Abstract
Maternal stress is a growing societal concern, with implications for both maternal wellbeing and infant development. One of the mechanisms by which maternal stress is thought to impact infant development is by shaping the development of the infant gut microbiome. Here, we examined how measures of maternal stress and social support were associated with alpha diversity, beta diversity, and relative abundance of individual bacterial taxa in the gut microbiota at 12 months of age in a community-based sample of infants and their biological mothers (n = 34) from New York. Maternal social support was negatively associated with alpha diversity of the infant gut microbiota and was associated with abundance of bacteria from several genera. We did not find associations between caregiver perceived stress and markers of infant gut microbiota diversity or composition. Results suggest that greater social support for new parents may be associated with infant health via changes in the diversity and composition of the infant gut microbiota.
Similar content being viewed by others
Introduction
The trillions of microorganisms living in the human gastrointestinal tract, collectively known as the gut microbiome, play an important role in human health throughout the lifespan. This system is especially sensitive to environmental influences in the first three to four years of life, which are thought to be a critical period in the development of connections between the gut microbiome and the brain, known as the microbiome-gut-brain axis1. One such environmental influence that has gained increasing attention in recent years is maternal mental health, particularly maternal stress2,3,4 and, to a lesser extent, maternal social support5. A robust body of animal research, and an emerging body of research in humans, has found that maternal stress can be transmitted to infants and shape infant development via alterations to the infant gut microbiome2. However, the specific features of the gut microbiome impacted by maternal stress and the role of maternal social support in shaping infant gut microbiome development are as yet unclear. To address these gaps, in this study we examine prospective associations between maternal stress and social support and the diversity and composition of the infant gut microbiota.
Maternal stress and the infant gut microbiome
Across both human and animal studies, maternal stress has been associated with differences in offspring microbiome. Most evidence in this area comes from studies of prenatal maternal stress and the infant gut microbiome2,6,7,8. Studies of postnatal maternal stress and the infant gut microbiome are fewer but provide some preliminary evidence of associations. Specifically, higher maternal stress has been associated with reduced infant gut microbiome diversity in human studies4,5. Likewise, postnatal maternal stress has been negatively associated with abundances of several species of Lactobacillus and Bifidobacterium in the infant gut in some studies3,4, but others have found no such associations5. These taxa represent some of the earliest colonizers of the infant gut microbiome and play an important role in the digestion of breastmilk and immune system development, among other key functions for infant health9. Taken together, available research in this area suggests that maternal perceived stress may be associated with differences in the infant gut microbiome, but mixed findings between studies suggest the need for more comprehensive studies of caregiver stress and the broader social environment to better understand these findings in context.
Maternal social support and the gut microbiome
Available evidence suggests that maternal stress is associated with the development of the infant gut microbiome, however the extent to which maternal social support influences the developing gut microbiome is as yet unclear. In one study, infants of mothers reporting lower levels of family support postpartum had reduced abundance of Bifidobacterium at two months of age5. There is also evidence for connections between maternal social support and markers of infant inflammatory activity10, which is strongly related to the gut microbiome11. Indirectly, there is ample evidence for the benefits of caregiver social support on child development in general. Several studies have shown that caregiver social support buffers the effects of socioeconomic disadvantage and caregiver depression on several child outcomes, including global measures of child development and mother-infant bonding12,13. These data collectively suggest that maternal social support may also be an important predictor of the infant gut microbiome.
The first year of life is a time marked by many changes and challenges as parents adapt to the demands of caring for a new infant, which can lead to high levels of stress and a strong need for social support. Higher maternal stress has been associated with reduced likelihood of breastfeeding and potentially poorer nutritional quality of breast milk14, which several studies have found to be associated with differences in the composition and diversity of the gut microbiome15,16,17,18. Specifically, breastfeeding is associated with lower gut microbiome diversity in infants as well as differences in the relative abundance of several strains of bifidobacteria17. This raises the possibility that higher maternal stress may be associated with higher infant gut microbiome diversity via reduced likelihood of breastfeeding, and that maternal stress may also influence the composition of the gut microbiome, both via breastfeeding behaviors and the composition of breastmilk. On the other hand, higher levels of perceived social support are associated with an increased likelihood of breastfeeding, plans to breastfeed for longer, and greater feelings of breastfeeding self-efficacy19,20. As such, higher caregiver social support may be associated with lower microbiome diversity via increased likelihood and longer duration of breastfeeding. Another potential mechanism by which maternal stress and social support may impact the infant gut microbiome is via the development of the hypothalamic-pituitary-adrenal axis (HPA-axis), which plays an important role in the body’s stress response. The HPA-axis is one of the main communication pathways of the microbiome-gut-brain axis and, along with the gut microbiome, undergoes a period of heightened sensitivity to environmental influence in the first year of life21. There is a large body of research demonstrating that maternal stress during infancy impacts the development of the HPA-axis5,22,23. This overlapping period of sensitivity between the HPA-axis and the gut microbiome makes the gut microbiome a particularly useful tool for studying the transmission of stress from parents to infants21.
The current study
In this prospective study we seek to understand the extent to which variations in caregiver stress and social support influence the developing gut microbiota in infancy. Specifically, we examine how stress and social support are associated with alpha and beta diversity of the gut microbiota measured at 12 months of age, and differential abundance of specific bacterial genera in the infant gut. Based on previous literature in early life stress and microbiome development, we hypothesize that higher caregiver stress will be associated with higher gut microbiota alpha diversity in infants, whereas higher caregiver social support will be associated with lower alpha diversity. We also hypothesize that stress and social support will be associated with variation in beta diversity and differential abundances of specific taxa in the infant gut microbiota, but we do not have hypotheses about specific taxa affected. Finally, given the importance of breastfeeding in the development of the infant gut microbiota, and theoretical associations between breastfeeding and caregiver stress and social support, we also examine the role of breastfeeding status in these associations. We hypothesize that caregivers reporting greater stress will be less likely to be breastfeeding at 12 months, but that caregivers reporting more social support will be more likely to still be breastfeeding at 12 months.
Methods
Participants and procedures
One hundred and six families were recruited for a longitudinal study from community events and flyers posted around the New York City metropolitan area. Inclusion criteria for children included birth at or after 37 weeks of gestation and no history of neurological or developmental delays. Infants and their primary caregivers participated in a lab visit when infants were 9 months of age and then participated in a phone screening and provided a stool sample when infants were 12 months old. In-lab data collection took place between May 2018 and December 2019 and remote stool sample collection took place between December 2018 and December 2020. To eliminate potential confounding of environmental changes due to COVID-19, for this analysis we only included infants whose caregivers provided a stool sample before March 22, 2020, which marked the beginning of stay-at-home orders in New York State24. Thus, the final analytic sample for this study consisted of 34 infants and their biological mothers (n = 34). Informed consent was obtained from all caregivers for their and the infants’ participation. All procedures were performed in accordance with relevant guidelines and regulations for human subjects research and approved by the Institutional Review Board at New York University. Demographic characteristics of the sample are summarized in Table 1.
Measures
Maternal stress
Mothers completed the Perceived Stress Scale (PSS)25 when the infant was 9 months old. The PSS is a 10-item self-report scale that assesses how often individuals have perceived situations as stressful within the last month. Respondents rated items on a 5-point Likert scale ranging from 0 (never) to 4 (very often). Total scores can range from 0 to 40 and are calculated as the sum of all responses (after 4 items are reverse scored). Scores from 0 to 13 are considered low perceived stress, 14–26 are considered moderate perceived stress, and 27–40 are considered high perceived stress. Sample items include: “In the last month, how often have you been upset because of something that happened unexpectedly” and “In the last month, how often have you been angered because of things that happened that were outside of your control?” The PSS showed good internal consistency in our sample (Cronbach’s ɑ=0.83).
Maternal social support
Mothers reported on social support at the 9-month laboratory visit using a modified version of the ENRICHD Social Support Inventory (ESSI)26. The ESSI asks caregivers how supported they felt in certain situations. These items were rated on a 5-point scale with options: none of the time, a little of the time, some of the time, most of the time, and all of the time. Sample items include: “Is there someone available to whom you can count on to listen to you when you need to talk,” “Is there someone to help you with your daily chores,” and “Do you have as much contact as you would like with someone you feel close to, someone in whom you can trust and confide in?” Two added items asked “Do you feel you are supported in your everyday life” rated on a 5-point likert scale, and asked caregivers to indicate their sources of social support in a question reading: “Where do you receive emotional support from? Check as many as apply” with answer options: mental health counseling, support from family, support from friends, support from community, support from religious practice, support groups (example - new parent group). The modified questionnaire demonstrated excellent internal consistency (Cronbach’s alpha = 0.94). We derive two metrics from this questionnaire: perceived social support, measured as the sum of the questions regarding how caregivers feel in certain situations, and sources of social support, measured by the number of options caregivers selected in the “where do you receive social support” question.
Breastfeeding
Caregivers reported on breastfeeding practices at infant ages of 3, 9, and 12 months via a questionnaire, including if they ever breastfed, if they are still breastfeeding, and the infant’s age, in months, when they stopped breastfeeding. We calculated a variable representing the amount of time, in months, that the infant had been breastfed at 12 months. Infants who were still breastfeeding at 12 months were assigned a value of 12, while infants who had never breastfed were assigned a value of 0. While we hypothesized that breastfeeding may serve as a mechanism by which caregiver stress and social support influence the infant gut microbiome, due to our small sample size we examine breastfeeding as a covariate, rather than as a mediator. We report results without controlling for breastfeeding and controlling for breastfeeding as a robustness check.
Demographic information
Caregivers reported on their and their infant’s race and ethnicity, sex, and information about caregiver education (primary and secondary), family income, and household composition on a demographics questionnaire at study entry. We then calculated a family income-to-needs ratio by dividing the family’s total reported income by the Federal Poverty Line for a family of that size. Values of 1 or below on this measure indicate a family is living in poverty, while values between 1 and 2 indicate a family is poor or near poverty. Using this information, we created a socioeconomic status composite by taking the average of z-scored values for income-to-needs ratio, maternal education (years) and paternal education (years).
Infant diet
Caregivers reported on the infant’s diet at 12 months using a food diary. Over two days, caregivers recorded everything the infant ate. Caregivers were instructed to provide as much detail about portions and brands as possible. Researchers then entered information from the diaries into an online calorie and macronutrient calculator to create estimates of daily fat, carbohydrate, and protein consumption in grams. We then took an average across the two days to create average daily fat, carbohydrate, and protein values.
Infant gut microbiota diversity and composition
Caregivers collected a stool sample from an infant diaper when the infant was 12 months of age (∓ 3 weeks) using the OMNIgene®•GUT at-home gut microbiome collection kit (OM-200; DNAgenotek) and completed a short questionnaire reporting on the date and time of sample collection. Samples were transferred to a tube filled with stabilizing liquid and mailed back to NYU, where they were stored at room temperature for no more than two weeks, and then frozen until processing. This method has shown to be effective at preserving bacteria concentrations in stool for up to 8 weeks27. The microbiota community composition measures of interest for this analysis include alpha diversity, measured via the Shannon and Chao 1 indices, beta diversity, measured via UniFrac distance, and differential abundances of specific bacterial genera in samples. Stool samples were only collected at one time point in this study, 12 months.
Alpha diversity is a within-subjects measure that assesses global diversity of species in the gastrointestinal tract, and has been used in a number of other microbiota and child development studies in humans28,29,30. The Chao 1 index of alpha diversity is a measure of the richness of the community - how many bacterial taxa (in this case - genera) are present in each individual’s sample. The Shannon index of alpha diversity, by contrast, balances richness with the evenness with which those taxa are distributed. Samples that have both high richness and evenness will have a higher Shannon value.
Beta diversity is a between-subjects metric that quantifies pairwise similarity of the entire gastrointestinal microbiota community across samples. The UniFrac distance metric uses phylogenetic relatedness to compare the samples and can either weight or not the distance matrix by low incidence microbes (weighted and unweighted UniFrac, respectively)31. Paired samples with more phylogenetic similarity have a low UniFrac distance (i.e., low beta diversity), and vice versa. Weighted UniFrac accounts for abundance of microbes in its measurement of community similarity, whereas unweighted UniFrac only accounts for the presence/absence of each microbe in its calculation. UniFrac distances have been used extensively in human microbiome studies, including developmental studies on stress effects4,28,32,33.
DNA extraction, 16 S rRNA gene sequencing and data processing
DNA extraction of the frozen stool samples was performed using the 96-well plate ZymoBIOMICS Magbead DNA/RNA Kit (Zymo Research, CA). The samples (< 200 mg) were homogenized with glass beads in 750 µl of DNA/RNA shield (Zymo Research, CA) using low-speed homogenizers (e.g. VortexGenie) for at least 15 min to ensure complete and even disruption of the material prior to DNA extraction. Homogenization was performed according to the kit manufacturer’s protocol. The centrifugation was performed at 10,000 x g for one minute to pellet debris during the extraction process. After centrifugation, the DNA was extracted from the supernatant using an Eppendorf epMotion 5705 automated system, following the manufacturer’s protocol. A commercially available mock community, ZymoBIOMICS Microbial Community Standard (Zymo Research, Cat. D6300), was included as a run control to monitor sequencing quality and taxonomic accuracy. Extracted DNA in the elution buffer was quantified using Quant-it (Thermo Fisher Scientific) and stored at −80 °C.
The 16 S rRNA gene V3-V4 region was amplified using Illumina adapter-ligated primers34, with 2.5 µl (5 ng) DNA template in a total reaction volume of 25 µl (12.5 µl KAPA HiFi HotStart ReadyMix, 5 µl each of forward and reverse primers) with the following cycling protocol: 95 °C for 3 min, 25 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and 72 °C for 5 min. The Illumina Nextera XT v2 index was used to barcode sequencing libraries. Libraries were sequenced on an Illumina MiSeq using the v3 reagent kit (600 cycles) and a loading concentration of 12 pM with 10% phiX spike-in.
Raw sequencing reads were adapter-trimmed and demultiplexed after FASTQ conversion in BaseSpace (Illumina). Divisive amplicon denoising algorithm version 2 (DADA2 1.12.1) in R 3.6.1 was used to trim, dereplicate, and filter chimeric sequences before generating amplicon sequence variant (ASV) Table (1). Taxonomic classification of ASVs was performed using the Greengenes 13_8 reference database. Based on the quality score profiles of sequencing reads, forward reads were truncated at 250 bp and reverse reads were truncated at 240 bp prior to merging, ambiguities in the overlap region were not allowed, and default parameters were otherwise applied in the filterAndTrim() function [truncLen = c(250,240); trimLeft = c(5,5), maxN = 0, maxEE = c(2,2), truncQ = 2]. After dereplication and merging of reads, chimeric reads were identified by consensus across samples using the DADA2 function removeBimeraDenovo(). All samples passed the imposed minimum of 7,500 reads after quality filtering for inclusion in this analysis. The MAFFT and FastTree modules in QIIME2 (v2020.6–2020.8.8) were used to generate a phylogenetic tree from the ASV sequences.
The ASV table, taxonomic classification table, and phylogenetic tree were imported into R 3.6.1 to generate microbial alpha diversity (Shannon, Chao indices) and beta diversity (weighted and unweighted UniFrac distances) metrics and differential abundances at the genus level of taxonomy, performed using functions from the phyloseq v1.28.0 package (2) and MaAslin2 package version 1.12.0 35.
Statistical analyses
We first performed a series of correlations between our measures of caregiver stress, social support, and both measures of alpha diversity. We then ran regressions with our measures of stress and social support as independent variables in separate models controlling for infant sex and diet, and then controlling for breastfeeding as a robustness check. Next, we examined associations between our measures of stress and social support and beta diversity of the gut microbiome. Using the phyloseq and vegan packages in R, we quantified between-subjects differences in the microbiota present in infant stool by calculating the phylogenetic dissimilarity between samples, using weighted and unweighted UniFrac distances. We first confirmed that our data did not violate homogeneity of dispersion, a necessary assumption for permutational multivariate analysis of variance (PERMANOVA). If data violated the assumption of homogeneity of dispersion, we did not proceed to the next step. Homogeneity of dispersion was violated for Bray-Curtis distances, so we do not report results using that distance metric. Next, we ran a PERMANOVA with 9,999 permutations using the adonis2 function from the vegan package to determine how much variance in beta diversity within our sample was explained by each of our predictors, controlling for infant sex, infant diet, and breastfeeding duration.
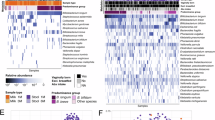
To examine how caregiver stress and social support are associated with differential abundance of individual taxa in the gut microbiota, we used Microbiome Multivariable Associations with Linear Models v2 (MaAsLin2 version 1.12.0)35 with our measures of stress or social support as predictors and default model type, transformation, and normalization parameters35. Because we lacked a priori hypotheses regarding differential abundance of specific taxa, and had limited power due to small sample size, we took a microbiome-wide approach to differential abundance using a filtered dataset that restricted comparisons to taxa that had non-zero abundance in at least 50% of samples (17/34 samples; min_prevalence parameter in MaAsLin2 set to 0.5). P-values were corrected for multiple comparisons within this set of analyses with a q-value threshold for significance of 0.25, as has been used in prior work and recommended for biomarker discovery approaches6,24,36. After identifying taxa significantly related to caregiver stress or social support, we then performed a series of regressions with taxon abundance as the outcome and caregiver stress and social support as predictors, controlling for infant sex, infant diet, and breastfeeding. All analyses were conducted in R.
Missing data and attrition
Out of 106 families recruited in the larger study, 55 infants became eligible for stool collection based on age before March 22, 2020 and were contacted to provide a stool sample before the start of the COVID-19 pandemic. Of the 55 families, 35 families returned a stool sample and of these 35 samples, 34 met processing requirements regarding sample quality and read depth and thus included in this analysis. A flowchart of data collection and attrition is depicted in Fig. 1. Families who returned a stool sample at 12 months tended to have higher incomes (t=−2.72, p = 0.01) and higher levels of maternal education (t=−2.74, p = 0.01). They did not differ on paternal education (t=−1.29, p = 0.20), total social support (t=−1.06, p = 0.31), sources of social support (t = 0.52, p = 0.61), or perceived stress (t = 1.79, p = 0.10). Missing data on other measures (surveys, etc.) were accounted for using full information maximum likelihood estimation using the Lavaan package in R for all regressions37.
Results
Descriptive statistics
In general, our sample reported low to moderate levels of stress on the PSS. About half of our sample fell into the “low stress” range on the PSS, about half in the “moderate stress” range and only one participant scored in the “high stress” range25. Our sample also reported generally moderate to high perceptions of social support, with a slight negative skew indicating that the sample is concentrated on the higher end of the spectrum. Finally, our sample reported a range of sources of social support. The median and modal number of social support sources that parents selected was 3, but with some variation across the spectrum. Full demographic characteristics of the sample and descriptive statistics can be found in Table 1.
Correlations
Perceived social support and sources of social support were significantly positively correlated (r = 0.42, p = 0.02), and perceived social support was negatively correlated with perceived stress at the trend level (r=−0.33, p = 0.07). Sources of social support and perceived stress were not correlated (r = 0.04, p = 0.81). Maternal perceived social support was negatively correlated with the Shannon diversity (r=−0.55, p = 0.001), and Chao1 richness (r=−0.39, p = 0.04) indices of the infant gut microbiota. Number of maternal sources of social support was significantly negatively correlated with Shannon diversity (r=−0.47, p = 0.01), but not with Chao 1 richness (r=−0.29, p = 0.12) of the infant gut microbiota. Neither measure of social support was correlated with duration of breastfeeding at 12 months (ps > 0.27), however those who were still breastfeeding at 12 months reported higher social support at 9 months (t=−2.26, p = 0.03), but not more sources of social support (t=−0.02, p = 0.98). Perceived stress was not correlated with duration of breastfeeding (r = 0.06, p = 0.78), nor were there differences in perceived stress between those who were vs. were not breastfeeding at 12 months (t = 0.32, p = 0.75).
Alpha diversity findings
A summary of regression models and results can be found in Table 2.
To test our hypotheses regarding potential associations between stress, social support, and gut microbiota diversity, we performed a series of correlations and regressions between our two measures of alpha diversity and our measures of stress and social support.
Social support
In a regression model controlling for infant sex and diet, the number of sources of caregiver support was significantly negatively related to Shannon diversity (β=−0.39, p = 0.02), but not with Chao 1 richness (β=−0.14, p = 0.44). When adding duration of breastfeeding to the model, the results were similar for both Shannon (β=−0.36, p = 0.02) and Chao 1 (β=−0.10, p = 0.57) indices. In regression models controlling for infant sex and diet, maternal perceived social support was negatively related to infant Shannon diversity (β=−0.55, p < 0.0001), and Chao1 richness (β=−0.35, p = 0.02). When adding duration of breastfeeding to the model, the results for Shannon diversity (β=−0.47, p = 0.002) were similar, but maternal perceived social support was no longer significantly related to Chao 1 richness (β=−0.24, p = 0.14). These findings are illustrated in Figs. 2 and 3.
Perceived stress
In regression models controlling for infant sex and diet, maternal perceived stress was not significantly related to infant Shannon diversity (β = 0.32, p = 0.17) or Chao1 richness (β = 0.37, p = 0.08). These results were similar when adding breastfeeding to the models: Shannon (β = 0.27, p = 0.22), Chao 1 (β = 0.31, p = 0.11).
Beta diversity findings
Social support
Neither sources of social support (F = 0.58, p = 0.52) nor perceived social support (F = 0.97, p = 0.34) explained significant variance in weighted UniFrac distance. The assumption of homogeneity of dispersion was violated for unweighted UniFrac distance for both social support measures, so we did not proceed with a PERMANOVA testing variation in unweighted UniFrac distance.
Perceived stress
Perceived stress did not explain significant variance in weighted UniFrac distance (F = 1.77, p = 0.18). The assumption of homogeneity of dispersion was violated for unweighted UniFrac, so we did not proceed with a PERMANOVA testing variation in unweighted UniFrac distance.
Relative abundance findings
Social support
Sources of social support
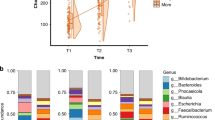
In a MaAslin2 model, the number of sources of social support was positively associated with the relative abundance of the following genera: Veillonella (B = 1.51, p < 0.001, q = 0.01), Clostridium sensu stricto 1 (B = 1.54, p < 0.001, q = 0.01), Ruminococcus gnavus group; B = 1.07, p = 0.002, q = 0.02). In regressions controlling for infant sex and diet, sources of social support was positively predictive of Veillonella (β = 0.39, p = 0.02), Clostridium sensu stricto 1 (β = 0.46, p = 0.01), and Ruminococcus gnavus group (β = 0.50, p = 0.01) abundances. These findings remained consistent when adding breastfeeding into these models: Veillonella (β = 0.38, p = 0.02), Clostridium sensu stricto (β = 0.46, p = 0.01), Ruminococcus gnavus group (β = 0.50, p = 0.003). These findings are illustrated in Figs. 4, 5 and 6.
Perceived social support
No genera were differentially abundant as a function of maternal perceived social support after adjustment for FDR in a MaAslin2 model.
Perceived stress
No genera were differentially abundant as a function of maternal perceived stress after adjustment for FDR.
Discussion
The results of this study indicate prospective associations between caregiver social support, measured at 9 months, and the composition and diversity of the infant gut microbiota at 12 months. We found that higher maternal social support was associated with lower infant gut microbial diversity. We did not find associations between maternal stress and gut microbiota diversity or composition, nor did we find associations between maternal stress or social support and gut microbiota beta diversity. This study adds to a growing body of literature highlighting the importance of maternal social support in promoting infant health and development5,10,13,38,39,40. Moreover, these findings emphasize the need for sensitive, robust measurement of maternal psychosocial wellbeing in infant gut microbiome research.
We found that infants of caregivers who reported feeling more supported and having more unique sources of social support had lower gut microbiota alpha diversity, measured via the Shannon index. The Shannon index accounts for both richness and evenness in the infant gut, while our other measure of alpha diversity, the Chao 1 index, only accounts for richness. As such, our findings suggest that higher maternal social support is associated with reduced evenness of the gut microbiome, but not necessarily richness. While higher alpha diversity in adults is thought to promote health, the role of alpha diversity in infancy is less clear. Alpha diversity is often much lower in breastfed infants, in part because breastfeeding has shown to encourage dominance in the infant gut by bifidobacteria, thereby reducing overall diversity of the infant gut15,16. Alpha diversity increases rapidly across the first few years of life, especially after infants stop breastfeeding, and as such higher alpha diversity in infancy is thought to reflect an accelerated maturation of the gut microbiome16,41. This may explain why some studies have found higher alpha diversity in infancy to be associated with poorer outcomes related to cognitive and emotional development29,30,32.
We also found that the number of sources of caregiver social support predicted greater abundances of Veillonella, Clostridium sensu stricto, and Ruminococcus gnavus in the infant gut. Veillonella abundance has previously been implicated in studies of prenatal maternal stress and infant gut microbiome5,42, though directionality has differed between studies, and is often enriched in the gut microbiota of breastfeeding infants due to its role in lactate metabolism18. Clostridium sensu stricto members are among the earliest producers of butyrate in the infant gut, a short-chain fatty acid important for immune system development. As such, a higher relative abundance of Clostridium sensu stricto in our sample of 12-month olds is developmentally appropriate in the context of the infant gut microbiome, and likely reflects a sign of health43. The Ruminococcus gnavus group is a group of bacteria found in the infant gut that has been previously associated with the transition from infant- to adult-like patterns of butyrate formation in the developing gut microbiota44. These taxa together suggest that the number of sources of caregiver social support might be particularly associated with microbes implicated in the developing immune system and the production of short-chain fatty acids.
Breastfeeding is often one of the most robust predictors of diversity and composition of the gut microbiome in infancy15,16. Here, we examined maternal stress and social support as predictors of infant gut microbiota with and without controlling for breastfeeding. While results were largely consistent between models, we did find some differences in results between alpha diversity models with and without breastfeeding. Specifically, while higher caregiver perceived social support predicted lower Chao 1 richness values in correlations and a regression without breastfeeding, including breastfeeding in this model weakened the association between perceived social support and Chao 1 richness. This suggests that some support-related variation in Chao 1 richness was explained by breastfeeding. As such, our findings suggest that breastfeeding may be one mechanism by which maternal social support influences the richness of the infant gut microbiota, but associations between maternal social support and other features of the infant gut microbiome emerge over and above the effects of breastfeeding.
There are several other potential mechanisms by which caregiver social support might influence the infant gut microbiota. One hypothesized mechanism is that having more social support reduces caregiver stress. We did find that higher caregiver social support was trending towards lower caregiver stress in our sample (r=−0.33, p = 0.07), but we did not find associations between caregiver stress and the infant gut microbiota. Another hypothesized mechanism by which caregiver social support influences the infant gut microbiota is via changes in the contents of breastmilk. There is some evidence to suggest that maternal psychosocial stress is associated with differences in energy density, fat content, and immune markers in breast milk45,46,47, and one study found that maternal social support was negatively associated with inflammatory markers in breast milk40. These findings suggest that maternal social support may influence the infant gut microbiota through both the duration of breastfeeding and changes in the nutritional and immunological composition of breastmilk.
Our finding that caregiver perceived stress was not associated with any features of the infant gut microbiota is inconsistent with previous studies of postnatal maternal stress and infant gut microbiome development. The limited variability in caregiver perceived stress in our sample may be one potential explanation for the discrepancies in findings between our study and others. Our sample reported overall low and moderate levels of stress (range 6–28 on the PSS), whereas the samples from previous studies of postnatal maternal stress and infant gut microbiota have come from samples with higher reported stress levels than ours3,4. This suggests that caregiver stress may exert a more meaningful influence on infant gut microbiota at higher levels of stress that we did not detect in this community-based sample.
This study, by focusing on measures of both caregiver stress and social support, adds nuance to a growing body of literature revealing the role of the early social environment in the development of the infant gut microbiota. A notable strength of this study is the diverse sample represented. Many studies of gut microbiome development have been done in more racially and ethnically homogenous samples, which has allowed for a foundation of research to draw from but limits generalization. Experiences of stress and social support are experienced differently and may have different impacts on physical and mental health based on cultural and demographic characteristics48,49 and, as such, the role of these experiences in infant development may differ based on social and structural conditions. An additional strength of our study comes from the longitudinal nature of our study design, which allowed us to capture prospective associations between caregiver social support and the infant gut microbiota, rather than cross-sectional associations.
Despite these strengths, there are several limitations worth addressing. We relied on a small sample (n = 34) for this study, which limited the types of analyses we could run, including testing breastfeeding as a mediator in associations between stress/social support and the infant gut microbiota. We included only one measurement of the gut microbiota in our study, but the infant gut microbiota undergoes substantial change across the first three to four years of life. Additionally, we observed some evidence of ceiling effects in our measure of perceived social support, with most participants reporting feeling highly supported. While our skewness (−1.2) and kurtosis values (0.29) were within a normal range for this measure, some skew in a small sample such as this may limit the statistical variability we are able to detect. A final limitation is in our use of 16 s rRNA gene sequencing for to profile the gut microbiota. While commonly used in studies of gut microbiome and child development, 16 s rRNA gene sequencing is limited in the level of detail it provides, typically only allowing for classification at the genus level, rather than the species of strain level50. Additionally, it only detects bacterial and archaeal DNA, excluding other microorganisms that may contribute to gut microbiome function. Future studies with larger samples, multiple measurements of the infant gut microbiome, more advanced sequencing technologies, and measurements of stress and caregiver support beginning prenatally and continuing throughout early life may help address these limitations and better characterize associations between the social environment and the development of the infant gut microbiome.
In this study, we have presented evidence that caregiver social support is associated with multiple markers of gut microbiota development in infancy. These results highlight the importance of promoting caregiver wellbeing as a means of supporting healthy infant development. Accessible and affordable mental health care, new parent support groups, paid parental leave, available and affordable childcare, and other services that promote social bonds and new parent emotional and social wellbeing may all play important roles in setting infants up for long-term health. This study expands our scientific understanding of how caregiver stress and social support in infancy shape the infant gut microbiota and sets the stage for a more nuanced understanding of the various social influences on the gut microbiota in early life.
Data availability
Data for this study can be found on the Open Science Framework at: https://osf.io/w6u5y/files/osfstorage.
References
Cowan, C. S. M., Dinan, T. G. & Cryan, J. F. Annual research review: critical windows–the microbiota–gut–brain axis in neurocognitive development. J Child psychology and psychiatry 5(3), 353–371 (2020).
Graf, M. D. et al. Maternal prenatal stress and the offspring gut microbiome: A Cross-Species systematic review. Dev. Psychobiol. 67, e70005 (2025).
Galley, J. D. et al. Maternal anxiety, depression and stress affects offspring gut Microbiome diversity and bifidobacterial abundances. Brain Behav. Immun. 107, 253–264 (2023).
Dutton, C. L. et al. Maternal Psychosocial Stress Is Associated with Reduced Diversity in the Early Infant Gut Microbiome. Microorganisms. 11, (2023).
Jahnke, J. R., Roach, J., Azcarate-Peril, M. A. & Thompson, A. L. Maternal precarity and HPA axis functioning shape infant gut microbiota and HPA axis development in humans. PLoS One. 16, e0251782 (2021).
Querdasi, F. R. et al. Multigenerational adversity impacts on human gut microbiome composition and socioemotional functioning in early childhood. Proc. Natl. Acad. Sci. U. S. A. 120, e2213768120 (2023).
Rojas, L. et al. Long-term and trimester-specific effects of prenatal stress on the child gut microbiota. Psychoneuroendocrinology 158, 106380 (2023).
Aatsinki, A. K. et al. Maternal prenatal psychological distress and hair cortisol levels associate with infant fecal microbiota composition at 2.5 months of age. Psychoneuroendocrinology 119, 104754 (2020).
Saturio, S. et al. Role Bifidobacteria Infant Health Microorganisms 9, 2415 (2021).
Nelson, B. W., Wright, D. B., Allen, N. B. & Laurent, H. K. Maternal stress and social support prospectively predict infant inflammation. Brain Behav. Immun. 86, 14–21 (2020).
Arrieta, M. C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M. & Finlay, B. The intestinal Microbiome in early life: health and disease. Front Immunol 5 (2014).
White, L. K. et al. The impact of postpartum social support on postpartum mental health outcomes during the COVID-19 pandemic. Arch Womens Ment Health 26, 1–11 (2023).
McDonald, S., Kehler, H., Bayrampour, H., Fraser-Lee, N. & Tough, S. Risk and protective factors in early child development: results from the all our babies (AOB) pregnancy cohort. Res. Dev. Disabil. 58, 20–30 (2016).
Fallon, V., Groves, R., Halford, J. C. G., Bennett, K. M. & Harrold, J. A. Postpartum anxiety and Infant-Feeding outcomes. J. Hum. Lact. 32, 740–758 (2016).
Kim, H., Sitarik, A. R., Woodcroft, K., Johnson, C. C. & Zoratti, E. Birth Mode, Breastfeeding, pet Exposure, and antibiotic use: associations with the gut Microbiome and sensitization in children. Curr. Allergy Asthma Rep. 19, 22 (2019).
Stewart, C. J. et al. Temporal development of the gut Microbiome in early childhood from the TEDDY study. Nature 562, 583–588 (2018).
Levin, A. M. et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut Microbiome structure and diversity. Sci. Rep. 6, 31775 (2016).
Matsuyama, M. et al. Breastfeeding: a key modulator of gut microbiota characteristics in late infancy. J. Dev. Orig Health Dis. 10, 206–213 (2019).
Mercan, Y. & Tari Selcuk, K. Association between postpartum depression level, social support level and breastfeeding attitude and breastfeeding self-efficacy in early postpartum women. PLoS One. 16, e0249538 (2021).
Lyons, G. C. et al. Social support and breastfeeding outcomes among a Racially and ethnically diverse population. Am. J. Prev. Med. 64, 352–360 (2023).
Callaghan, B. Nested sensitive periods: how plasticity across the microbiota-gut-brain axis interacts to affect the development of learning and memory. Curr. Opin. Behav. Sci. 36, 55–62 (2020).
Blair, C. Stress and the development of Self-Regulation in context. Child. Dev. Perspect. 4, 181–188 (2010).
Gunnar, M. R. & Donzella, B. Social regulation of the cortisol levels in early human development. Psychoneuroendocrinology 27, 199–220 (2002).
Querdasi, F. R., Vogel, S. C., Thomason, M. E., Callaghan, B. L. & Brito, N. H. A comparison of the infant gut Microbiome before versus after the start of the covid-19 pandemic. Sci. Rep. 13, 13289 (2023).
Cohen, S., Kamarck, T. & Mermelstein, R. Perceived stress scale (PSS). J. Health Soc. Behav. 24, 285 (1983).
Mitchell, P. et al. A short social support measure for patients recovering from myocardial infarction: the ENRICHD social support inventory. J. Cardiopulm. Rehabil. 23, 398–403 (2003).
Song, S. J. et al. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitability for Field Studies. mSystems, 1, (2016).
Callaghan, B. L. et al. Mind and gut: associations between mood and Gastrointestinal distress in children exposed to adversity. Dev. Psychopathol. 32, 309–328 (2020).
Carlson, A. L. et al. Infant gut Microbiome associated with cognitive development. Biol. Psychiatry. 83, 148–159 (2018).
Gao, W. et al. Gut Microbiome and brain functional connectivity in infants-a preliminary study focusing on the amygdala. Psychopharmacology 236, 1641–1651 (2019).
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Carlson, A. L. et al. Infant gut Microbiome composition is associated with non-social fear behavior in a pilot study. Nat. Commun. 12, 3294 (2021).
Michels, N. et al. Gut Microbiome patterns depending on children’s psychosocial stress: reports versus biomarkers. Brain Behav. Immun. 80, 751–762 (2019).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021).
Kelsey, C. M. et al. Gut microbiota composition is associated with newborn functional brain connectivity and behavioral temperament. Brain Behav. Immun. 91, 472–486 (2021).
Rosseel, Y. Lavaan: an R package for structural equation modeling. J. Stat. Softw. 48, 1–36 (2012).
Giesbrecht, G. F. et al. The buffering effect of social support on hypothalamic-pituitary-adrenal axis function during pregnancy. Psychosom. Med. 75, 856–862 (2013).
MacMillan, K. K., Lewis, A. J., Watson, S. J., Bourke, D. & Galbally, M. Maternal social support, depression and emotional availability in early mother-infant interaction: findings from a pregnancy cohort. J. Affect. Disord. 292, 757–765 (2021).
Kim, E. S. et al. Maternal psychosocial factors that affect breastfeeding adaptation and immune substances in human milk. Korean J. Women Health Nurs. 20, 14–28 (2014).
Derrien, M., Alvarez, A. S. & de Vos, W. M. The gut microbiota in the first decade of life. Trends Microbiol. 27, 997–1010 (2019).
Warner, B. B. et al. Social and psychological adversity are associated with distinct mother and infant gut Microbiome variations. Nat. Commun. 14, 1–19 (2023).
Appert, O. et al. Initial butyrate producers during infant gut microbiota development are endospore formers. Environ. Microbiol. 22, 3909–3921 (2020).
Nilsen, M. et al. Butyrate levels in the transition from an infant- to an adult-like gut microbiota correlate with bacterial networks associated with Eubacterium rectale and Ruminococcus gnavus. Genes (Basel). 11, 1245 (2020).
Ziomkiewicz, A. et al. Psychosocial stress and cortisol stress reactivity predict breast milk composition. Sci. Rep. 11, 11576 (2021).
Groer, M., Davis, M. & Steele, K. Associations between human milk SIgA and maternal immune, infectious, endocrine, and stress variables. J. Hum. Lact. 20, 153–158 (2004). quiz 159-63.
Zagoory-Sharon, O. et al. Breast milk Oxytocin and s-IgA modulate infant biomarkers and social engagement; the role of maternal anxiety. Compr. Psychoneuroendocrinol. 17, 100219 (2024).
Shavitt, S. et al. Culture moderates the relation between perceived Stress, social Support, and mental and physical health. J. Cross Cult. Psychol. 47, 956–980 (2016).
Abdou, C. M. et al. Communalism predicts prenatal affect, stress, and physiology better than ethnicity and socioeconomic status. Cultur Divers. Ethnic Minor. Psychol. 16, 395–403 (2010).
Janda, J. M. & Abbott, S. L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764 (2007).
Acknowledgements
Research reported in this manuscript was supported by the National Institutes of Health (NIH) under award numbers T32MH015750 and T32GM086270 to FRQ and award numbers R00HD086255, R01MH125870, R01MH126468, R01DA059415 to NHB. We would like to thank the students and staff who contributed their time to collecting and processing data for this study, including Maggie Zhang, Karina Kozak, and Amy Hume. We would also like to extend our thanks to the Columbia University Medical Center Microbiome Core and their team for their sequencing and informatics support. Finally, we would like to thank the families who participated in this research for their time.
Author information
Authors and Affiliations
Contributions
S.C.V. contributed conceptualization, data collection, data analysis, writing, and editing. F.R.Q. contributed data analysis, writing, and editing. B.L.C. contributed writing and editing. N.H.B. contributed conceptualization, data collection, writing, and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vogel, S.C., Querdasi, F.R., Callaghan, B.L. et al. Examining associations among caregiver stress, social support, and the infant gut microbiota. Sci Rep 16, 946 (2026). https://doi.org/10.1038/s41598-025-30553-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-30553-w