an anticoagulant and was immediately transport to the laboratory. The experimental procedure was approved by the Institutional Research Ethics Committee of UNTRM in concordance with Peruvian National Law No. 30,407: “Animal Protection and Welfare”. All procedures were conducted in compliance with relevant institutional guidelines and regulations. Furthermore, the experimental methods adhered to the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. High molecular weight DNA extraction, PacBio HiFi Library and SMRTcell PacBio sequencing was prepared in Biotechnology Center of the University of Wisconsin-Madison, USA. Briefly, Nanobind CBB DNA Kit (PacBio) was used to extract high molecular weight DNA. The quality of the extracted DNA was measured on a NanoDrop™ One instrument (ThermoFisher Scientific). Quantification of the extracted DNA was measured using the Qubit™ dsDNA High Sensitivity kit (ThermoFisher Scientific). The HiFi library was prepared according to PN 102-166-600 Version 04 (Pacific Biosciences), and its quality was assessed using the Agilent FemtoPulse System. The library was quantified using the Qubit™ dsDNA High Sensitivity kit. Sequencing was performed on a PacBio Revio system.
Assembly and annotation
A total of 14,549,851 PacBio Circular Consensus Sequencing (CCS) reads were generated from whole-genome PacBio HiFi sequencing, yielding 239.2 Gb of data. The mean HiFi read length was 16,318 bp and an N50 of 15,533 bp. The average read quality was Q35, with 93.6% of bases scoring ≥ Q30. The mitochondrial genome was assembled, circularized, and annotated using MitoHiFi v3.2.357, which extracted mitochondrial-specific reads from the whole-genome dataset. To distinguish mitochondrial DNA (mtDNA) sequences from nuclear mitochondrial DNA segments (NUMTs), MitoHiFi applies a combination of criteria including contig circularity, read depth, and alignment consistency. Only the contig showing typical mitochondrial features is retained as the final mitochondrial genome assembly. Gene and RNA annotations of the assembled mitochondrial genome were generated automatically by MitoHiFi v3.2.3, which integrates external tools such as MITOS2 and tRNAscan-SE for the identification of protein-coding genes, rRNAs, and tRNAs, using Equus caballus reference genome (NCBI: NC_001640.1)27 as a reference. A graphical representation of the mitochondrial genome was generated using the OGDRAW web server (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html, accessed 9 December 2024).
Codon usage and tRNA analysis
A CUB analysis was done using python script from https://github.com/rhondene/Codon-Usage-in-python. To predict the secondary structure of each tRNA, we used the tRNAscan-SE website server (http://lowelab.ucsc.edu/tRNAscan-SE/).
Heteroplasmy, repetitive sequences, and comparative mitogenomic analysis
To assess the presence of heteroplasmy in the assembled mitochondrial genome, we employed the MitoRSaw software v0.2.1(https://github.com/PacificBiosciences/mitorsaw) using the following parameters: a minimum of three supporting reads per variant (--min-read-count 3), a minimum alternative allele frequency of 10% (--min-maf 0.1), and a minimum mapping fraction of 0.9. Repeat sequences within the mitochondrial genome were identified using Tandem Repeats Finder v4.07b58 applying the following scoring parameters: match = + 2, mismatch = − 7, and indel = − 7, match probability = 80, indel probability = 10, minimum alignment score = 50, and maximum period size = 500. Only repeat elements with a score greater than 50 were considered for downstream analysis. Additionally, short tandem repeats (STRs), also known as simple sequence repeats (SSRs), were detected using MISA v2.159, with the repeat thresholds set to: mononucleotide ≥ 10, dinucleotide ≥ 5, trinucleotide ≥ 4, tetranucleotide ≥ 3, pentanucleotide ≥ 3, and hexanucleotide ≥ 3. To explore structural variation across horse mitochondrial genomes, the Peruvian Paso Horse (PPH) mitogenome was aligned with 14 mitochondrial genomes from other representative horse breeds retrieved from GenBank (including Akhal-Teke, Arabian, Barb, Holsteiner, Shagya Arab, Welsh Pony, Thoroughbred, Feral Horse, Mongolian, Marwari, Caribbean Colonial Horse, Chinese Ancient Horse, and two unspecified breeds). Whole-genome alignment was performed using progressiveMauve v2.4.0, which identifies conserved Locally Collinear Blocks (LCBs) and detects potential rearrangements or divergent regions. The resulting alignment was visually inspected and exported for comparative analysis of genome structure and variability.
Phylogenetic analysis
Phylogenetic analyses were conducted to determine the evolutionary position and haplogroup affiliation of the Peruvian Paso Horse (PPH) mitochondrial genome. All available mitochondrial genomes (n = 681) assigned to taxid 9796 (Equus caballus), with lengths ranging from 16,000 to 18,000 bp, retrieved from the NCBI database (Supplementary Table S5). This range was selected to exclude incomplete or abnormally long sequences, which are often the result of low sequencing quality or annotation errors. Given that the typical length of the horse mitochondrial genome is approximately 16,660 bp60, this threshold ensured that only complete and biologically relevant mitogenomes were retained. As an outgroup, we included E. zebra, a species of the genus Equus. Full mitochondrial genomes including both coding and non-coding intergenic regions were aligned using MAFFT v7.205b61. Gaps in the alignment, including those within repetitive regions, were handled using MAFFT’s standard gap-penalty settings. As the dataset consisted of complete mitogenomes of similar lengths, the alignment showed minimal ambiguous or gap-rich regions. Therefore, no additional trimming was applied. Instead, the alignment was manually reviewed, and regions with clear misalignments, particularly in repetitive elements, were examined to minimize the introduction of phylogenetic noise62. A maximum likelihood (ML) tree was inferred under the GTR + GAMMA model of nucleotide substitution. The best-scoring ML tree was selected, and node support was assessed using 1,000 nonparametric bootstrap replicates implemented in RAxML v8.2.1163. The resulting topology was visualized in FigTree v1.4.5, and compared to the major haplogroups (A–F) defined by Achilli et al. (2012).
Data availability
The complete mitochondrial genome sequence has been deposited in the GenBank database under accession number PQ663616. The corresponding BioProject and BioSample identifiers are PRJNA1149496 and SAMN43249583 respectivelyOther data from the study are available from the corresponding authors.
References
Kelly, L. et al. Genetic characterisation of the Uruguaayan Creole horse and analysis of relationships among horse breeds. Res. Vet. Sci. 72 (1), 69–73 (2002).
Gonzales, R., Li, R., Kemper, G., Del Carpio, C. & Ruiz, E. An algorithm for estimating the variation of the joint angles of the limbs of Peruvian Paso horse. Proc. IEEE 25th Int. Conf. Electron. Electr. Eng. Comput. (INTERCON) (2018). https://doi.org/10.1109/INTERCON.2018.8526449
Ministerio de Comercio Exterior y Turismo del Perú (MINCETUR). Resolución Ministerial N.º 381-MINCETUR/DM: Declaratoria del Caballo Peruano de Paso Como Producto Bandera (2012). https://www.gob.pe/institucion/mincetur/normas-legales/27285-381-2012-mincetur-dm
Del Carpio Ramos, P. A. & Del Carpio Hernández, S. R. B. El Caballo Peruano: Mitos y Realidades. Universidad Nacional Pedro Ruiz Gallo (2019). https://isbn.bnp.gob.pe/catalogo.php?mode=detalle&nt=107117 (physical copy owned).
Palacios, P. et al. L-carnitine enhances the kinematics and protects the sperm membranes of chilled and frozen-thawed Peruvian Paso horse spermatozoa. Cryobiology 115, 104884 (2024).
Tamay, E. et al. Effect of melatonin and caffeine supplementation to freezing medium on cryosurvival of Peruvian Paso horse sperm using a two-step accelerating cooling rate. Biopreserv Biobank. 21, 561–568 (2023).
Vilela, J. L. L. et al. PSXII-3 Estimation of heritability and correlation of functional traits in the Peruvian Paso horse (preliminary studies). J. Anim. Sci. 100 (Suppl. 3), 211–212 (2022).
Larrea Izurieta, C. O. L. et al. Evaluation of inbreeding and genetic variability of the Peruvian Paso horse registered in Ecuador. Rev. Investig Vet. Perú. 33 (5), e21672. https://doi.org/10.15381/rivep.v33i5.21672 (2022).
Vilela, J. L. et al. Preliminary analysis of the development of the breeding program of Peruvian Paso Horse under field conditions. Preprint (2024). https://www.preprints.org/manuscript/202403.0549.
Cothran, E. G., Canelon, J. L., Luis, C., Conant, E. & Juras, R. Genetic analysis of the Venezuelan criollo horse. Genet. Mol. Res. 10 (4), 2394–2403 (2011).
Allio, R., Donega, S., Galtier, N. & Nabholz, B. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 34, 2762–2772 (2017).
Librado, P. et al. Ancient genomic changes associated with domestication of the horse. Science 356, 442–445 (2017).
Achilli, A. et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA. 109, 2449–2454 (2012).
Delsol, N. et al. Analysis of the earliest complete MtDNA genome of a Caribbean colonial horse (Equus caballus) from 16th-century Haiti. PLoS One. 17 (7), e0270600 (2022).
Ahlawat, S. et al. Unraveling the maternal heritage: identifying the complex origins of Indigenous Indian horse and pony breeds through mitochondrial genome analysis. Mamm. Genome. 35, 1–11 (2024).
Khrabrova, L. A., Blohina, N. V., Chysyma, R. B., Bazaron, B. Z. & Khamiruev, T. N. Assessment of MtDNA variability and phylogenetic relationships of Siberian local horse breeds. IOP Conf. Ser. Earth Environ. Sci. 839 (5), 052009 (2021).
Engel, L. et al. Exploring the origin and relatedness of maternal lineages through analysis of mitochondrial DNA in the Holstein horse. Front. Genet. 12, 632500 (2021).
Machmoum, M. et al. Genetic diversity and maternal phylogenetic relationships among populations and strains of Arabian show horses. Animals 13 (12), 2021 (2023).
Khrabrova, L. A., Nikolaeva, A. A., Blohina, N. V. & Sorokin, S. I. Genetic diversity of mitochondrial DNA haplogroups in the don horse breed. Sib J. Life Sci. Agric. 15 (4), 278–290 (2023).
Nikbakhsh, M., Varkoohi, S. & Seyedabadi, H. R. Mitochondrial DNA D-loop hyper‐variable region 1 variability in Kurdish horse breed. Veterinary Med. Sci. 9 (2), 721–728 (2023).
Weingarten, M. et al. Mitochondrial genomes of middle pleistocene horses from the open-air site complex of Schöningen. Nat. Ecol. Evol. 9, 1485–1493. https://doi.org/10.1038/s41559-025-02859-5 (2025).
Poullet, M. Epigenomic study on the domestication of the horse using ancient DNA. In Doctoral Dissertation, Université Paul Sabatier–Toulouse III (2022).
Zhu, S. et al. Ancient genomes reveal a rare maternal lineage of domestic horse in China. J. Archaeol. Sci. Rep. 57, 104592 (2024).
Formenti, G. et al. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 22 (1), 120 (2021).
Zhang, T. et al. Complex genome assembly based on long-read sequencing. Brief. Bioinform. 23 (5), bbac305 (2022).
Warburton, P. E. & Sebra, R. P. Long-read DNA sequencing: recent advances and remaining challenges. Annu. Rev. Genom Hum. Genet. 24 (1), 109–132 (2023).
Kalbfleisch, T. S. et al. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun. Biol. 1, 10 (2010).
Tešija, T. & Safner, T. Analyses of wild ungulates mitogenome. Agric. Conspec Sci. 86 (1), 1–12 (2021).
Jain, K. et al. The evolution of contemporary livestock species: insights from mitochondrial genome. Gene 148728 (2024).
Librado, P. & Orlando, L. Genomics and the evolutionary history of equids. Annu. Rev. Anim. Biosci. 9 (1), 81–101 (2021).
Behura, S. K. & Severson, D. W. Comparative analysis of codon usage bias and codon context patterns between dipteran and hymenopteran sequenced genomes. PLoS One 7(8), e43111 (2012).
Kamatani, T. & Shirota, T. Analysis of factors affecting codon usage bias in human papillomavirus. J. Bioinform Seq. Anal. 9 (1), 1–9 (2018).
Zhao, Y. et al. Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution. BMC Genom. 17, 1–10 (2016).
Lamelas, L. et al. Complete mitochondrial genome of three species of the genus microtus (Arvicolinae, Rodentia). Animals 10 (11), 2130 (2020).
Arbizu, C. I. et al. The complete mitochondrial genome of a neglected breed, the Peruvian Creole cattle (Bos taurus), and its phylogenetic analysis. Data 7 (6), 76 (2022).
Das, P. J. et al. Characterization of the complete mitochondrial genome and identification of signature sequence of Indian wild pig. Gene 897, 148070 (2024).
Jia, X. et al. Characterization and phylogenetic evolution of mitochondrial genome in Tibetan chicken. Anim. Biotechnol. 33 (6), 1371–1377 (2022).
Cieslak, M. et al. Origin and history of the domestic horse: the evidence of mitochondrial DNA. J. Hered. 101 (5), 449–458. https://doi.org/10.1093/jhered/esq005 (2010).
Ojala, D., Montoya, J. & Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474 (1981).
Bratic, A. et al. Mitochondrial polyadenylation is a one-step process required for mRNA integrity and tRNA maturation. PLoS Genet. 12 (5), e1006028 (2016).
Sharp, P. M. & Li, W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 24, 28–38 (1986).
Plotkin, J. B. & Kudla, G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12 (1), 32–42 (2011).
Liu, X. & Shan, G. Mitochondria encoded non-coding RNAs in cell physiology. Front. Cell. Dev. Biol. 9, 713729 (2021).
Abhyankar, A., Park, H. B., Tonolo, G. & Luthman, H. Comparative sequence analysis of the non-protein-coding mitochondrial DNA of inbred rat strains. PLoS One 4(12), e8148 (2009).
Yoon, S. H. et al. Complete mitochondrial genome sequences of Korean native horse from Jeju Island: Uncovering the spatio-temporal dynamics. Mol. Biol. Rep. 44, 233–242 (2017).
Taanman, J. W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta Bioenerg. 1410 (2), 103–112 (1999).
Zhao, Q. et al. Primer effect in the detection of MtDNA heteroplasmy: insights from horse cytochrome b gene. Mitochondrial DNA. 26, 178–181 (2015).
Li, M. et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 87, 237–249. https://doi.org/10.1016/j.ajhg.2010.07.014 (2010).
Lippold, S. et al. Discovery of lost diversity of paternal horse lineages using ancient DNA. Nat. Commun. 2, 450. https://doi.org/10.1038/ncomms1447 (2011).
Moridi, M. et al. Mitochondrial DNA diversity of Persian horses belonging to different horse breeds in Iran. J. Anim. Breed. Genet. 130, 113–120. https://doi.org/10.1111/jbg.12003 (2013).
Ministerio de Desarrollo Agrario y Riego del Perú. Caballos de Paso. https://www.midagri.gob.pe/portal/40-sector-agrario/situacion-de-las-actividades-de-crianza-y-producci/305-caballos-de-paso (s.f.).
Xu, X. & Árnason, Ú. The complete mitochondrial DNA sequence of the horse, Equus caballus: extensive heteroplasmy of the control region. Gene 148, 357–362 (1994).
Giontella, A., Cardinali, I., Sarti, F. M., Silvestrelli, M. & Lancioni, H. Y-chromosome haplotype report among eight Italian horse breeds. Genes 14 (8), 1602 (2023).
Welker, V., Stock, K. F., Schöpke, K. & Swalve, H. H. Genetic parameters of new comprehensive performance traits for dressage and show jumping competitions performance of German riding horses. Livest. Sci. 212, 93–98 (2018).
Spooner, D. M., McLean, K., Ramsay, G., Waugh, R. & Bryan, G. J. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl. Acad. Sci. USA 102(41), 14694–14699 (2005). https://doi.org/10.1073/pnas.0507400102
Iorizzo, M. et al. A high-quality Carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48 (6), 657–666. https://doi.org/10.1038/ng.3565 (2016).
Uliano-Silva, M. et al. MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinform. 24 (1), 288 (2023).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 (2), 573–580 (1999).
Thiel, T., Michalek, W., Varshney, R. & Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L). Theor. Appl. Genet. 106, 411–422 (2003).
Xiufeng, X. & Árnason, Ú. The complete mitochondrial DNA sequence of the horse, equus caballus: extensive heteroplasmy of the control region. Gene 148 (2), 357–362 (1994).
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. https://doi.org/10.1080/10635150701472164 (2007).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780 (2013).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313 (2014).
Acknowledgements
We thank Elías Carranza and Maribel Vásquez for their valuable assistance in the care of the mare Amunet and for their support during the blood sample collection. We also thank Ángel Pilco and Mario Ernesto for their help with data submission. Our gratitude extends to the National Association of Peruvian Paso Horse Breeders (ANCPCPP) for their continued support and collaboration. In addition, we are grateful to Stefany Lobato and Adriana Díaz for their assistance in editing the figures included in this manuscript, and to Maili Muñoz for her support with the logistical activities of the project. Also, the authors acknowledge the High-Performance Computational Cluster of the National University Toribio Rodríguez de Mendoza of Amazonas (UNTRM–DATA SCIENCE) and the Bioinformatics High-Performance Computing Server of the Universidad Nacional Agraria La Molina for providing computational resources for data analysis. The authors thank the University of Wisconsin – Madison Biotechnology Center’s DNA Sequencing Facility (Research Resource Identifier – RRID: SCR_017759) for extracting HMW DNA and generating PacBio libraries.
Funding
We thank to the proyect “Creación del servicio de investigación y enseñanza en Equinos en la Universidad Nacional Toribio Rodríguez de Mendoza de Amazonas” (Grant ID CUI N° 2314510) of the Peruvian Goverment and “Tesis de Pregrado y Postgrado en Ciencia, Tecnologia e Innovacion Tecnologica” (Grant ID PE501085176-2023) Programa Nacional de Investigacion Cientifica y Estudios Avanzados, PROCIENCIA.
Author information
Authors and Affiliations
Contributions
C.L.S.: Designed the study, sample collection, sample processing, analyzed the data, writing-original draft, writing-review & editing, S.J.: Analyzed the data, writing-original draft, writing-review & editing, L.M.: Sample collection, sample processing, writing-original draft, writing-review & editing, H.V.V: Writing-original draft, writing-review & editing, J.L.M: Writing-original draft, writing-review & editing, C.I.A.: Designed the study, sample collection, sample processing, analyzed the data, writing-original draft, writing-review & editing, W.B: Designed the study, sample collection, writing-original draft, writing-review & editing . All authors discussed the methodologies, results, and read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Saldaña, C.L., Justo, S., Murga, L. et al. Mitochondrial genome assembly of the Peruvian Paso horse through PacBio long-read sequencing. Sci Rep 16, 85 (2026). https://doi.org/10.1038/s41598-025-29107-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-29107-x