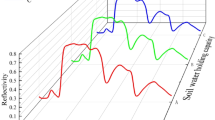
Abstract
Segmentation of hyperspectral image data is a well-established technique in remote sensing. While it is commonly applied to individual field crops, its use for individual trees is less prevalent. Conifers are crucial in forestry, and assessing physiological status, or genetic diversity is required for effective early-age treatment in nurseries and hyperspectral imaging (HSI) combined with high-throughput phenotyping (HTP) offers faster and non-destructive evaluation. NDVI-based thresholding is sufficient for detection of leaves with large projection areas, but needles of conifers present challenges due to spatial resolution constraints and increased proportion of border pixels. This study monitored the offspring of three locally adapted Scots pine (Pinus sylvestris L.) populations, representing distinct upland and lowland ecotypes. This study presents a hyperspectral image processing pipeline for segmenting and isolating individual Scots pine seedlings. Using a K-means algorithm, 23 hyperspectral centroids were successfully derived and subsequently classified into ten biologically distinct groups. Random forest classification model effectively differentiated Scots pine seedlings based on origin during water stress and recovery periods. This study highlights the potential of hyperspectral imaging and machine learning in evaluating the physiological state of conifer seedlings, demonstrating promising applications in forest tree physiology research and tree breeding.
Similar content being viewed by others
Data availability
Demonstration sample data and their accompanying descriptions are available in the Zenodo repository (https://doi.org/10.5281/zenodo.17167809).The scripts developed for this study are available on GitHub at: https://github.com/JCepl/Pine-hyperspectral-image-segmentaionComplete experimental data and other materials will be made available to third-party academic researchers upon reasonable request.
Abbreviations
- DAT:
-
Days after transplantation
- SRWC:
-
Soil relative water content
- HSI:
-
Hyperspectral imaging
- VI:
-
Vegetation index
- PCA:
-
Principal component analysis
- LDA:
-
Linear discriminant analysis
- RF:
-
Random forest
References
Hallingbäck, H. R. et al. Managing uncertainty in Scots pine Range-Wide adaptation under climate change. Front. Ecol. Evol. 9, 724051. https://doi.org/10.3389/fevo.2021.724051 (2021).
Taeger, S., Sparks, T. H. & Menzel, A. Effects of temperature and drought manipulations on seedlings of Scots pine provenances. Plant. Biol. J. 17, 361–372. https://doi.org/10.1111/plb.12245 (2015).
Palmroth, S. et al. Structural adaptation rather than water conservation was observed in Scots pine over a range of wet to dry climates. Oecologia 121, 302–309. https://doi.org/10.1007/s004420050932 (1999).
González-Díaz, P. et al. Neighbouring Scots pine populations from contrasting Climatic regions show substantial variability but consistent response to warming. Environ. Exp. Bot. 218, 105603. https://doi.org/10.1016/j.envexpbot.2023.105603 (2024).
Miettinen, I. et al. Hyperspectral imaging reveals differential carotenoid and chlorophyll Temporal dynamics and Spatial patterns in Scots pine under water stress. Plant. Cell. Environ. 48, 1535–1554. https://doi.org/10.1111/pce.15225 (2025).
Hornero, A. et al. Modelling hyperspectral- and thermal-based plant traits for the early detection of Phytophthora-induced symptoms in oak decline. Remote Sens. Environ. 263, 112570. https://doi.org/10.1016/j.rse.2021.112570 (2021).
Coops, N. et al. Assessment of Dothistroma needle blight of Pinus radiata using airborne hyperspectral imagery. Phytopathology® 93, 1524–1532. https://doi.org/10.1094/PHYTO.2003.93.12.1524 (2003).
Yu, R., Ren, L. & Luo, Y. Early detection of pine wilt disease in Pinus tabuliformis in North China using a field portable spectrometer and UAV-based hyperspectral imagery. For. Ecosyst. 8, 44. https://doi.org/10.1186/s40663-021-00328-6 (2021).
Yu, R. et al. A machine learning algorithm to detect pine wilt disease using UAV-based hyperspectral imagery and lidar data at the tree level. Int. J. Appl. Earth Obs. Geoinf. 101, 102363. https://doi.org/10.1016/j.jag.2021.102363 (2021).
Zhang, S., Huang, H., Huang, Y., Cheng, D. & Huang, J. A GA and SVM classification model for pine wilt disease detection using UAV-Based hyperspectral imagery. Appl. Sci. 12, 6676. https://doi.org/10.3390/app12136676 (2022).
Campbell, P. K. E. et al. Detection of initial damage in Norway Spruce canopies using hyperspectral airborne data. Int. J. Remote Sens. 25, 5557–5583. https://doi.org/10.1080/01431160410001726058 (2004).
Mišurec, J. et al. Utilization of hyperspectral image optical indices to assess the Norway Spruce forest health status. J. Appl. Remote Sens. 6, 063545–063541. https://doi.org/10.1117/1.JRS.6.063545 (2012).
Mišurec, J., Kopačková, V., Lhotáková, Z., Campbell, P. & Albrechtová, J. Detection of spatio-temporal changes of norway spruce forest stands in ore mountains using landsat time series and airborne hyperspectral imagery. Remote Sens. 8, 92. https://doi.org/10.3390/rs8020092 (2016).
Kopačková, V., Mišurec, J., Lhotáková, Z., Oulehle, F. & Albrechtová, J. Using multi-date high spectral resolution data to assess the physiological status of macroscopically undamaged foliage on a regional scale. Int. J. Appl. Earth Obs. Geoinf. 27, 169–186. https://doi.org/10.1016/j.jag.2013.09.009 (2014).
D’Odorico, P., Besik, A., Wong, C. Y. S., Isabel, N. & Ensminger, I. High-throughput drone‐based remote sensing reliably tracks phenology in thousands of conifer seedlings. New. Phytol. 226, 1667–1681. https://doi.org/10.1111/nph.16488 (2020).
Santini, F., Kefauver, S. C., Resco de Dios, V., Araus, J. L. & Voltas, J. Using unmanned aerial vehicle-based multispectral, RGB and thermal imagery for phenotyping of forest genetic trials: A case study in Pinus halepensis. Ann. Appl. Biol. 174, 262–276. https://doi.org/10.1111/aab.12484 (2019).
Santini, F. et al. Bridging the genotype–phenotype gap for a mediterranean pine by semi-automatic crown identification and multispectral imagery. New Phytol. 229, 245–258. https://doi.org/10.1111/nph.16862 (2021).
Haagsma, M. et al. Using hyperspectral imagery to detect an invasive fungal pathogen and symptom severity in Pinus strobiformis seedlings of different genotypes. Remote Sens. 12, 4041. https://doi.org/10.3390/rs12244041 (2020).
Pandey, P. et al. Hyperspectral imaging combined with machine learning for the detection of fusiform rust disease incidence in loblolly pine seedlings. Remote Sens. 13, 3595. https://doi.org/10.3390/rs13183595 (2021).
Xing, D. et al. Non-destructive Estimation of needle leaf chlorophyll and water contents in Chinese Fir seedlings based on hyperspectral reflectance spectra. f. ;4. (2024). https://doi.org/10.48130/forres-0024-0021
Masaitis, G., Mozgeris, G. & Augustaitis, A. Spectral reflectance properties of healthy and stressed coniferous trees. iForest 6, 30–36 (2013).
Lu, Y. et al. Hyperspectral imaging with Cost-Sensitive learning for High-Throughput screening of loblolly pine (Pinus Taeda L.) seedlings for freeze tolerance. Trans. ASABE. 64, 2045–2059. https://doi.org/10.13031/trans.14708 (2021).
Long, T. et al. Visible-near-infrared hyperspectral imaging combined with ensemble learning for the nutrient content of Pinus Elliottii × P. caribaea canopy needles detection. Front. Glob Change. 6. https://doi.org/10.3389/ffgc.2023.1203626 (2023).
Czyż, E. A. et al. Intraspecific genetic variation of a Fagus sylvatica population in a temperate forest derived from airborne imaging spectroscopy time series. Ecol. Evol. 10, 7419–7430. https://doi.org/10.1002/ece3.6469 (2020).
Stejskal, J. et al. Making the genotypic variation visible: hyperspectral phenotyping in Scots pine seedlings. Plant. Phenomics 2023;Article in Press:0111. https://doi.org/10.34133/plantphenomics.0111
Danusevicius, D., Masaitis, G. & Mozgeris, G. Visible and near infrared hyperspectral imaging reveals significant differences in needle reflectance among Scots pine provenances. Silvae Genetica. 63, 169–180. https://doi.org/10.1515/sg-2014-0022 (2014).
Williams, D. et al. A method for automatic segmentation and splitting of hyperspectral images of raspberry plants collected in field conditions. Plant. Methods. 13, 74. https://doi.org/10.1186/s13007-017-0226-y (2017).
Li, X., Chen, Z., Wang, J. & Jin, J. LeafSpec-Dicot: an accurate and portable hyperspectral imaging device for Dicot leaves. Sensors 23, 3687. https://doi.org/10.3390/s23073687 (2023).
Grubinger, S. et al. Seasonal vegetation dynamics for phenotyping using multispectral drone imagery: genetic differentiation, climate adaptation, and hybridization in a common-garden trial of interior Spruce (Picea engelmannii × glauca). Remote Sens. Environ. 317, 114512. https://doi.org/10.1016/j.rse.2024.114512 (2025).
Neuwirthová, E. et al. Drought response and genetic variation in Scots pine seedlings’ provenances: insights from High-Throughput phenotyping for Climate-Resilient forestry. Evol. Appl. 18, e70157. https://doi.org/10.1111/eva.70157 (2025).
Miao, C. et al. Semantic Segmentation of Sorghum Using Hyperspectral Data Identifies Genetic Associations. Plant. Phenomics ;2020:1–11. https://doi.org/10.34133/2020/4216373. (2020).
Findurová, H. et al. Phenotyping drought tolerance and yield performance of barley using a combination of imaging methods. Environ. Exp. Bot. 209, 105314. https://doi.org/10.1016/j.envexpbot.2023.105314 (2023).
Paulus, S. & Mahlein, A-K. Technical workflows for hyperspectral plant image assessment and processing on the greenhouse and laboratory scale. GigaScience 9, giaa090. https://doi.org/10.1093/gigascience/giaa090 (2020).
Brichta, J. et al. Importance and potential of Scots pine (Pinus sylvestris L.) in 21st century. Cent. Eur. Forestry J. 69, 3–20. https://doi.org/10.2478/forj-2022-0020 (2023).
Moncholi-Estornell, A. et al. Impact of Structural, photochemical and instrumental effects on leaf and canopy reflectance variability in the 500–600 Nm range. Remote Sens. 14, 56. https://doi.org/10.3390/rs14010056 (2021).
Haboudane, D., Miller, J. R., Pattey, E., Zarco-Tejada, P. J. & Strachan, I. B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: modeling and validation in the context of precision agriculture. Remote Sens. Environ. 90, 337–352. https://doi.org/10.1016/j.rse.2003.12.013 (2004).
Abdelhakim, L. O. A. et al. High throughput Image-Based phenotyping for determining morphological and physiological responses to single and combined stresses in potato. JoVE 66255. https://doi.org/10.3791/66255 (2024).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL (2021). https://www.R-project.org/. https://www.r-project.org/. Accessed 24 Nov 2020.
Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory. 28, 129–137. https://doi.org/10.1109/TIT.1982.1056489 (1982).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S, Springer, New York: ISBN 0-387-95457-0. (2002).
Liaw, A. & Wiener, M. Classification and regression by randomforest. R News. 2, 18–22 (2002).
Baret, F., Jacquemoud, S., Guyot, G. & Leprieur, C. Modeled analysis of the biophysical nature of spectral shifts and comparison with information content of broad bands. Remote Sens. Environ. 41, 133–142. https://doi.org/10.1016/0034-4257(92)90073-S (1992).
Gitelson, A. A. & Merzlyak, M. N. Remote Estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 18, 2691–2697. https://doi.org/10.1080/014311697217558 (1997).
Ely, K. S., Burnett, A. C., Lieberman-Cribbin, W., Serbin, S. P. & Rogers, A. Spectroscopy can predict key leaf traits associated with source–sink balance and carbon–nitrogen status. J. Exp. Bot. 70, 1789–1799. https://doi.org/10.1093/jxb/erz061 (2019).
Blackburn, G. A. Relationships between spectral reflectance and pigment concentrations in stacks of deciduous broadleaves. Remote Sens. Environ. 70, 224–237. https://doi.org/10.1016/S0034-4257(99)00048-6 (1999).
Neuwirthová, E., Lhotáková, Z. & Albrechtová, J. The effect of leaf stacking on leaf reflectance and vegetation indices measured by contact probe during the season. Sensors 17, 1202. https://doi.org/10.3390/s17061202 (2017).
Einzmann, K., Ng, W-T., Immitzer, M., Pinnel, N. & Atzberger, C. Method analysis for collecting and processing in-situ hyperspectral needle reflectance data for monitoring Norway Spruce methodenanalyse Zur erfassung und prozessierung hyperspektraler in-situ nadelreflexionsdaten zum monitoring von Fichten. Photogrammetrie - Fernerkundung -Geoinformation. 2014, 423–434. https://doi.org/10.1127/1432-8364/2014/0234 (2014).
Jacquemoud, S. et al. PROSPECT + SAIL models: A review of use for vegetation characterization. Remote Sensing of Environment. ;113, Supplement 1:S56–66. (2009). https://doi.org/10.1016/j.rse.2008.01.026
Lai, Y. et al. Bidirectional reflectance factor measurement of conifer needles with microscopic spectroscopy imaging. Agric. For. Meteorol. 330, 109311. https://doi.org/10.1016/j.agrformet.2023.109311 (2023).
Neuwirthová, E., Lhotáková, Z., Lukeš, P. & Albrechtová, J. Leaf surface reflectance does not affect biophysical traits modelling from VIS-NIR spectra in plants with sparsely distributed trichomes. Remote Sens. 13, 4144. https://doi.org/10.3390/rs13204144 (2021).
Yang, B. et al. Analyses of impact of needle surface properties on Estimation of needle absorption spectrum: case study with coniferous needle and shoot samples. Remote Sens. 8, 563. https://doi.org/10.3390/rs8070563 (2016).
O’Neill, A. L., Kupiec, J. A. & Curran, P. J. Biochemical and reflectance variation throughout a Sitka Spruce canopy. Remote Sens. Environ. 80, 134–142. https://doi.org/10.1016/S0034-4257(01)00294-2 (2002).
Homolová, L. et al. Measurement methods and variability assessment of the Norway Spruce total leaf area: implications for remote sensing. Trees 27, 111–121. https://doi.org/10.1007/s00468-012-0774-8 (2013).
Lhotáková, Z. et al. Foliage biophysical trait prediction from laboratory spectra in Norway Spruce is more affected by needle age than by site soil conditions. Remote Sens. 13, 391. https://doi.org/10.3390/rs13030391 (2021).
Gitelson, A. A. Nondestrictive estimation of foliar pigment (chlorophylls, carotenoids, and anthocyanins) contents: evaluating a semianalytical three-band model. In: Hyperspectral Remote Sensing of Vegetation. 1st Edition. CRC Press; p. 141. (2011).
Guzicka, M., Marek, S., Gawlak, M. & Tomaszewski, D. Micromorphology of pine needle primordia and young needles after bud dormancy breaking. Plants 12, 913. https://doi.org/10.3390/plants12040913 (2023).
Lewis, F. J. & Dowding, E. S. The anatomy of the buds of coniferae. Ann. Botany. os-38, 217–228. https://doi.org/10.1093/oxfordjournals.aob.a089891 (1924).
Soukupova, J., Rock, B. N. & Albrechtova, J. Spectral characteristics of lignin and soluble phenolics in the near infrared - a comparative study. Int. J. Remote Sens. 23, 3039–3055. https://doi.org/10.1080/01431160110104683 (2002).
Dillen, S. Y., de Beeck, M. O., Hufkens, K., Buonanduci, M. & Phillips, N. G. Seasonal patterns of foliar reflectance in relation to photosynthetic capacity and color index in two co-occurring tree species, Quercus rubra and betula papyrifera. Agric. For. Meteorol. 160, 60–68. https://doi.org/10.1016/j.agrformet.2012.03.001 (2012).
Polák, T., Albrechtová, J. & Rock, B. N. Bud development types as a new macroscopic marker of Norway Spruce decline and recovery processes along a mountainous pollution gradient. Forestry (Lond). 79, 425–437. https://doi.org/10.1093/forestry/cpl009 (2006).
Eitel, J. U. H., Gessler, P. E., Smith, A. M. S. & Robberecht, R. Suitability of existing and novel spectral indices to remotely detect water stress in Populus spp. For. Ecol. Manag. 229, 170–182. https://doi.org/10.1016/j.foreco.2006.03.027 (2006).
Seelig, H. D. et al. The assessment of leaf water content using leaf reflectance ratios in the visible, near‐, and short‐wave‐infrared. Int. J. Remote Sens. 29, 3701–3713. https://doi.org/10.1080/01431160701772500 (2008).
Zhang, Y., Wang, A., Li, J. & Wu, J. Water content Estimation of conifer needles using leaf-level hyperspectral data. Front. Plant. Sci. 15, 1428212. https://doi.org/10.3389/fpls.2024.1428212 (2024).
Carter, G. A. Primary and secondary effects of water content on the spectral reflectance of leaves. Am. J. Bot. 78, 916–924. https://doi.org/10.2307/2445170 (1991).
Stasinski, L., White, D. M., Nelson, P. R., Ree, R. H. & Meireles, J. E. Reading light: leaf spectra capture fine-scale diversity of closely related, hybridizing Arctic shrubs. New Phytol. 232, 2283–2294. https://doi.org/10.1111/nph.17731 (2021).
Li, C. et al. Evaluating potential of leaf reflectance spectra to monitor plant genetic variation. Plant. Methods. 19, 108. https://doi.org/10.1186/s13007-023-01089-9 (2023).
Tirado, S. B., Dennis SSt, Enders, T. A. & Springer, N. M. Utilizing Spatial variability from hyperspectral imaging to assess variation in maize seedlings. Plant. Phenome J. 4, e20013. https://doi.org/10.1002/ppj2.20013 (2021).
Gao, D. et al. Improvement of chlorophyll content Estimation on maize leaf by vein removal in hyperspectral image. Comput. Electron. Agric. 184, 106077. https://doi.org/10.1016/j.compag.2021.106077 (2021).
Ren, S., He, K., Girshick, R., Sun, J. & Faster, R-C-N-N. Towards Real-Time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39, 1137–1149. https://doi.org/10.1109/TPAMI.2016.2577031 (2017).
Kong, W., Zhang, C., Huang, W., Liu, F. & He, Y. Application of hyperspectral imaging to detect sclerotinia sclerotiorum on oilseed rape stems. Sensors 18, 123. https://doi.org/10.3390/s18010123 (2018).
Shahrimie, M. A. M. et al. Modeling effects of illumination and plant geometry on leaf reflectance spectra in close-range hyperspectral imaging. In: 2016 8th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS). Los Angeles, CA: IEEE; 1–4. https://doi.org/10.1109/WHISPERS.2016.8071753. (2016).
Ji, F. et al. Unveiling the transferability of PLSR models for leaf trait estimation: lessons from a comprehensive analysis with a novel global dataset. New Phytol. 243, 111–131. https://doi.org/10.1111/nph.19807 (2024).
Semerci, A. et al. Morphological and physiological responses to drought stress of European provenances of Scots pine. Eur. J. For. Res. 136, 91–104. https://doi.org/10.1007/s10342-016-1011-6 (2017).
Acknowledgements
We thank to Dr. Jan Kaňák, Arboretum Sofronka, Pilsen for providing plant materials and consultation throughout the cultivation process of pine trees. We thank to Jakub Dvořák, Daniel Provazník and Jiří Chuchlík for help with field sampling; Anna Pilská and Monika Pilátová for DNA extraction (all from CULS); and Miroslav Barták (CUNI) for field sampling, sample processing and graphical help. We also appreciate the time and effort that the Associate Editor and both reviewers dedicated to reviewing our manuscript, and we are grateful for the constructive and inspirative feedback provided. We acknowledge that feedback, which greatly contributed to improving the content of our manuscript and enhancing its quality.
Funding
This research was funded mainly by the Ministry of Education, Youth and Sports of the Czech Republic, scheme INTER-EXCELLENCE, INTER-ACTION, grant number LTAUSA19113, titled “Genetic variability of hyper-spectral reflectance in Scots pine ecotypes for selection of drought-resistant individuals.” This work was partially supported by the Ministry of Education, Youth and Sports of the Czech Republic with the European Regional Development Fund-Project “SINGING PLANT” (no. CZ.02.1.01/0.0/0.0/16_026/0008446). The final phase of manuscript writing was supported for JA and ZL by the Národní agentura pro zemědělský výzkum (NAZV) grant no. QL24010275.
Author information
Authors and Affiliations
Contributions
Conceptualisation ML, JA; Methodology JČ, MP, EN, JCH, IK, KP, JS; Software JČ, MP, EN; Validation JČ, EN; Writing - Original Draft EN, JČ, MP, IK, KP, ZL; Writing – Review & Editing EN, JČ, MP, IK, JCH, ZL, JS, JA, ML.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Neuwirthová, E., Chuchlík, J., Pikl, M. et al. Overcoming difficulties in segmentation of hyperspectral plant images with small projection areas using machine learning. Sci Rep (2026). https://doi.org/10.1038/s41598-025-31952-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-31952-9