Abstract
Establishing an environmental DNA (eDNA) reference library at regional and local scales is essential not only for accurate biodiversity assessment but also for comprehensive long-term monitoring. To date, genetic diversity studies of the Chinese minnow (Rhynchocypris oxycephalus) have largely been restricted to China, leaving substantial knowledge gaps across its broader distribution, including South Korea. Hence, the present study identified suitable regions for guiding eDNA surveys and non-invasive sampling, based on documented occurrences retrieved from the IUCN Geospatial Conservation Assessment Tool (GeoCAT). The newly designed primer pairs successfully amplified long mitochondrial fragments (~ 1 kb) of the cytochrome b (Cytb) and 16S ribosomal RNA (16S rRNA) genes. The generated sequences revealed 29 haplotypes from 41 Cytb sequences and 13 haplotypes from 21 16S rRNA sequences, corresponding to high intraspecific genetic diversity (5.57% for Cytb and 2.46% for 16S rRNA), thereby indicating potential cryptic diversity of R. oxycephalus in South Korea. The phylogenetic analyses, combined with multiple species delimitation methods, resolved several putative molecular operational taxonomic units and highlighted a distinct genetic clade in the Seomjin River basin, likely driven by microhabitat-specific evolutionary processes. In addition, the shared haplotypes across catchments of different river basins indicate either ongoing gene flow or anthropogenic influences contributing to genetic admixture of R. oxycephalus. The time-calibrated phylogenetic analyses also suggest that historical geographic changes and ancient river networks, from the Early Miocene to the Late Pliocene, likely facilitated the diversification of R. oxycephalus across China, the Korean Peninsula, and Japan. Overall, this study represents the first eDNA-based assessment of R. oxycephalus diversity in South Korea, while also providing new evolutionary insights from a broader geographic context in China and Japan. Given the complexity of multiple river networks in South Korea, further investigations using multiple genetic markers are recommended to enhance understanding of this cyprinid species phylogeography in the region.
Similar content being viewed by others
Introduction
Fish monitoring plays a critical role in understanding freshwater biodiversity, assessing ecosystem health, and guiding conservation and resource management strategies1,2. It provides baseline data for detecting population trends, habitat degradation, and the impacts of climate change or anthropogenic disturbances3,4. Traditionally, monitoring has relied on methods such as electrofishing, netting, and visual surveys, which can be labor-intensive, taxonomically constrained, and potentially harmful to both target and non-target organisms and their associated habitats5. In contrast, environmental DNA (eDNA) has emerged as a transformative and non-invasive tool for fish monitoring in aquatic ecosystems6. This method enables detection of multiple species from a single water sample by analyzing trace DNA shed by organisms into the environment, significantly reducing disturbance to aquatic ecosystems7,8. This is particularly valuable in sensitive or protected areas, where minimizing ecological impact is essential9. Moreover, eDNA also improves detection sensitivity for rare, elusive, or low-abundance species, which are rarely detected by conventional methods10,11. eDNA metabarcoding further enhances this approach by enabling fine-scale spatial and temporal assessments of fish community composition, supporting an efficient and repeatable long-term biodiversity monitoring12,13. However, accurate species identification depends on the availability of comprehensive local or regional reference databases containing verified genetic sequences linked to taxonomically validated specimens. Such databases are essential for assigning unknown sequences with high confidence to the species level14.
Freshwater fish often exhibit greater genetic variation compared to their marine counterparts due to historical isolation driven by terrestrial barriers and watershed boundaries15. The Chinese minnow (Rhynchocypris oxycephalus) is a small riverine cyprinid fish that typically inhabits cold, well-oxygenated headwaters at high elevations, and is widely distributed across East Asia, including China, Japan, and the Russian Federation16,17,18. In South Korea, it occurs in various river systems and tributaries where temperatures remain below 20 ℃ and dissolved oxygen levels are high19. Due to these habitat preferences, R. oxycephalus is also regarded as a bioindicator species for assessing water quality and river ecosystem health20,21,22,23,24. Several taxonomic and phylogenetic studies have highlighted complex evolutionary relationships within the genus Rhynchocypris25,26,27. In particular, the morphological similarity and overlapping distribution range of R. oxycephalus and R. lagowskii in the Amur River and its tributaries complicate their differentiation26,28. Thus, several molecular studies based on the partial and complete mitochondrial genome of R. oxycephalus have been conducted in recent years23,27,29,30. In South Korea, only a limited number of molecular studies on Rhynchocypris have been conducted to reassess their systematics and diversity31,32. Mitogenomic comparisons indicate a closer genetic affinity between Korean and Japanese R. oxycephalus populations than with those from China, a pattern likely shaped by historical land bridges and vicariance events23. The molecular data have also provided new insights into the genetic diversity of various Rhynchocypris species, including R. oxycephalus, contributing to the development of sustainable conservation strategies29. Furthermore, mitochondrial DNA based investigation also evidenced that the introgression between closely related species, such as R. czekanowskii and R. lagowskii, further complicates species boundaries and highlights the dynamic evolutionary processes within the genus25.
Currently, most genetic data for R. oxycephalus are derived from China, with only one complete mitochondrial genome from South Korea and a few partial gene sequences from Japan archived in the GenBank database. Given this limited reference coverage, a comprehensive nationwide assessment of R. oxycephalus genetic variation in South Korea is needed to clarify its genetic diversity and phylogenetic placement. Such data are also essential for supporting eDNA-based biodiversity monitoring, particularly for South Korean populations. However, most previous eDNA-based studies have used universal MiFish primers targeting a short fragment of the mitochondrial 12S ribosomal RNA gene, which often lacks sufficient resolution to distinguish closely related species and their populations33,34,35. This limitation further highlights the need for comprehensive reference database with longer sequences to ensure reliable taxonomic assignments as well as evolutionary or biogeographical inference. The prior genetic ivestigation of R. oxycephalus was accomplished from China and Japan by using two well-known markers cytochrome b (Cytb) gene and 16S ribosomal RNA (16S rRNA) genes, but both studies relies on their native population18,25. However, as the Korean Peninsula also accommodates R. oxycephalus and other indigenous Rhynchocypris species (e.g., R. keumkang and R. semotilus), thus warranting to design a new primer sets by accommodating South Korean species for amplification of both Cytb and 16S rRNA for the detection of R. oxycephalus from eDNA. Therefore, the present study aims to (i) assess the genetic diversity of R. oxycephalus across different river basins in South Korea using longer mitochondrial gene fragments derived from eDNA samples and (ii) investigate the phylogenetic relationships and potential lineage diversification of South Korean R. oxycephalus populations across different riverine systems in comparison with conspecific populations from China and Japan. These genetic data will provide a critical foundation for developing the first eDNA-based detection strategies and for enhancing our understanding of the phylogeographic structure of this cyprinid species in South Korea. Moreover, this genetic information will aid in elucidating the evolutionary history of R. oxycephalus within the distinct biogeography of the Korean Peninsula, shaped by its complex riverine systems.
Materials and methods
Environmental DNA sampling, and pre-treatment
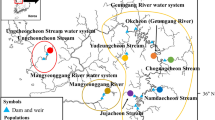
Based on previously documented 237 occurrences records retrieved from the IUCN Geospatial Conservation Assessment Tool (GeoCAT) assessed on 2nd January 2025, the sampling sites were selected in the different river basins including Han, Geum, Nakdong, Seomjin, and Yeongsan River basins in South Korea, that may support distinct populations of the targeted fish species19,36,37,38,39,40. A total of 45 water samples were collected from 21 sampling sites to assess the potential occurrence of eDNA for investigating the genetic diversity of R. oxycephalus (Fig. 1). From each site, 4 L of water was collected in a disposable plastic bottle and immediately placed in an icebox for transport to the laboratory for subsequent filtration. A 500 ml aliquot of each water sample was filtered through a 0.45 μm pore-sized GN-6 membrane filter (47 mm, Pall Corporation, New York, NY, USA). To prevent cross-contamination, the vacuum filtration system was treated with a 10% sodium hypochlorite bleach solution before each filtration procedure. The filtered membranes were then stored in 2 ml centrifuge tubes at −80 ℃. Prior to genomic DNA extraction, each membrane filter was pretreated with 630 µL of ATL buffer and 70 µL of Proteinase K (20 g/L) and homogenized using the FastPrep-2 system (MP Biomedicals™, Irvine, CA, USA).
(A) The map representing the river networks (blue lines) and the occurrence records of R. oxycephalus in South Korea (red dots) acquired from secondary resources (IUCN GeoCAT). (B) Sampling sites of eDNA samples in five different river basins in South Korea. The map was generated by using ArcGIS software version 10.6 (https://www.arcgis.com/index.html) and edited manually in Adobe Photoshop CS 8.0. The river basins shape file was retrieved from VWorld Data Center (VDC) (https://www.vworld.kr/).
Primer designing of the targeted loci
The present study designed two new primer pairs by aligning all available Rhynchocypris species sequences retrieved from the global GenBank database. For Cytb, (Rhox_Cytb_F: 5′-CAGCKCTTTGAAATTTCGGCTCCCTA-3′; Rhox_Cytb_R: 5′-GRCGAAAGGTTAGTCCTCGTTGTTTTGAM-3′) and for 16S rRNA (Rhox_16S_F: 5′-RCACACCCCRAACCAARRRATATTATCRC-3′; Rhox_16S_R: 5′-CCTTGGGTAACTTGGTYCGTTGG-3′). The newly designed primer pairs can amplify the target genes, Cytb (~ 818 bp) and 16S rRNA (~ 908 bp) of the dominant haplotype. The feasibility of the newly designed primer pairs was further validated by amplifying the target genes from genomic DNA extracted from morphologically identified R. oxycephalus collected from a South Korean River.
DNA extraction, PCR, and sequencing
The genomic DNA was extracted from each homogenized filtrate using DNeasyⓇ Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols. Both quantification and integrity of the genomic DNA was checked through NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The PCR mixture (30 µL) contained 1 µL of template DNA, 3 µL of 10x EX Taq buffer, 3 µL of dNTPs (each 2.5 mM), 1 µL of forward and reverse primers (10 pmol), 0.3 µL of EX Taq Hot Start polymerase enzyme (Takara, Tokyo, Japan), and the remaining quality DNase/RNase-free water maintaining the final volume. The PCR conditions for both Cytb and 16S rRNA gene amplification included an initial denaturing at 94 ℃ for 3 min, followed by 40 cycles of 94 ℃ for 30 s, 64 ℃ for 30 s, 72 ℃ for 1 min, and a final extension of 72 ℃ for 5 min. The PCR products were visualized by 1.5% agarose gel electrophoresis. The PCR products were further purified using AccuPrepⓇ PCR/Gel DNA Purification Kit (Bioneer, Daejeon, Republic of Korea). Furthermore, sequencing procedures were conducted bi-directionally by using Sanger sequencing at Macrogen (https://dna.macrogen.com/) (Daejeon, Republic of Korea).
Dataset construction, genetic distance, and haplotype analyses
To make a consensus sequence of each sample, the bi-directional chromatograms were screened through MEGA v1141. The low-quality bases were trimmed from the 5′ and 3′ ends of both forward and reverse sequences using SeqScanner v 1.0 (Applied Biosystems Inc., CA, USA) to ensure unambiguous base calls. The generated sequences were examined through nucleotide BLAST search (https://blast.ncbi.nlm.nih.gov) to avoid the insertion/deletions. Both forward and reverse complements of reverse sequences were aligned through ClustalX software to make a consensus sequence, ensuring a Phred quality score of ≥ 3042. The final generated sequences were submitted to the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/). To examine the genetic diversity, population structure, phylogenetic analyses, and MOTUs estimation, two different datasets were built-up: (i) 209 sequences (41 generated and 168 database) for Cytb, and (ii) 73 sequences (21 generated and 52 database) for 16S rRNA gene (Supplementary Tables S1, S2, S3). The DNA sequences (Accession Nos. NC_018817, AP011418) of Oreoleuciscus potanini and Pseudaspius brandtii (Cypriniformes: Cyprinidae) were used as the outgroup in both the datasets. The inter- and intra- genetic distances were calculated using Kimura 2 parameter (K2P) by MEGA v11. The genetic diversity indices, polymorphic sites (P), number of haplotypes (H), average number of nucleotide differences (K), haplotype diversity (Hd), and nucleotide diversity (π) were computed by using DnaSP v6. 12.0343. The Templeton Crandall Singh (TCS) networks of all haplotypes were constructed by using Popart44,45.
Phylogenetic analyses and MOTUs estimation
The best-fit model (GTR + G + I) was selected based on the lowest Bayesian Information Criterion (BIC) scores for both Cytb and 16S rRNA dataset through PartitionFinder 2 and JModelTest v246,47. All three tree building methods, Neighbor-joining (NJ), Maximum likelihood (ML), and Bayesian inference (BI) were adopted to visualize the cladistic relationship of Rhynchocypris species. The NJ and ML phylogenies were estimated in MEGA v11 and PhyML 3.0 web server respectively with 1000 bootstrap supports48. However, the BI was performed using MrBayes 3.2.7, with a model configuration of nst = 6, involving one cold and three hot Metropolis-coupled Markov Chain Monte Carlo (MCMC) chains. The analysis was conducted over 1,000,000 generations, with trees sampling at every 100 generations, and 25% of the samples were excluded as burn-in49. The final topologies (NJ, ML, and BI) were visualized using the iTOL v4 web server (https://itol.embl.de/login.cgi)50.
In addition, to identify the molecular operational taxonomic units (MOTUs), four species delimitation methods, Automatic Barcode Gap Discovery (ABGD), Assemble Species by Automatic Partitioning (ASAP), Poisson Tree Process (PTP) and Generalized Mixed Yule Coalescent (GMYC) were further applied for the datasets51,52,53,54. The ABGD and ASAP use clustering approaches based on genetic distance for MOTUs estimation, while the PTP and GMYC are the coalescent-based species delimitation methods which execute model-based approaches on ultra-metric gene tree. The ABGD, ASAP, and PTP analyses were performed by using iTaxoTools 0.1 tool with default parameters55. Both Kimura (K80) as well as Jukes–Cantor (JC69) models were tested for ABGD and ASAP analyses. The ultra-metric ML tree was used in PTP analysis, while, the GMYC analyses were performed by using both BEAST v1.10.4 for conducting the ultra-metric BI phylogeny and the R package ‘splits’ for the species delimitation analyses56,57.
Divergence time estimation
The RelTime ML method was employed to calculate the divergence times of lineages within the Rhynchocypris species and population58. It is evidenced that RelTime offers both computational efficiency and accuracy in estimating TimeTrees, making it particularly suitable for large datasets by reducing substantial computational time59,60,61. As the present study aimed to elucidate the divergence time of R. oxycephalus from South Korea in comparison with China and Japan, as well as among different populations within South Korean riverine systems, we utilized divergence time estimates from a previous study18. The TimeTree was constructed using the unique haplotypes of Cytb dataset with multiple known calibration constraints of R. oxycephalus, particularly from Chinese populations (2.56 Ma for clade B1 and B2 and 2.98 Ma for clade C1 and C2) resulted in the previous study18. After inputting the sequence data, the ML topology (newick format) served as the baseline tree. After assigning the outgroup taxa, the TimeTree computation integrated calibration constraints using the calibration editor in MEGA v11.
Results
Primer efficacy and similarity search
The present study successfully amplified substantial fragments of the Cytb (~ 818 bp) and 16S rRNA (~ 908 bp) genes from environmental DNA (eDNA) samples using newly designed primer pairs. Out of 45 samples, 41 samples showed successful amplification of the Cytb genes and 21 samples exhibited amplification of 16S rRNA genes. Chromatograms showed clear trace signals with no baseline noise or mixed base peaks, indicating that the single sequence for the dominant haplotype was amplified in each water sample62. The generated sequences have been deposited in GenBank under accession numbers PQ330261-PQ330301 for the Cytb gene and PQ333020-PQ333040 for the 16S rRNA gene, respectively (Supplementary Table S1). Further, the nucleotide BLAST (BLASTn) results of the generated sequences showed a wide range of similarity with published sequences for both Cytb (94.37% to 99.15%) and 16S rRNA (96.57% to 99.34%) genes (Supplementary Tables S4, S5).
Genetic diversity
The studied species, R. oxycephalus showed 5.57% and 2.46% intra-species genetic distances in Cytb and 16S rRNA gene, respectively (Table 1). Further, R. oxycephalus also maintained inter-species genetic distance ranging from 7.46% (R. jouyi) to 18.77% (R. keumkang) in Cytb and 2.37% (R. jouyi) to 6.78% (R. keumkang) in 16S rRNA (Table 1). Comparing the genetic distance of R. oxycephalus across different countries, the intra-population genetic distance of the South Korean R. oxycephalus was 4.08% for the Cytb gene and 1.81% for the 16S rRNA gene (Table 2). Additionally, the inter-population genetic distance, estimated among geographically distinct populations from China, Korea, and Japan, ranged from 5.54% to 7.65% for Cytb and 2.27% to 2.89% for the 16S rRNA gene, indicating moderate genetic divergence across its range countries. (Table 2). Furthermore, comparing the genetic distance of R. oxycephalus populations across different river basins in South Korea, the Han River basin exhibited the highest intra-population genetic distance for the Cytb gene (4.34%), followed by the Seomjin River basin (0.59%) and the Nakdong River basin (0.03%). A similar pattern was also observed for the 16S rRNA gene, with the Han River basin showing the highest genetic distance (2.09%) compared to the Nakdong River basin (Supplementary Table S6). Moreover, the inter-population genetic distance ranged from 3.62% (Han River basin vs. Nakdong River basin) to 5.68% (Geum River basin vs. Yeongsan River basin) for the Cytb dataset and from 1.85% (Han River basin vs. Nakdong River basin) to 3.53% (Han River basin vs. Seomjin River basin) for the 16S rRNA dataset (Supplementary Table S6). Moreover, across the surveyed catchment sites, R. oxycephalus exhibited substantial genetic divergence based on both Cytb and 16S rRNA genes (Table 3). The Yangyang-namdaecheon (YN) population showed the highest intra-population genetic distance for the Cytb gene (4.46%), followed by Samcheok-osipcheon (SO; 2.83%). In contrast, comparatively low intra-population divergence was observed in the Seom River (SR; 0.90%), Oknyeodongcheon (OD; 0.42%), Sincheon (SN; 0.37%) and Gilancheon (GA; 0.03%). Due to limited sample sizes, intra-specific genetic distances for the 16S rRNA gene could be estimated for all catchment sites, but only two SO and GA showed measurable values of 1.30% and 0.08%, respectively (Table 3). The inter-population genetic distances varied widely among catchment sites, ranging from 0.67% (Hwangjeoncheon, HJ vs. OD) to 6.00% (SN vs. YN) for the Cytb gene, and from 0.88% (Hotancheon, HT vs. YN) to 4.51% (OD vs. SN) for the 16S rRNA gene (Table 3). More specifically, the catchment sites on the eastern slope of the Taebaek mountains (YN and SO), although administratively classified under the Han River basin, exhibited substantial genetic differentiation from the SN and SR catchment sites on the western slope of the Taebaek mountains, with Cytb distances ranging from 4.28% to 6.00% and 16S rRNA distances from 3.89% to 4.37%. The two eastern slope catchments (YN and SO) also showed pronounced divergence from each other (4.99% for Cytb and 2.57% for 16S rRNA). The GA catchment of the Nakdong River basin showed moderate divergence from SO (2.80%) and the highest distance from Yeongsan River (YR) in the Yeongsan River basin (5.03%). Within the Seomjin River basin, the HJ and OD populations were closely related, with only 0.67% Cytb divergence. Meanwhile, SR in the Han River basin exhibited relatively close genetic affinity (1.43%) with HT in the Geum River basin for the Cytb gene. Nevertheless, the YR catchment in the Yeongsan River basin showed comparatively close distances (3.82%) with YN in the Han River basin (Table 3).
Country-level and Korean river systems haplotype diversity
On country-level analyses of R. oxycephalus, 42 Cytb sequences from South Korea revealed 30 haplotypes with Hd = 0.95 and π = 0.037 (Table 2, Supplementary Table S7). However, 146 sequences from China and four sequences from Japan revealed 92 and 3 haplotypes, respectively. Further, considering 16S rRNA gene, 22 sequences from South Korea revealed 14 haplotypes with Hd = 0.92 and π = 0.013 (Table 2, Supplementary Table S8). However, 28 sequences from China and nine sequences from Japan depicted 27 and eight haplotypes, respectively. In different river basins in South Korea, 29 Cytb sequences of R. oxycephalus from the Han River basin revealed 25 haplotypes with Hd = 0.99 and π = 0.039 (Supplementary Table S6). Further, eight sequences from the Nakdong River basin and three sequences from the Seomjin River basin revealed a single haplotype and three haplotypes, respectively. In addition, 11 sequences of 16S rRNA gene from the Han River basin and nine sequences from the Nakdong River basin revealed 10 and four haplotypes, respectively (Supplementary Table S6). Furthermore, comparing each catchment sites in different river basins in South Korea, eight sequences from YN, 12 sequences from SO and two sequences from SN revealed eight, 10, and two haplotypes, respectively (Table 3). Also, six sequences from SR and two sequences from OD revealed four and two haplotypes, respectively (Table 3). In addition, eight sequences of 16S rRNA gene from SO revealed seven haplotypes with Hd = 0.96 and π = 0.010 (Table 3).
The TCS haplotype networks revealed nine distinct clusters for the Cytb dataset and seven clusters for the 16S rRNA dataset, each corresponding to populations from different riverine systems across the Korean Peninsula (Figs. 2 and 3). For the Cytb gene, Cluster-1 comprised shared haplotypes, including sequences from the SO in the Han River basin and the GA in the Nakdong River basin (Fig. 2). In contrast, Cluster-2 included sequences from the YN and the SR, both within the Han River basin, as well as from the HT in the Geum River basin. Further, the Cluster-3 encompassed sequences from the YN and the Mt. Suri region of the Han River basin, along with those from the Yeongsan River basin. Meanwhile, Cluster-4 formed distinct group with sequences originating from the HJ and OD of the Seomjin River basin. Furthermore, the Cluster-5 to Cluster-9 formed distinct groups with sequences originating from different streams (SN, YN, and SO) in the Han River basin (Fig. 2). For the 16S rRNA gene, Cluster-1 similarly represented shared haplotypes, including sequences from the SO in the Han River basin and the GA in the Nakdong River basin (Fig. 3). Similarly, the Cluster-2 comprised sequences from the YN of the Han River basin and the HT of the Geum River basin. In contrast, Cluster-3 is defined by unique haplotypes originating from the OD in the Seomjin River basin, whereas the Cluster-4 to Cluster-7 were distinguished by haplotypes specific to SO, the Mt. Suri region, and SN of the Han River basin (Fig. 3).
The Templeton Crandall Singh (TCS) haplotype network demonstrates the relationship among the Cytb sequences revealed haplotypes of R. oxycephalus in South Korea. Each circle indicates the haplotype of R. oxycephalus and is colored based on the river basin. Circle sizes are proportional to the haplotype frequencies. The number of mutations is represented by the hatch marks. The black circles denote the median vectors. The red dashed lines indicate clade and labeled with the river basin included in each clade. The circle points of map indicate the river location in this study. The map was generated by using ArcGIS software version 10.6 (https://www.arcgis.com/index.html). The map and haplotypic network were merged manually in Adobe Photoshop CS 8.0.
The TCS haplotype network demonstrates the relationship among the 16S rRNA haplotypes of R. oxycephalus from South Korea. Each circle indicates the haplotype of R. oxycephalus from Korea and is colored based on the river basin. Circle sizes are proportional to the haplotype frequencies. The number of mutations is represented by the hatch marks. The black circles denote the median vectors. The red dashed lines indicate clade and labeled with the river basin included in each clade. The circle points of map indicate the river location in this study. The map was generated by using ArcGIS software version 10.6 (https://www.arcgis.com/index.html). The map and haplotypic network were merged manually in Adobe Photoshop CS 8.0.
Species delimitation and phylogenetic interpretation
All four species delimitation analyses identified multiple MOTUs for the Cytb and 16S rRNA datasets of R. oxycephalus sampled from China, South Korea, and Japan (Figs. 4 and 5, Supplementary Tables S9-S16). The clustering patterns of MOTUs inferred from Cytb and 16S rRNA were largely congruent and corresponded well with the haplotype clusters identified across different river basins in South Korea (Supplementary Figs. 1, 2). All delimitation methods detected more than one MOTU within populations from the Han River basin in both Cytb and 16S rRNA datasets (Supplementary Figs. 1, 2). The Cytb sequence from the Geum River basin was resolved as a distinct MOTU by ABGD, ASAP, and PTP, but grouped with Han River sequences under GMYC, in agreement with the haplotype clustering pattern (Supplementary Fig. 1). A similar trend was observed in the 16S rRNA dataset, supported by both GMYC and PTP (Supplementary Fig. 2). The generated sequences from the Nakdong River basin were resolved as a single MOTU by at least two methods and exhibited close genetic similarity with a subset of Han River sequences in both Cytb and 16S rRNA (Supplementary Figs. 1, 2). By contrast, all sequences from the Seomjin River basin formed a distinct cluster in the haplotype network, which was consistently supported by all delimitation methods across both markers. Additionally, the Cytb sequence from the Yeongsan River basin clustered with those from the Han River basin, a relationship further supported by more than three delimitation methods (Supplementary Fig. 1). The ML phylogenetic trees based on both Cytb and 16S rRNA confirmed the monophyly of R. oxycephalus, which was distinctly separated from other Rhynchocypris species with strong bootstrap support (Figs. 4 and 5). Furthermore, NJ and BI analyses yielded concordant topologies, corroborating the phylogenetic relationships among Rhynchocypris species (Supplementary Figures S3-S6). Notably, R. jouyi clustered unexpectedly with R. oxycephalus, forming a species complex in the present analyses.
The Maximum-Likelihood tree based on Cytb sequences showed the phylogenetic relationship of R. oxycephalus with other congeners. The sequences of R. oxycephalus are colored based on the country; Green indicates Korea, pink indicates China, and gray indicated Japan. Bootstrap values of each node are indicated by orange dots. The topology was constructed through PhyML 3.0 web server (http://www.atgc-montpellier.fr/phyml/) and edited by web based iTOL tool (https://itol.embl.de/itol.cgi). The MOTUs estimation by different species delimitation methods (GMYC, PTP, ABGD and ASAP) are presented by different color side bars. Species delimitation analyses were performed by R package ‘splits’ and iTaxoTools 0.1 tool with default parameters and edited manually in Adobe Photoshop CS 8.0.
The Maximum-Likelihood tree based on 16S rRNA sequences showed the phylogenetic relationship of R. oxycephalus with other congeners. The sequences of R. oxycephalus are colored based on the country; Green indicates Korea, pink indicates China, and gray indicated Japan. Bootstrap values of each node are indicated by orange dots. The topology was constructed through PhyML 3.0 web server (http://www.atgc-montpellier.fr/phyml/) and edited by web based iTOL tool (https://itol.embl.de/itol.cgi). The MOTUs estimation by different species delimitation methods (GMYC, PTP, ABGD and ASAP) are presented by different color side bars. Species delimitation analyses were performed by R package ‘splits’ and iTaxoTools 0.1 tool with default parameters and edited manually in Adobe Photoshop CS 8.0.
Divergence time and lineage diversification
The time tree analysis indicates that R. oxycephalus diverged from other Rhynchocypris species during the Pliocene, approximately 4.8 million years ago (Ma) (Fig. 6). The divergence between South Korean populations of R. oxycephalus and the matrilineal populations from China (C1 and C2) is estimated to have split around 3.2 Ma, while the other matrilineal populations from China (B1 and B2) diverged earlier. Furthermore, the estimated divergence time between the South Korean populations of R. oxycephalus and the complex group comprising R. oxycephalus and R. jouyi from Japan is approximately 1.7 Ma (Fig. 6). Further, in this dataset, R. keumkang, endemic to the South Korea, diverged in the early Miocene (~ 16 Ma), making it one of the oldest lineages in this species group (Fig. 6). Conversely, other congeners, such as R. czekanowskii, R. lagowskii, R. percnurus, R. semotilus, and R. steindachneri, show divergence times from the late Pliocene to late Miocene (~ 2.3 to 6.5 Ma) in the present time tree analyses, with broad geographic distributions in Kazakhstan, Mongolia, Russia, China, Japan, and the Korean Peninsula.
The ML TimeTree based on RelTime approach illustrating the approximate divergence times of R. oxycephalus at both species and population levels. The estimated divergence times are indicated on each branch by black numerical values. Red rhombus symbols represent calibration points derived from previous studies18. Side bars are color-coded according to the country of origin: green for Korea, pink for China, and gray for Japan. Each circle adjacent to the sequence accession numbers generated in this study denotes the respective river basin catchment. The upper panel map illustrates the hypothesized ancient river networks linking the Korean Peninsula and China, based on previous studies73,74,87 and the East Sea opening tectonic model described in the National Atlas of Korea Ⅱ (2020) (http://nationalatlas.ngii.go.kr/). The lower panel map, created using ArcGIS software version 10.6 (https://www.arcgis.com/index.html), shows distinct catchment sites across major river basins in South Korea. The final figure was manually refined and edited using Adobe Photoshop CS 8.0.
Discussion
Past geological events and current bioclimatic variations have profoundly shaped the genetic diversity and distribution of freshwater species and their populations worldwide18,63,64. Specifically, mountain uplift and the formation of riverine systems in a given region often create substantial geological barriers, promoting genetic diversity within native fish resources and driving speciation over time65,66. The Korean Peninsula has a complex topographic history since Cenozoic and the terrains in this region gradually slope from east to west forming a unique inland riverine trajectory67,68. These geographical and ecological dynamics impose ecological isolation on the diverse freshwater systems in this region19,69. For instance, the demersal fish Koreocobitis naktongensis, endemic to South Korea and restricted to the Nakdong River, exhibits a historical vicariance pattern driven by geographic isolation and subsequent speciation70. Currently, South Korea is hydrologically divided into five principal river basins viz., Han, Geum, Nakdong, Seomjin, and Yeongsan based on the administrative division of the Korean territory (National Geography Information Institute (NGII), http://nationalatlas.ngii.go.kr/). Past geological studies indicate that during the Pleistocene, the southwestern Korean Peninsula was connected to China through ancient river networks and prehistoric lakes64,71,72,73,74. Thereafter, the fluctuations in sea level, particularly during glacial and interglacial periods, contributed to the eventual formation of the Yellow Sea as rising waters inundated the exposed landmass. Similarly, prior to the last glacial maximum, the Japanese archipelago was connected to mainland Asia, enabling the exchange of freshwater fauna between South Korea, China, and Japan. This connection played a crucial role in shaping the evolutionary and biogeographical relationships among these regions’ aquatic ecosystems69,75. Thus, in addition to molecular systematics and population genetic structure, examining the potential evolutionary processes, dispersal patterns, and biogeographical interpretations of any fish species is important. The present genetic investigation of R. oxycephalus across various riverine systems in South Korea, alongside comparative analyses with populations from China and Japan, provides a clearer understanding of the genetic diversity and evolutionary history shaped by prehistoric biogeographic patterns.
Among the 45 water samples analyzed, Cytb sequences were successfully obtained from 41 samples, whereas 16S rRNA sequences were recovered only from 21 samples. This difference likely reflects variation in amplification efficiency, possibly due to differences in amplicon size between the two targeted genes. Inspection of all long-read chromatograms (> 800 bp) revealed no ambiguous base calls or nuclear-mitochondrial DNA segments, indicating that each PCR reaction yielded a single dominant haplotype represented by each generated sequence. This is consistent with the understanding that, although eDNA contains genetic material from multiple individuals, intact long fragments of a dominant haplotype can still be recovered and used for biodiversity assessment76. Furthermore, the BLASTn searches showed a broad range of similarity with public sequences (94.37–99.15% for Cytb and 96.57–99.34% for 16S rRNA). This is likely due to the lack of R. oxycephalus sequences from diverse localities in South Korea, with only one mitogenome (Accession No. MK208924) from Mt. Suri represented in GenBank, while most reference sequences are originated from Japan or China18,25. These results underscore the need to expand local and regional reference sequence databases to improve genetic diversity estimation, cryptic diversity detection, and phylogeographic interpretation77.
Further, the detected high intraspecific genetic diversity (5.57% for Cytb and 2.46% for 16S rRNA), highlighting the presence of multiple populations or potential cryptic diversity of R. oxycephalus in South Korea. A comparable pattern of high intraspecific genetic diversity was also observed in the Chinese (4.33%) and Japanese (5.02%) populations of R. oxycephalus, based on the present analysis of available Cytb sequences from GenBank, as well as in earlier studies that reported values ranging from 6.5% to 7.4%18. The elevated genetic divergence may be attributed to its fragmented distribution across geologically isolated landmasses in China, Korean Peninsula, and Japan during the distant past78,79. Notably, high intra-population genetic distance was also observed in the Han River basin (4.34%), comparable to values reported for populations in the Yangtze River basin, China (5.5% to 5.6%)18. This pronounced intra-population divergence may resulted from microhabitat differentiation among tributaries within these two large river basins in China and South Korea.
It can also be assumed that the high intraspecific genetic distances observed among populations across geographically distant localities within the Han River basin, particularly those separated by the two slopes of the Taebaek Mountain range, are likely influenced by ecological barriers that promote the development of unique genetic traits in freshwater taxa80,81. Specifically, populations from YN and SO, originating from the eastern slope of the Taebaek Mountains and flowing toward the East Sea, exhibit pronounced genetic differentiation (4.28–6.00% in Cytb; 3.89–4.34% in 16S rRNA) from populations such as SR and SN, which originate from the western slope and form the main Han River basin before discharging into the Yellow Sea. The substantial genetic divergence observed between YN and SO, both located on the eastern slope, may result from additional ecological barriers imposed by intervening mountain ranges within the Taebaek Mountain. Conversely, minimal genetic differentiation is observed between SO (Han River basin) and GA (Nakdong River basin), which may reflect historical connectivity facilitated by the Osipcheon fault during the Calabrian stage of the Pleistocene, allowing gene flow between Han and Nakdong River populations, followed by subsequent isolation due to later tectonic events80,82.
In contrast, low genetic diversity was observed in R. oxycephalus populations in the Nakdong River basin (0.03% in Cytb and 0.08% in 16S rRNA), with a similar pattern detected in the Seomjin River basin based on Cytb data. Although the sampling sites in the Seomjin River basin are separated by more than 75 km, the inter-site distances within the Nakdong River basin are comparatively smaller (14 km), suggesting that population differentiation in the Nakdong may not have been adequately captured due to the limited spatial coverage of the sampling design. While the present study provides preliminary insights into the genetic diversity of R. oxycephalus in South Korea using eDNA analysis, it highlights the need for more comprehensive sampling, particularly in the Nakdong, Geum, and Yeongsan River basins, to more accurately assess the genetic diversity of this species.
Nevertheless, the phylogenetic analyses based on Cytb and 16S rRNA genes clearly delineated R. oxycephalus from other congeners, consistent with previous study25. Notably, R. jouyi, considered a valid species from Japan, exhibited ambiguous clustering and a strong genetic affinity with R. oxycephalus in the current phylogenetic analyses83,84. Thus, the taxonomic status of R. jouyi warrants further investigation using integrated taxonomic approaches, including the examination of topotypic samples. Consequently, based on the present phylogenetic evidence, this study refutes the validity of R. jouyi as a distinct species and supports earlier hypotheses suggesting that it represents the easternmost population of R. oxycephalus, potentially expanding into Japan25,29,85,86. The present study also indicates that diversification of R. oxycephalus populations across river basins in South Korea began in the late Pliocene (~ 2.7 Ma) and continues, as reflected by current phylogenies.
Furthermore, as the SO population in the Han River basin and the GA population in the Nakdong River basin share a common haplotype, this gene flow may have been facilitated by crustal fluctuations along the fault zone between the Nakdong River basin and the eastern region of the Han River basin80,87,88. The YN and SR population of the Han River basin also shows potential connections with the HT in Geum and YR in Yeongsan River basins, as reflected in the haplotype networks and time tree. These connections likely indicate the historical links between these river basins through ancient riverine networks that once connected the Han, Geum, and Yeongsan River basins before the Last Glacial Maximum72,87. A similar phylogeographic pattern has been observed in other freshwater fishes through population genetic analyses previously72,73. Based on the mixed haplotype clustering observed across distant localities in different river basins of South Korea in this study, it can be further assumed that R. oxycephalus may be experiencing ongoing anthropogenic pressures. The national news channels frequently highlight that fish larvae are often released into various riverine systems to enhance population recovery, support aquaculture practices and excellent water quality, as well as provide human benefits (https://m.metroseoul.co.kr/article/20250924500612). Although these releases are generally conducted through several fishery management project, the extent of molecular-level assessment or genetic monitoring prior to stocking remains poorly understood. This lack of genetic evaluation may partly explain the observed genetic admixture of R. oxycephalus across geographically distant riverine and drainage systems in South Korea. Remarkably, the Seomjin River basin population exhibits distinct haplotypes and forms a separate clade in the time tree, diverging from populations in the Han, Geum, and Yeongsan River basins around 2.6 Ma. However, given the restricted populations in the HJ and OD of the Seomjin River basin, it can be inferred that R. oxycephalus may have established a distinct population there, likely due to the utilization of specific ecological envelop. In addition, based on the phylogenetic placement and divergence times, the present study supports earlier hypotheses, confirming that R. jouyi is the easternmost population of R. oxycephalus distributed in Japan25,29,85,86. Since the Japanese archipelago separated from the mainland (Korean Peninsula) during the early Miocene, it is presumed that the ancestor of both the South Korean and Japanese populations of R. oxycephalus separated during that period and subsequently evolved independently, leading to distinct populations in these isolated biogeographic regions.
Conclusion
This study represents the first population genetic investigation of R. oxycephalus using eDNA across different river basins in South Korea. The findings revealed a distinct population of R. oxycephalus in the Seomjin River basin, while populations from other river basins exhibited substantial genetic mixing. The phylogenetic analysis based on two mitochondrial genes further supported the monophyletic grouping of R. oxycephalus and highlighted its phylogenetic relationships with other congeners. These results could be valuable for refining the conservation status of R. oxycephalus in East Asia, specifically for the IUCN/SSC Freshwater Fish Specialist Group (FFSG). Additionally, the study provided insights into the divergence time and potential evolutionary patterns of R. oxycephalus across a broad geographical range, including China, the Korean Peninsula, and Japan. The time tree analysis suggested that ancient river networks before early Miocene to Late Pliocene likely facilitated gene flow between China, the Korean Peninsula, and Japan, contributing over time to the formation of distinct populations or species complexes, particularly in Japan. Considering the complex river network in South Korea, this study recommends further investigations of the genetic diversity of R. oxycephalus through expanded sampling and increased replication, to enhance our understanding of the phylogeography of this species in the region.
Data availability
The generated sequencing data has been deposited in NCBI GenBank (https://www.ncbi.nlm.nih.gov/) with the following accession numbers PQ330261 to PQ330301 and PQ333020 to PQ333040.
References
Karr, J. R. Assessment of biotic integrity using fish communities. Fisheries 6, 21–27. https://doi.org/10.1577/1548-8446(1981)006%3C0021:AOBIUF%3E2.0.CO;2 (1981).
Reynolds, J. D., Dulvy, N. K., Goodwin, N. B. & Hutchings, J. A. Biology of extinction risk in marine fishes. Proc. R Soc. B-Biol Sci. 272, 2337–2344. https://doi.org/10.1098/rspb.2005.3281 (2005).
Dudgeon, D. et al. Freshwater biodiversity: importance, threats, status and conservation challenges. Biol. Rev. 81, 163–182. https://doi.org/10.1017/S1464793105006950 (2006).
Jackson, D. A., Peres-Neto, P. R. & Olden, J. D. What controls who is where in freshwater fish communities the roles of biotic, abiotic, and Spatial factors. Can. J. Fish. Aquat. Sci. 58, 157–170. https://doi.org/10.1139/f00-239 (2001).
Bonar, S. A., Hubert, W. A. & Willis, D. W. Standard Methods for Sampling North American Freshwater Fishes (American Fisheries Society, 2009).
Deng, J. et al. eDNA metabarcoding reveals differences in fish diversity and community structure in Danjiang river. Sci. Rep. 14, 29460. https://doi.org/10.1038/s41598-024-80907-z (2024).
Deiner, K. et al. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. https://doi.org/10.1111/mec.14350 (2017).
Thomsen, P. F. et al. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS One. 7, e41732. https://doi.org/10.1371/journal.pone.0041732 (2012).
Goldberg, C. S. et al. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307. https://doi.org/10.1111/2041-210X.12595 (2016).
Evans, N. T. Fish community assessment with eDNA metabarcoding: effects of sampling design and bioinformatic filtering. Can. J. Fish. Aquat. Sci. 74, 1362–1374. https://doi.org/10.1139/cjfas-2016-0306 (2017).
Jerde, C. L., Mahon, A. R., Chadderton, W. L. & Lodge, D. M. Sight-unseen detection of rare aquatic species using environmental DNA. Conserv. Lett. 4, 150–157. https://doi.org/10.1111/j.1755-263X.2010.00158.x (2011).
Bista, I. et al. Annual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nat. Commun. 8, 14087. https://doi.org/10.1038/ncomms14087 (2017).
Valentini, A. et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 25, 929–942. https://doi.org/10.1111/mec.13428 (2016).
Hubert, N. & Hanner, R. DNA barcoding, species delineation and taxonomy: a historical perspective. DNA Barcodes. 3, 44–58 (2015). https://hal.science/hal-01958691
Bernatchez, L. & Wilson, C. C. Comparative phylogeography of Nearctic and Palearctic fishes. Mol. Ecol. 7, 431–452. https://doi.org/10.1046/j.1365-294x.1998.00319.x (1998).
Bogutskaya, N. Rhynchocypris oxycephalus. The IUCN Red List of Threatened Species e.T159709916A159709987 (2022). https://doi.org/10.2305/IUCN.UK.2022-1.RLTS.T159709916A159709987.en (2022).
Xu, W., Chen, A., Xia, R. & Fu, C. Complete mitochondrial genome of Rhynchocypris cf. lagowskii (Cypriniformes: Cyprinidae). Mitochondrial DNA. 25, 379–380. https://doi.org/10.3109/19401736.2013.809432 (2014).
Yu, D., Chen, M., Tang, Q., Li, X. & Liu, H. Geological events and pliocene climate fluctuations explain the phylogeographical pattern of the cold water fish Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae) in China. BMC Evol. Biol. 14, 1–12. https://doi.org/10.1186/s12862-014-0225-9 (2014).
Jang, M. H., Lucas, M. C. & Joo, G. J. The fish fauna of mountain streams in South Korean National parks and its significance to conservation of regional freshwater fish biodiversity. Biol. Conserv. 114, 115–126. https://doi.org/10.1016/S0006-3207(03)00016-8 (2003).
Bogutskaya, N. G., Naseka, A. M., Shedko, S. V., Vasileva, E. D. & Chereschnev, I. A. The fishes of the Amur river: updated check-list and zoogeography. Ichthyol. Explor. Freshw. 19, 301–366 (2008).
Jang, M. H. et al. The current status of the distribution of introduced fish in large river systems of South Korea. Int. Rev. Hydrobiol. 87, 319–328. https://doi.org/10.1002/1522-2632(200205)87:2/3%3C319::AID-IROH319%3E3.0.CO;2-N (2002).
Liang, Y., Sui, X., Chen, Y., Jia, Y. & He, D. Life history traits of the Chinese minnow Rhynchocypris oxycephalus in the upper branch of Yangtze River, China. Zool. Stud. 53, 1–10. https://doi.org/10.1186/s40555-014-0036-0 (2014).
Park, J., Kim, Y. & Xi, H. The complete mitochondrial genome sequence of Chinese minnow in Korea, Rhynchocypris oxycephalus (Sauvage and Dabry de Thiersant, 1874). Mitochondrial DNA Part. B-Resour. 4, 662–663. https://doi.org/10.1080/23802359.2019.1572472 (2019).
Yu, D. et al. Global climate change will severely decrease potential distribution of the East Asian Coldwater fish Rhynchocypris oxycephalus. Cyprinidae) Hydrobiologia. 700, 23–32. https://doi.org/10.1007/s10750-012-1213-y (2013). Actinopterygii.
Sakai, H. et al. Phylogenetic and taxonomic relationships of Northern Far Eastern Phoxinin minnows, Phoxinus and Rhynchocypris (Pisces, Cyprinidae), as inferred from allozyme and mitochondrial 16S rRNA sequence analyses. Zool. Sci. 23, 323–331. https://doi.org/10.2108/zsj.23.323 (2006).
Yang, S. & Min, M. Sympatry and species status of Moroco lagowskii and M. oxycephalus (Cyprinidae). Korean J. Zool. 31, 56–61 (1988).
Zhang, Z., Cheng, Q. & Ge, Y. The complete mitochondrial genome of Rhynchocypris oxycephalus (Teleostei: Cyprinidae) and its phylogenetic implications. Ecol. Evol. 9, 7819–7837. https://doi.org/10.1002/ece3.5369 (2019).
Takai, N. et al. Species identification of upstream fatminnow Rhynchocypris oxycephalus and downstream fatminnow Rhynchocypris lagowskii, based on PCR-RFLP of mitochondrial DNA. Ichthyol. Res. 59, 156–163. https://doi.org/10.1007/s10228-011-0266-7 (2012).
Nishida, K. et al. Genetic evidence of the native Easternmost distribution limit of Rhynchocypris oxycephala (Actinopterygii: Cypriniformes) and its introduction to rivers in Eastern Japan, based on mitochondrial DNA D-loop analysis. Biogeography 25, 45–54. https://doi.org/10.11358/biogeo.25.45 (2023).
Sui, X., Liang, Y. & He, D. The complete mitochondrial genome of Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae). Mitochondrial DNA Part. A. 27, 3367–3369. https://doi.org/10.3109/19401736.2015.1018224 (2016).
Min, M. & Yang, S. Morphology and distribution of Moroco percnurus (Cyprinidae). Korean J. Zool. 35, 508 (1992).
Park, I. S., Choi, Y., Kim, Y. H., Nam, Y. K. & Kim, D. S. Flow cytometric and cytogenetic studies in Rhynchocypris oxycephalus and R. steindachneri. J. Aquaculture. 13, 193–196 (2000).
Miya, M. et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R Soc. Open. Sci. 2, 150088. https://doi.org/10.1098/rsos.150088 (2015).
Alam, M. J. et al. Assessment of fish biodiversity in four Korean rivers using environmental DNA metabarcoding. PeerJ 8, e9508. https://doi.org/10.7717/peerj.9508 (2020).
Sato, Y., Miya, M., Fukunaga, T., Sado, T. & Iwasaki, W. MitoFish and mifish pipeline: a mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol. Biol. Evol. 35, 1553–1555. https://doi.org/10.1093/molbev/msy074 (2018).
Choi, J. W. & An, K. G. Characteristics of fish compositions and longitudinal distribution in Yeongsan river water-shed. Korean J. Ecol. Environ. 41, 301–310 (2008).
Choi, K. S., Han, M. S., Yoon, J. D. & Ko, M. H. Characteristics of fish community and the effects of water quality on river health in Sincheon, Imjin river, Korea. Korean J. Environ. Ecol. 35, 265–276. https://doi.org/10.13047/KJEE.2021.35.3.265 (2021).
Amin, M. H. F. Comparative analysis of benthic invertebrates and fish communities in three Korean streams using environmental DNA metabarcoding. PhD thesis, Pukyong National University (2022).
Lee, H. J. et al. Seasonal variation in longitudinal connectivity for fish community in the Hotancheon from the geum River, as assessed by environmental DNA metabarcoding. J. Ecol. Environ. 48, 5. https://doi.org/10.5141/jee.23.067 (2024).
Park, S. K. & Joo, H. S. Fish fauna in the Seomjin River, Korea. Korean J. Environ. Biol. 33, 314–329. https://doi.org/10.11626/KJEB.2015.33.3.314 (2015).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. https://doi.org/10.1093/molbev/msab120 (2021).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. https://doi.org/10.1093/nar/25.24.4876 (1997).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. https://doi.org/10.1093/molbev/msx248 (2017).
Clement, M., Posada, D. & Crandall, K. A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. https://doi.org/10.1046/j.1365-294x.2000.01020.x (2000).
Leigh, J. W., Bryant, D. & Nakagawa, S. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. https://doi.org/10.1111/2041-210X.12410 (2015).
Miller, M. A. et al. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. 11, EBO-S21501; (2015). https://doi.org/10.4137/EBO.S21501
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T. & Calcott, B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. https://doi.org/10.1093/molbev/msw260 (2017).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. https://doi.org/10.1080/10635150390235520 (2003).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. https://doi.org/10.1093/bioinformatics/btg180 (2003).
Letunic, I. & Bork, P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. https://doi.org/10.1093/bioinformatics/btl529 (2007).
Pons, J. et al. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. https://doi.org/10.1080/10635150600852011 (2006).
Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x (2012).
Puillandre, N., Brouillet, S. & Achaz, G. ASAP: assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620. https://doi.org/10.1111/1755-0998.13281 (2021).
Zhang, J., Kapli, P., Pavlidis, P. & Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876. https://doi.org/10.1093/bioinformatics/btt499 (2013).
Vences, M. et al. iTaxoTools 0.1: kickstarting a specimen-based software toolkit for taxonomists. Megataxa 6, 77–92. https://doi.org/10.11646/megataxa.6.2.1 (2021).
Drummond, A. J. & Rambaut, A. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. https://doi.org/10.1186/1471-2148-7-214 (2007).
Ezard, T., Fujisawa, T. & Barraclough, T. G. SPLITS: species’ limits by threshold statistics. R package version 1.0–18/r45. (2009). http://R-Forge.R-project.org/projects/splits
Mello, B. Estimating timetrees with MEGA and the timetree resource. Mol. Biol. Evol. 35, 2334–2342. https://doi.org/10.1093/molbev/msy133 (2018).
Tamura, K., Tao, Q. & Kumar, S. Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol. Biol. Evol. 35, 1770–1782. https://doi.org/10.1093/molbev/msy044 (2018).
Mello, B., Tao, Q., Tamura, K. & Kumar, S. Fast and accurate estimates of divergence times from big data. Mol. Biol. Evol. 34, 45–50. https://doi.org/10.1093/molbev/msw247 (2017).
Tamura, K. et al. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. U S A. 109, 19333–19338. https://doi.org/10.1073/pnas.1213199109 (2012).
Crossley, B. M. et al. Guidelines for Sanger sequencing and molecular assay monitoring. J. Vet. Diagn. Invest. 32, 767–775. https://doi.org/10.1177/1040638720905833 (2020).
Chen, W., Zhong, Z., Dai, W., Fan, Q. & He, S. Phylogeographic structure, cryptic speciation and demographic history of the sharpbelly (Hemiculter leucisculus), a freshwater habitat generalist from Southern China. BMC Evol. Biol. 17, 1–13. https://doi.org/10.1186/s12862-017-1058-0 (2017).
Won, H., Jeon, H. B. & Suk, H. Y. Evidence of an ancient connectivity and biogeodispersal of a Bitterling species, Rhodeus notatus, across the Korean Peninsula. Sci. Rep. 10, 1011. https://doi.org/10.1038/s41598-020-57625-3 (2020).
Liu, J. Q., Wang, Y. J., Wang, A. L., Hideaki, O. & Abbott, R. J. Radiation and diversification within the Ligularia–Cremanthodium–Parasenecio complex (Asteraceae) triggered by uplift of the Qinghai-Tibetan plateau. Mol. Phylogenet Evol. 38, 31–49. https://doi.org/10.1016/j.ympev.2005.09.010 (2006).
Rundle, H. D. (ed, P.) Ecological speciation. Ecol. Lett. 8 336–352 https://doi.org/10.1111/j.1461-0248.2004.00715.x (2005).
Chough, S., Kwon, S. T., Ree, J. H. & Choi, D. Tectonic and sedimentary evolution of the Korean peninsula: a review and new view. Earth-Sci. Rev. 52, 175–235. https://doi.org/10.1016/S0012-8252(00)00029-5 (2000).
Kwon, S., Kim, J., Lee, E. H. & Jung, C. Y. Geography of Korea: the Understanding Korea Series (UKS) 7 (The Academy of Korean Studies, 2016).
Yoon, J. D., Kim, J. H., Park, S. H. & Jang, M. H. The distribution and diversity of freshwater fishes in Korean Peninsula. Korean J. Ecol. Environ. 51, 71–85. https://doi.org/10.11614/KSL.2018.51.1.071 (2018).
Kim, I. S., Park, J. Y. & Nalbant, T. T. A new species of Koreocobitis from Korea with a redescription of K. rotundicaudata. Korean J. Ichthyol. 12, 89–95 (2000).
Jang, J. E. et al. Genetic diversity and genetic structure of the endangered Manchurian trout, Brachymystax Lenok tsinlingensis, at its Southern range margin: conservation implications for future restoration. Conserv. Genet. 18, 1023–1036. https://doi.org/10.1007/s10592-017-0953-7 (2017).
Jeon, H. B., Song, H. Y., Suk, H. Y. & Bang, I. C. Phylogeography of the Korean endemic Coreoleuciscus (Cypriniformes: Gobionidae): the genetic evidence of colonization through Eurasian continent to the Korean Peninsula during late Plio-Pleistocene. Genes Genom. 44, 709–719. https://doi.org/10.1007/s13258-022-01243-y (2022).
Jeon, H. B. & Suk, H. Y. Genetic structure of Rhodeus sinensis reflects historical imprints of East Asian paleo-drainages. Anim. Cells Syst. 27, 353–365. https://doi.org/10.1080/19768354.2023.2285829 (2023).
Zhang, J. et al. History of yellow river and Yangtze river delivering sediment to the yellow sea since 3.5 Ma: tectonic or climate forcing? Quat Sci. Rev. 216, 74–88. https://doi.org/10.1016/j.quascirev.2019.06.002 (2019).
Taniguchi, S., Bertl, J., Futschik, A., Kishino, H. & Okazaki, T. Waves out of the Korean Peninsula and inter-and intra-species replacements in freshwater fishes in Japan. Genes 12, 303. https://doi.org/10.3390/genes12020303 (2021).
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C. & Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. https://doi.org/10.1111/j.1365-294X.2012.05470.x (2012).
Jense, C. et al. Cryptic diversity within two widespread diadromous freshwater fishes (Teleostei: Galaxiidae). Ecol. Evol. 14, e11201. https://doi.org/10.1002/ece3.11201 (2024).
Chen, T., Jiao, L. & Ni, L. The phylogeographical pattern of the Amur minnow Rhynchocypris lagowskii (Cypriniformes: Cyprinidae) in the Qinling mountains. Ecol. Evol. 12, e8924. https://doi.org/10.1002/ece3.8924 (2022).
Oh, C. W., Sajeev, K., Kim, S. W. & Santosh, M. Tectonic evolution of Korean Peninsula and adjacent crustal fragments in asia: introduction. Gondwana Res. 9, 19–20. https://doi.org/10.1016/j.gr.2005.07.001 (2006).
Kim, D., Hirt, M. V., Won, Y. J. & Simons, A. M. Small fishes crossed a large mountain range: quaternary stream capture events and freshwater fishes on both sides of the Taebaek mountains. Integr. Zool. 12, 292–302. https://doi.org/10.1111/1749-4877.12228 (2017).
Yun, B. H., Kim, Y. H., Han, H. S. & Bang, I. C. Population genetics analysis based on mitochondrial cytochrome c oxidase subunit I (CO1) gene sequences of Cottus Koreanus in South Korea. Genes Genom. 47, 207–221. https://doi.org/10.1007/s13258-024-01600-z (2025).
Kim, D. E., Seong, Y. B., Weber, J. & Yu, B. Y. Unsteady migration of Taebaek Mountain drainage divide, Cenozoic extensional basin margin, Korean Peninsula. Geomorphology 352, 107012; (2020). https://doi.org/10.1016/j.geomorph.2019.107012
Fricke, R., Eschmeyer, W. N. & Van der Laan, R. (eds) Eschmeyer’s Catalog of Fishes: Genera, Species, References. (2025). http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp accessed 11 March 2025).
Hata, H., Fujita, T., Watanabe, S. & Hosoya, K. Current status of two closely related Phoxinus species, in the Ishi river (Yamato river basin), Osaka Prefecture, Japan. Jpn J. Ichthyol. 66, 15–22. https://doi.org/10.11369/jji.18-30 (2019).
Fujita, T. & Hosoya, K. Biochemical and morphological comparison between two Japanese daces, Phoxinus lagowskii steindachneri and P. oxycephalus Jouyi in the sympatric sites. Jpn J. Ichthyol. 50, 55–62. https://doi.org/10.11369/jji1950.50.55 (2003).
Imoto, J. M. et al. Phylogeny and biogeography of highly diverged freshwater fish species (Leuciscinae, Cyprinidae, Teleostei) inferred from mitochondrial genome analysis. Gene 514, 112–124. https://doi.org/10.1016/j.gene.2012.10.019 (2013).
Jeon, H. B. Evolutionary history of Acheilognathid fish inferred based on genetic and genomic analyses. PhD thesis, Yeungnam University (2018).
Jeon, H. B., Kim, D. Y., Lee, Y. J., Bae, H. G. & Suk, H. Y. The genetic structure of Squalidus multimaculatus revealing the historical pattern of serial colonization on the tip of East Asian continent. Sci. Rep. 8, 10629. https://doi.org/10.1038/s41598-018-28340-x (2018).
Acknowledgements
The authors wish to express their profound gratitude to the laboratory colleagues of the Molecular Physiology Laboratory, Department of Marine Biology, Pukyong National University for their support during this research.
Funding
This work was supported by Dongwon Research Foundation in 2024 (202404170001) and the corresponding author (Hyun-Woo Kim) was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2021-NR060118).
Author information
Authors and Affiliations
Contributions
S.K. and H.-W.K. conceived and supervised the study. G.B., S.R.L., and H.H. performed the experiments. G.B., H.-E.K., and A.R.K. performed the data analyses. M.Y., H.J.K., I.A. and M.H.F.A. contributed to the data analyses. G.B., S.K., and H.-W.K. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical accordance for procedures
The study is entirely based on environmental water samples; therefore, no animal samples were collected from the wild, and no special permissions were required for this research.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bang, G., Lee, S.R., Kang, HE. et al. Environmental DNA reveals the distinct genetic diversity and evolutionary pathways of the Chinese Minnow Rhynchocypris oxycephalus in Korean freshwater systems. Sci Rep 16, 2323 (2026). https://doi.org/10.1038/s41598-025-32073-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-32073-z