Abstract
Neuropathic pain (NP) is a complex and prevalent chronic pain condition that affects millions of individuals worldwide. Previous studies have shown that prior exercise protects against NP caused by nerve injury. However, the underlying mechanisms of this protective effect remain to be uncovered. Therefore, the purpose of this study is to investigate how prior exercise affects protein expression in NP model rats and thus gain comprehensive insights into the molecular mechanisms involved. To achieve this objective, 6-week-old male Sprague–Dawley rats were randomly assigned into three groups, named as chronic constriction injury (CCI) of the sciatic nerve, CCI with prior 6-week swimming training (CCI_Ex), and sham operated (Sham). The CCI_Ex group underwent 6 weeks of swimming training before CCI surgery, while the CCI and sham groups had no intervention. Mechanical withdrawal threshold (MWT) and thermal withdrawal latency (TWL) were used as the main observation indicators to evaluate the behavioral changes associated with pain. Tissues from the spinal dorsal horn of the rats in the three groups were collected at 4 weeks after operation. LC–MS/MS proteomic analysis based on the label-free approach was used to detect protein profiles, and volcano plots, Venn diagrams, and clustering heatmaps were used to identify differentially expressed proteins (DEPs). Gene Ontology (GO) annotations, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and protein–protein interaction networks were employed to explore the biological importance of DEPs. At 14, 21, and 28 days following CCI, CCI rats with prior exercise showed a significant increase in the MWT and TWL of the injured lateral hind paw compared with those without exercise. A total of 122 proteins with significant changes in abundance were detected after CCI surgery, and 55 proteins were detected in the comparison between the CCI_Ex and CCI groups. GO and KEGG enrichment analysis revealed that oxygen transport capacity and the complement and coagulation cascades may be the critical mechanism by which prior exercise protects against NP. Serpina1, DHX9, and Alb are the key proteins in this process and warrant further attention, as confirmed by the results of Western blot analysis. In conclusion, this study provides new evidence that active physical activity can accelerate the relief of hyperalgesia after NP. Proteomic analyses revealed the potential target proteins and pathways for this process, offering valuable data resources and new insights into the pathogenesis and therapeutic targets of NP.
Similar content being viewed by others
Introduction
Neuropathic pain (NP) is defined as chronic pain caused by lesion or diseases of the somatosensory system, affecting 6.9–10% of the general population1,2. Complex and diverse symptoms are NP characteristics, which seriously affect the quality of life of patients. In a survey on the quality of life of patients with NP, 17% rated their quality of life as “worse than death”3. Currently, the treatment of NP includes drug and non-drug treatments. However, evidence suggests that treatment effectiveness based on existing intervention is limited when NP occurred4. Hence, exploring a program that could accelerate pain relief after NP and promote clinical treatment effectiveness is of great importance for improving the quality of life of patients and saving medical expenses.
In recent years, safe and low-cost exercise has been concerned in the exploration of accelerating pain relief after NP. Animal research found that rats with prior exercise had enhanced improvements in pain behavior after peripheral nerve damage5,6. According to the results of a meta-analysis, rats that started exercising before suffering from peripheral nerve damage can alleviate hyperalgesia symptoms faster7. The results were also supported by clinical research8,9. These studies revealed, from a positive perspective, that maintaining exercise habits may be an important factor in protecting against NP. However, the mechanisms by which exercise preconditioning protects against NP are still unknown.
The spinal dorsal horn (SDH) is the primary center of pain signal transduction and local processing, playing an important role in the development of NP10. Proteins are the primary agents of cellular function. Abnormal protein expression or protein modification changes in the cells may trigger a series of pathological changes in SDH and induce or aggravate NP11. Proteomics, which can analyze protein changes in target tissues, is an important tool for exploring new biomarkers and therapeutic targets for diseases12. Proteomics based on label-free approach was used to detect protein expression profiles in the SDH of rats in the exercise, chronic constriction injury (CCI), and sham groups to determine the differentially expressed proteins (DEPs) involved in prior-exercise protection against NP. Subsequently, bioinformatic analysis was combined to explore the function of DEPs and the key signaling pathways involved. In summary, this work provides a foundation for promoting the clinical efficacy of NP and the clinical application of prior-exercise.
Results
Animal characteristics
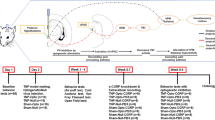
Figure 1 shows the animal characteristics, including body weight, Mechanical withdrawal threshold (MWT) and thermal withdrawal latency (TWL). The timepoints for recording the animal characteristics were 0, 7, 14, 21, 28, 35, 42, and 47 days before the CCI surgery and 3, 7, 14, 21, and 28 days after the operation (Fig. 1A). In terms of weight, no significant difference was found among the three groups at any time point (Fig. 1B). Moreover, no significant differences were observed in the MWT between the groups before the operation. After the operation, the MWT in the CCI and CCI_Ex groups were significantly lower than that in the sham group on day 3. The data on day 7 were similar to those on day 3, but the pain level in the CCI group worsened. On day 14, a significant difference was still observed between the two groups and the sham group. A notable detail that the CCI_Ex group showed significant pain reduction, whereas the CCI group reached its peak in pain intensity, and a remarkable difference was observed between the two groups. Then, the data for day 21 showed that the CCI group began to show pain reduction. On day 28, no significant difference was found in the MWT between the CCI_Ex group and the sham group, whereas a significant difference was still observed between the CCI group and the other two groups (Fig. 1C). The TWL in the exercise and CCI groups significantly reduced compared with that in the sham group at day 3. Meanwhile, no significant difference was observed between the CCI_Ex group and the sham group on days 21 and 28. Similar to MWT, swimming increased the TWL in the CCI_Ex group on days 7, 14, 21, and 28 compared with that in the CCI group (Fig. 1D).
Experimental workflow and results of behavioral assessment. (A) Schematics showing the labeling and recording of study design in Ex_CCI group and the experimental strategy of proteomics. (B) Time courses of body weight, (C) MWT, and (D) TWL. Data were analyzed by two-way ANOVA for repeated measures. Data are presented as mean ± standard error of the mean (SEM, n = 8 animals/group). *p < 0.05 and **p < 0.01, for comparisons between sham group and CCI group or Ex_CCI group. #p < 0.05 and ##p < 0.01 for comparisons between Ex_CCI group and CCI group.
Identification of DEPs
Through label-free quantitative proteomics, the information of protein changes in SDH was quantified to explore the DEPs that may respond to the neuroprotection of prior exercise for sudden NP. DEPs were defined as when the expression of a protein was 1.5 times higher or 2/3 lower in the group than in another group (fold change > 1.5 or < 0.66) and the p-value was < 0.05. Ultimately, 121 DEPs (73 upregulated and 48 downregulated) were detected after CCI surgery (Fig. 2A), and 55 DEPs (27 upregulated and 28 downregulated) were detected in the CCI_Ex group compared with the CCI group (Fig. 2B).
The overlapping proteins among the DEPs from different comparisons were analyzed by Venn diagram to further screen key proteins. The results showed that 5 overlapping DEPs (protein accession: Q811T3, D4ABP4, P61314, D4A7V6 and M0R439) were significantly downregulated after CCI, while they were significantly upregulated in the CCI_Ex group compared to the CCI group. A total of 13 overlapping DEPs (protein accession: B1H216, B0BNN3, O55158, D4A9D6, P17475, Q63910, Q62669, P06866, A0A0G2JU, P06399, P02091, P02680 and P02770) were upregulated after CCI and downregulated in prior exercise (Fig. 2C and Table 1). Table 1 shows the information of the 18 common DEPs. According to the results of disease annotation in the Rat Genome Database (RGD) and literature review, three proteins were directly related to NP, four proteins were related to hyperalgesia, and 11 proteins were related to inflammation in the final 18 common DEPs. Euclidean distance and hierarchical clustering were used to cluster the DEPs, and the dynamic changes in DEPs were compared (Fig. 2D). The results showed that the DEPs abundance intensity of the CCI group was significantly different from that of the exercise and sham groups.
Analysis of DEPs. (A) Volcano plots showing DEPs between CCI and sham groups and (B) between CCI_Ex and CCI groups. (C) Venn diagrams and bar chart displaying the number of upregulated and downregulated DEPs in the comparisons between CCI and sham groups and CCI_Ex and CCI groups. (D) Cluster heatmap showing the expression intensity of DEPs. Each row represents a gene, and each column represents a sample dimension. Colors represent relative expression levels of proteins, with red and green representing upregulation and downregulation, respectively.
Gene ontology (GO) annotations and kyoto encyclopedia of genes and genomes (KEGG) pathways analysis of DEPs
The proteomic data were analyzed using GO and KEGG pathway analyses to further investigate the neuroprotective effect of prior exercise on NP. GO analysis was conducted to evaluate the critical terms of the DEPs by exploring their biological process (BP), cellular component (CC), and molecular function (MF) in two comparisons (comparison 1: CCI group vs. sham group, DEPs = 122 proteins; comparison 2: CCI_Ex group vs. CCI group, DEPs = 55 proteins). Figure 3A shows the top 10 GO terms in comparison 1 (p < 0.05). The top three enriched BPs related to NP include platelet aggregation, response to calcium ion, and cellular macromolecular complex assembly. They were mainly derived from CCs, such as synapse, cytoplasm, and blood microparticle, and had MFs, such as protein binding, oxygen binding, and calcium-dependent phospholipid binding. The top 10 GO terms in comparison 2 are shown in Fig. 3B. The most significantly enriched BPs were oxygen transport, acute-phase response, nitric oxide transport, carbon dioxide transport, and cellular oxidant detoxification. The DEPs were mainly derived from CCs, such as haptoglobin–hemoglobin complex, hemoglobin complex, blood microparticle, and axon. The most significantly enriched MFs were oxygen binding, oxygen transporter activity, and haptoglobin binding.
In addition, KEGG enrichment analysis was conducted to evaluate the important signaling pathways of the DEPs. The DEPs of comparison 1 were significantly enriched in the complement and coagulation cascades, bile secretion, African trypanosomiasis, and sphingolipid signaling pathway (Fig. 3C). The DEPs of comparison 2 were significantly enriched in KEGG pathways, such as African trypanosomiasis, malaria, coronavirus disease, and complement and coagulation cascades (Fig. 3D). The results of the two comparisons showed that complement and coagulation cascades was a potentially critical pathway. Fgb, Fga, Serpina1, and Fgg were enriched in this pathway.
GO and KEGG pathway analysis of DEPs. (A) GO analysis of DEPs in the comparisons of CCI and sham groups and (B) CCI_Ex and CCI groups. The X-axis denotes the number of genes that engage in the GO term, and the p-value of the GO analysis is represented by the colormap. (C) KEGG pathway scatterplots of DEPs in the comparisons between CCI and sham groups and (D) CCI_Ex and CCI groups. The color of the point corresponds to different p value ranges, and the size of the point represents the number of target protein in the pathway.
Protein–protein interaction (PPI) network analysis
After the DEPs obtained from two comparisons (DEPs in comparison 1 = 122 proteins; DEPs in comparison 2 = 55 proteins) were imported into the STRING website, two networks of PPIs were obtained, and Cytoscape was used to analyze and visualize the importance of target proteins.
First, a PPI network (78 nodes and 162 edges) was constructed containing 122 DEPs identified from CCI group vs. sham group (Fig. 4A). Four algorithms (MNC, MCC, EPC, and Degree) based on CytoHubba were used to analyze 122 proteins with interactions to identify the hub proteins in PPI networks. The intersection of the top 10 proteins of each algorithm was shown in the Venn diagram. Five hub proteins (Fgg, Fga, Serpina1, Apoa1, and Map2k1) were obtained (Fig. 4B), and they were closely implicated with positive regulation of ERK1 and ERK2 cascade and the complement and coagulation cascade pathway (Fig. 4C). Subsequently, MCODE was applied for clustering proteins to further validate the hub proteins and pathways, and the two clusters with the highest scores are shown in Fig. 4D. GO enrichment analysis (Fig. 4E) was performed for the two clusters separately. The results of the two GO analyses indicated that the transport capacity of oxygen in the blood may be an important enrichment pathway after CCI.
PPI analysis of DEPs in the comparisons of CCI and sham groups. (A) PPI network construction of 122 DEPs in comparison between CCI and sham groups. (B) Five hub proteins identified by CytoHubba plugins of Cytoscape based on four algorithms: MNC, MCC, EPC, and Degree. (C) BP and KEGG analysis based on five hub proteins. (D) Most significant MCODE cluster with 10 nodes and 41 edges. (E) BP and KEGG analysis based on proteins in the cluster.
A PPI network consisting of 20 nodes and 36 edges was constructed using 55 DEPs to explore the mechanism of prior exercise on neuroprotection after CCI (Fig. 5A). Nine common DEPs (Hba-a1, Hbb, Hba-a2, Fgg, Mug1, Fga, Serpina1, Hp, and Alb) were obtained in the same manner (Fig. 5B). The GO and KEGG analysis results of nine DEPs (Hba-a1, Hbb, Hba-a2, Fgg, Mug1, Fga, Serpina1, Hp, and Alb) showed that acute-phase response and cellular oxidant detoxification were the main BPs, and complement and coagulation cascade pathways were significantly enriched (Fig. 5C). MCODE was applied for clustering proteins, and only one cluster was shown (Fig. 5D). The GO analysis showed that DEPs were closely related to oxygen transport capacity and significantly enriched in complement and coagulation cascades (Fig. 5E).
PPI analysis of DEPs in the comparisons of CCI and sham groups. (A) PPI network construction of 55 DEPs in comparison between CCI group and CCI_Ex group. (B) Nine hub proteins identified by CytoHubba plugins based on four algorithms: MNC, MCC, EPC, and Degree. (C) BP and KEGG analysis based on nine hub proteins. (D) Most significant MCODE cluster with 10 nodes and 25 edges. (E) BP and KEGG analysis based on proteins in the cluster.
Confirmation of Serpina1, DHX9, and Alb
According to the results of disease annotation in the RGD and PPI analysis, Serpina1, DHX9, and Alb constitute the core proteins within the interaction network among DEPs and are highly correlated with the pathophysiology of NP. The western blot validation was performed to further confirm the expression of three key proteins in each group. As shown in Fig. 6, compared with the sham group, the expression levels of Serpina1, DHX9, and Alb in the CCI group were significantly increased, while they decreased in the CCI_Ex group compared to the CCI group (P < 0.05).
The western blot results of Serpina1, DHX9, and Alb. (A–C) Serpina1, DHX9, and Alb expression in the SDH of three (sham, CCI, and CCI_Ex) groups confirmed through western blotting and statistical histogram (n = 3 in each group). Data are presented as mean ± standard error of the mean (SEM). *p < 0.05 and **p < 0.01, for comparisons between sham group and CCI group. #p < 0.05 and ##p < 0.01 for comparisons between Ex_CCI group and CCI group. The original blots were displayed in Additional File 2.
Discussions
The behavioral results of rats suggested that prior regular swimming played an important role to accelerate the relief of NP induced by CCI. These results were supported by previous animal research5,6 and have important public health implications because they suggest that an active lifestyle could protect against NP. However, the pathways and molecular signatures that mediate the functions remain ambiguous. Our objective is to identify key proteins with therapeutic potential by using proteomic analysis. Although exercise can alleviate NP, not all patients with chronic pain can tolerate exercise. Exploring the mechanisms of pain relief from prior exercise can provide new insights for developing NP therapeutics, benefiting patients who cannot exercise. We identified 18 DEPs through proteomics, with Serpina1, ALB, and Dhax9 being recognized as potential key proteins for alleviating NP. Moreover, the results of bioinformatics analysis indicated that the complement and coagulation cascades were noteworthy pathways. To our knowledge, this study is the first to apply proteomics to explore the potential biomarkers and pathways of prior-exercise protection against NP in rats.
Serpina1 (alpha-1-antiproteinase), also known as alpha-1-proteinase inhibitor or α1-antitrypsin, is a member of the serpin superfamily13. Although its primary target is elastase, it also has a moderate affinity for plasmin and thrombin14. Previous studies focused on the relationship between Serpina1 and NP. Muley et al.15 deemed that neutrophil elastase can activate proteinase-activated receptor-2 to trigger acute inflammation, hyperalgesia, and pain. The increase in the proteolytic activity of neutrophil elastase was effectively reduced by the injection of Serpina1. Bäckryd et al.16 reported that Serpina1 is involved in pathophysiological processes in the spinal cord for chronic peripheral neuropathic pain and regarded as a potential biomarker. A recent study also focused on the association between Serpina1 and NP17. The authors induced mechanical hypersensitivity in mice by constructing partial nerve ligation to simulate NP18. SP16, an innovative peptide derived from the carboxyl terminus of Serpina1, prevents mechanical hyperalgesia by exerting strong anti-inflammatory activity on damaged peripheral nerves. In the present study, the results of proteomic analysis showed that Serpina1 was not only closely related to NP after CCI but was also the hub protein in the protective effect of prior exercise against NP. In the comparison between the CCI_Ex and CCI groups, the fold change of Serpina1 in SDH was 0.00313, revealing that prior exercise significantly reduced the expression of Serpina1 in the SDH of CCI rats. This finding was confirmed by the results of Western blot analysis. Given the relationship between Serpina1 and NP, Serpina1 may be a key protein for the protective effect of prior exercise against NP.
Alb occupied the central position in the PPI networks of the CCI group versus the sham group (Fig. 4A) and the CCI_Ex group versus the CCI group (Fig. 5A) and was identified in hub protein analysis based on Cytohubbo and MCODE. As early as 2004, researchers have been interested in the link between Alb and NP. Pérez et al.19 found that the consumption of Alb protein produced high levels of allodynia. A recent study explained the relationship between Alb and NP20. NP caused by L5–6 nerve constriction resulted in pain hypersensitivity that was progressively associated with the breakdown of the blood–spinal cord barrier into large molecules, such as Alb, within the spinal cord parenchyma. The leakage of Alb across the BSCB led to edema formation within the spinal cord21. The author also observed that Alb, other proteins, and inflammatory factors were upregulated in spinal cord tissue. The decrease in proteins, such as Alb, by cerebrolysin prevented spinal cord pathology and hyperalgesia. In addition, a recent research22 demonstrated that the expression changes in ciRNA-Fmn1, which regulates production by binding to DHX9 and DNA tandem repeats23, in SDH neurons were closely related to the expression of Alb and that the decreased expression of Alb in SDH alleviated the pain associated with hypersensitivity. DHX9 is one of the 18 DEPs screened in the present study. Recent research on the correlation between DHX9 and NP has indicated that following peripheral nerve injury, the levels of DHX9 in the hippocampus declined. The increased expression of DHX9 in the hippocampus considerably alleviated nociceptive responses and enhanced anxiolytic behavior24. The findings of another study are similar to those of ours. An increase in DHX9 expression in the dorsal root ganglion following peripheral nerve injury may contribute to the downregulation of circ-Ankib1, which promotes NP25. The variations in findings across studies may stem from distinct sampling locations, necessitating further exploration to validate the correlation between DHX9 and NP. In summary, these studies suggested that Alb and DHX9 are closely associated with the progression of NP. In conjunction with our research findings showing that prior exercise can remarkably downregulate the expression of Alb and DHX9 in SDH induced by CCI surgery, DHX9 and Alb may be the key proteins involved in the protective effect of prior exercise against NP, although further validation is required.
A series of bioinformatic analyses was conducted to investigate the function of DEPs. According to GO and KEGG enrichment analysis, the DEPs after CCI surgery mainly enriched platelet aggregation, metal ion (calcium/sodium ion) transport, blood coagulation, fibrin clot formation, oxygen transporter activity, protein binding, and complement and coagulation cascades. This finding was consistent with the fact that NP induced by peripheral nerve injury was mainly related to ion transport and inflammatory immune response26,27. Further GO and KEGG enrichment analyses based on hub proteins obtained similar results. The enrichment analysis results of DEPs from the CCI_Ex group versus the CCI group suggested that oxygen transport, cellular oxidant detoxification, platelet aggregation, oxygen binding, metal ion binding, oxygen transporter activity, and complement and coagulation cascades may be the potential mechanisms to explain the accelerated recovery after CCI by prior exercise. Previous studies showed that the energy required for cellular metabolism in tissues, such as the repair process of peripheral nerve damage, is mostly obtained by oxidation of nutrients28. Therefore, efficient oxygen uptake and transportation are the key factors in energy metabolism of tissues. Research found that gabapentin and transcranial direct-current stimulation can alleviate pain responses by regulating glucose metabolism29,30. Long-term exercise has been proven to promote oxygen uptake and transportation by strengthening the functions of the respiratory and circulatory systems31,32. Therefore, the accelerated recovery of rats in prior exercise after CCI may be based on stronger respiratory and circulatory systems, which promote cellular metabolism of damaged nerves. In addition, studies reported that complement and coagulation cascades play a crucial role in innate immunity and are involved in the pathogenesis of inflammatory-related diseases, such as NP33. Evidence based on a scoping review34 suggested that long-term exercise and the accompanying increases in muscle mass and cardiorespiratory function appear to downregulate C3 family proteins (an important complement protein) in the blood of healthy people. This finding is consistent with the viewpoint that regular exercise has an anti-inflammatory effect and may have implications for the prevention and treatment of diseases related to inflammation and complement disorders35.
The relief of pain hypersensitivity induced by prior swimming was not caused by exercise-induced stress responses or negative emotions. First, instead of beginning at the final intensity, the rats were provided with a week of gradually increasing adaptive training before the start of formal training. Second, the rats swam in a spacious and warm pool as a group. This condition differed from the stressful environment of the forced swimming test. After swimming, we would carefully dry the rats’ fur and blow it dry with gentle warm air. In summary, we provided a highly comfortable environment for the rats. Finally, numerous studies have proven that swimming with high motor complexity may be particularly beneficial for patients with depression because it requires high levels of attention, memory, and motor difficulty, producing greater cortical activation than exercises with low motor complexity36,37,38. In fact, the weight changes in each group of rats that we previously reported also verified this result. When rats are under stress or in a depressive state, they typically exhibit conditions, such as decreased appetite and reduced food intake, that lead to weight loss. Of course, objective evaluations like serum cortisol level, forced swimming, and tail suspension behavior tests can offer a straightforward explanation for this issue. This situation is one limitation of our study.
This study has some other limitations. First, we only selected the fourth week after CCI as the time point for detecting proteome changes. However, the research results revealed remarkable differences in behavioral tests at multiple time points. Therefore, the present study cannot reflect all the important protein regulatory processes involved in NP response to prior exercise. Additionally, long-term behavioral observation after CCI can help deepen our understanding of how pre-exercise prevents the chronicization of pain caused by peripheral nerve injury. Therefore, research with prolonged observation periods and additional time points is necessary to address the above limitation. Second, our study only focused on male rats and found that six weeks of swimming before CCI could reduce subsequent pain hypersensitivity in male rats. Green-Fulgham et al.6 showed that prior exercise has similar effects on relieving NP in female rats. However, whether the key proteins in female rats are the same as those in male rats is not yet known and must be verified through further experiments in the future.
In conclusion, this study was the first to apply proteomics to explore the potential biomarkers and pathways of prior-exercise protection against NP in rats. The results showed that prior exercise can accelerate the relief of hyperalgesia after CCI surgery. Moreover, maintaining positive exercise habits could contribute to protect against NP caused by sudden peripheral nerve injury, and this finding has important public health implications. Inflammatory immune response, especially the oxygen transport capacity and complement and coagulation cascades, may be the critical mechanism by which prior exercise protects against NP. And Serpina1, Alb, and DHX9 have been preliminarily validated as hub proteins for this process. However, further experiments are needed to verify the role of hub proteins and potential pathways in this process. In summary, this work provided new evidence that active physical activity can prevent NP and preliminarily explained its potential molecular biological mechanisms.
Materials and methods
Animals and grouping
A total of 24 male Sprague–Dawley rats (age: 6 weeks; weight: 180–200 g) were obtained from the Experimental Animal Center of Xuzhou Medical University and raised in an SPF barrier system. The rats were raised in cages of four per cage, with unlimited food and water, at a temperature of 24 °C ± 1 °C with a 12/12-h cycle of darkness/light. An adaptation period of 1 week was given to the animals before the experiment. All animal procedures were in strict accordance with relevant international laboratory animal use guidelines and approved by the Scientific Research Ethics Committee of Xuzhou Medical University (No: 202207S014).
The SD rats were randomly assigned to one of three groups: (1) CCI of the sciatic nerve; (2) CCI with prior 6-week swimming training (CCI_Ex); (3) sham operated (Sham).
Experimental design
The study design is shown in Fig. 1A. The experiment was divided into three periods. During the period of adaptive feeding, the baseline behavior of rats was evaluated through the MWT and TWL. Next, the CCI_Ex group received 1 week of adaptive exercise training and 6 weeks of formal exercise training. The CCI and sham groups were fed normally without any intervention during this period. Behavioral testing was conducted once a week. Then, the CCI and CCI_Ex groups received modeling to simulate symptoms of NP after completing the exercise training stage. After the modeling operation, all rats were fed normally for 4 weeks and subjected to behavioral testing at different time points (3, 7, 14, 21, and 28 days).
Exercise intervention procedure
Swimming was used as the form of exercise intervention, including a 1-week adaptive training stage and a 6-week formal training stage39,40. In the adaptive training stage, rats in the CCI_Ex group underwent 5 days of adaptive swimming. The rats trained for 10 min on the first day, and the training time was then increased by 10 min every day until it reached 50 min. The formal training stage included 6 weeks of training session, with 50 min of training per day and 5 days of training per week. The training was applied at 18:00 to 20:00 p.m. The rats were placed in a plastic container (length = 82 cm, width = 60 cm, and height = 59 cm) filled with approximately 200 L water (37 °C). The researchers supervised the training to prevent floating behavior in the rats. Once floating behavior occurred, the researchers tapped the rats on the back of the neck with a pen or stirred the water to create a current to keep them swimming39,41. The rats were gently dried after the exercise with a cloth towel.
Behavioral testing
Behavioral test was conducted during the adaptive feeding period, the exercise period, and the observation period after the operation (13 tests). Awake, unrestrained rats were subjected to behavioral tests by a single experienced investigator (JB Guo) who was blinded to the group assignments. Before each behavioral test, the rats were placed in individual glass boxes (11 cm × 22 cm × 13 cm) to adapt to the environment for 20 min.
MWT
The up-and-down method42 with von Frey hairs (Aesthesio, Danmic Global, USA) was used to measure the MWT in the hind paws of rats. The paw was touched with one of a series of nine von Frey hairs with logarithmically incremental stiffness (0.6, 1.0, 1.4, 2.0, 4.0, 6.0, 8.0, 10.0, and 15.0 g). The upper limit of 15 g was selected for testing because stiffer hairs tended to raise the whole limb of rats instead of buckling. The test area was located in the center of the hind paw sole, ensuring the accuracy and stability of the test. First, a 4.0 g von Frey filament was selected to thrust at the sole of hind paw of rats to cause slight buckling against the paw and maintained for approximately 6–8 s43. If a positive reaction of paw withdrawal was triggered, the next weaker von Frey filament was used. The next stronger von Frey filament was applied without triggering a positive reaction. The interval between each stimulation occurred within 2 min. PWT (50%) was calculated in accordance with the methods of Dixon44. The von Frey filaments were calibrated between each assay.
TWL
A thermal plantar algesimeter (Cat. No. 37370, Ugo Basile, Italy) was utilized to test the TWL. The test apparatus was calibrated before TWL measurement. The rats were placed sequentially above a transparent glass plate. A radiant heat was applied by aiming a beam of light (intensity of 20 I.R. and cutoff time of 30) onto the plantar surface of the hind paw through the glass plate. The latency from onset of the light to brisk withdrawal of the stimulated paw was measured as TWL. This step was repeated three times, with a 5-min interval between stimuli, and then the mean TWL was calculated45.
CCI models
CCI of the sciatic nerve was used as a model of NP induced by peripheral nerve injury and constructed in accordance with a previous description46. First, the rats were quickly anesthetized with 5% isoflurane in O2 in an induction chamber and then fully anesthetized with 2% isoflurane in O2 continuously by using a facemask of the animal anesthesia machine. After the rat’s right hind leg was shaved and disinfected, a parallel incision was made into the skin 3–4 mm below the middle femur. Then, the sciatic nerve was exposed after the connective tissue between the superficial gluteal muscle and the biceps femoris muscle was bluntly separated. A 4–0 chrome catgut was used to ligate four loose segments on the sciatic nerve at distances of 1 mm apart. The ligation strength had no effect on the blood transport of the nerve trunk, and a slight twitch of the calf muscle was observed during the operation. Silk sutures with 5 –0 were used to close the skin incision. In the sham group, the same procedure was used to expose the sciatic nerve of rats without ligation.
Proteomic analysis
All rats were deeply anesthetized with 5% isoflurane in O2 in an induction chamber. After being anesthetized, rats were perfused with normal saline at 4 °C. The L4–L6 spinal cord of the rat is approximately located between the T13 at the 13th rib level and L1 pyramids. After localization, quickly dissect the bilateral pedicles to expose the spinal cord level along the coronal plane. Quickly cut the spinal cord transversely with a sharp blade, sever the surrounding spinal nerves, and then use the sharp blade to take the quarter of the spinal cord where the SDH lies. After carefully removing the white matter, the remaining gray matter is the L4–L6 SDH. Take the sample and put it in 2 mL RNAse-free centrifuge tubes. Place it in liquid nitrogen quickly and then transfer it to a − 80 °C freezer for storage.
Sample preparation
Five samples in each group were used for proteomic analysis. First, the SDH tissues were converted into protein lysates and transferred into a micro centrifuge tube for further use. The protein was extracted using a high-intensity ultrasonic processor after adding the lysis buffer (8 M urea and 1% protease inhibitor) into the centrifugal tube. Finally, the supernatant was collected after centrifugation at 12,000g for 10 min at 4 °C. BCA kit was used to detect the protein concentration in the supernatant.
Trypsin digestion
The protein solution was reduced with 5 mM dithiothreitol at 56 °C for 30 min and then alkylated in the dark with 11 mM iodoacetamide at room temperature for 15 min for digestion. Next, the protein solution was diluted with 100 mM triethylammonium bicarbonate to ensure that the urea concentration was less than 2 M. Finally, a 1:50 enzyme-to-protein ratio was used to digest sweat gland proteins at 37 °C overnight with trypsin. For the second digestion, the enzyme-to-protein ratio was adjusted to 1:100, and the digestion was carried out for 4 h. The reaction was stopped by adding 1 µl trifluoroacetic acid. The sample was centrifuged at 5000g for 3 min, and the peptide filtrate was retained. The peptide solution underwent STAGETIP-C18 desalting, and then the sample was dried in SpeedVac. Finally, the sample was resuspended in 20 µl sample buffer (0.1% FA solution).
Liquid chromatography tandem mass spectrometry (LC–MS/MS) analysis
Capillary C18 RPLC columns (inner dimeter = 150 μm and length = 300 mm) were installed on an EasyLC (Thermo Fisher, Sunnyvale, CA). During the first installation, the mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). Peptides were separated by a column at 1000 nL/min flow rate and a 60-min gradient program. As peptides eluted, they were entered into an Orbitrap Velos Pro (Thermo Scientific, San Jose, CA). Survey scans (MS) were obtained from the Orbitrap with a resolution of 30,000 at 400 m/z with a target for the automatic gain control of 1 × 106. Data were acquired for MS/MS analysis for the top 20 ions. MS/MS was performed using the LTQ Orbitrap Velos (Thermo) instrument.
Database search
Raw data files were processed using MaxQuant (version 1.6.15.0)47 against a rat FASTA database (downloaded on March 17, 2021, 36,189 entries). The default settings were used during the search. Briefly, trypsin/P was specified as the cleavage enzyme, allowing up to two missing cleavages. The minimum peptide length was seven amino-acid residues. The mass tolerance values for the primary precursor ion search and main search were 20 and 5 ppm, respectively. The mass tolerance for fragment ions was 0.5 Da. The quantitative method was set to label free, and the FPR for peptide-spectrum match identification was 1%48.
Bioinformatic analysis
Proteomic analysis was performed using the “Statistical Analysis [one factor]” module from Metaboanalyst (https://www.metaboanalyst.ca/MetaboAnalyst/ModuleView.xhtml)49. The module can perform missing value estimation, sample normalization, and data transformation for raw data. Next, a volcano plot was used to identify DEPs, which were defined by fold change > 1.5 or < 0.66 and p-value < 0.05, from two comparisons (CCI group vs. sham group and CCI_Ex group vs. CCI group). The Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to identify common DEPs. A hierarchical clustering heatmap was plotted to obtain an overview of common DEPs in all the samples. The DEPs were entered into DAVID Bioinformatics Resources (https://david.ncifcrf.gov/tools.jsp) to obtain GO annotations and KEGG pathways50,51,52. p values < 0.05 were set as enrichened significance. The figure of GO annotations and KEGG pathways were plotted by https://www.bioinformatics.com.cn, an online platform for data analysis and visualization53.
The STRING database (https://cn.string-db.org/cgi/input?sessionId=bcrNj2ChbPBcinput page_show_search = on) was used to predict the PPI network of DEPs. The PPI network was imported into Cytoscape version 3.10.0 for visualization. The cytoHubba plugin was applied to identify the hub genes on the basis of DEGs from two comparisons in Cytoscape by using four algorithms (MNC, MCC, EPC, and Degree). In addition, the MCODE plugin was used to identify the potential vital modules54.
Western blotting
After the protein concentrations were measured, the samples were heated for 10 min at 100 °C, and 30–60 µg protein was loaded onto 10% SDS-polyacrylamide gels (PG111, Shanghai Epizyme Biomedical Technology Co., Ltd, China). Subsequently, the proteins were electrophoretically transferred onto PVDF membranes (Roche Diagnosties, Lot. 16916700). The blotting membranes were blocked with 5% BSA for 2 h and incubated overnight at 4 °C with the primary antibody. The following primary antibodies were used for western blotting: DHX9 (1:1000, ab183731, Abcam), Albumin (1:1000, ab207327, Abcam), Serpina1 (1:1000, 16382-1-AP, Proteintech), β-actin (1:5000, 66009-1-LG, Proteintech) and GAPDH (1:1000, 60004-1-LG, Proteintech). Then, goat anti-rabbit IgG HRP-conjugated antibody (1:3000, SA00001-2, Proteintech) and goat anti-mouse IgG HRP-conjugated antibody (1:5000, SA00001-1, Proteintech) were used for incubation for 1 h. The proteins were visualized using the chemiluminescence reagents provided with the ECL kit (Millipore, WBKLS0100) and detected by a machine of ChemiDocTM Touch Imaging System (Bio-Rad Laboratories, USA). The gray value of each protein band is analyzed with Image J software, and the results were normalized with corresponding loading control.
Statistical analysis
GraphPad Prism 8 statistical software (GraphPad Software, Inc., San Diego, CA, USA) was used to analyze the data from behavioral test and western blotting. Values were expressed as the mean ± standard error of the mean (SEM). Two-way repeated ANOVA was used to analyze behavioral data for statistical differences followed by Tukey’s post hoc tests. The data from western blotting among different groups were conducted using one-way ANOVA followed by Tukey’s multiple comparison test. P_values < 0.05 were considered as significant difference on statistics.
Data availability
The mass spectrometry proteomics data had been deposited to the ProteomeXchange Consortium with the dataset identifier PXD054342.
References
Smith, B. H., Hebert, H. L. & Veluchamy, A. Neuropathic pain in the community: prevalence, impact, and risk factors. Pain 161, S127–S137 (2020).
Finnerup, N. B., Kuner, R. & Jensen, T. S. Neuropathic pain: from mechanisms to treatment. Physiol. Rev. 101, 259–301 (2021).
Torrance, N. et al. Estimating the burden of disease in chronic pain with and without neuropathic characteristics: does the choice between the EQ-5D and SF-6D matter? Pain 155, 1996–2004 (2014).
Hussain, G. et al. Current status of therapeutic approaches against peripheral nerve injuries: a detailed story from injury to recovery. Int. J. Biol. Sci. 16, 116–134 (2020).
Green-Fulgham, S. M. et al. Preconditioning by voluntary wheel running attenuates later neuropathic pain via nuclear factor E2-related factor 2 antioxidant signaling in rats. Pain 163, 1939–1951 (2022).
Green-Fulgham, S. M. et al. Voluntary wheel running prevents formation of membrane attack complexes and myelin degradation after peripheral nerve injury. Brain Behav. Immun. 115, 419–431 (2024).
Guo, J. B. et al. Meta-analysis of the effect of exercise on neuropathic pain induced by peripheral nerve injury in rat models. Front. Neurol. 10, 636 (2019).
Landmark, T., Romundstad, P. R., Borchgrevink, P. C., Kaasa, S. & Dale, O. Longitudinal associations between exercise and pain in the general population—the HUNT pain study. PLoS One. 8, e65279 (2013).
Muller, J. et al. Preventive effect of sensorimotor exercise and resistance training on chemotherapy-induced peripheral neuropathy: a randomised-controlled trial. Br. J. Cancer. 125, 955–965 (2021).
Woolf, C. J. Central sensitization: implications for the diagnosis and treatment of pain. Pain 152, S2–S15 (2011).
Liao, M. F. et al. The role of autophagy and apoptosis in neuropathic pain formation. Int. J. Mol. Sci. 23, 2685 (2022).
Goeminne, L. J. E., Gevaert, K. & Clement, L. Experimental design and data-analysis in label-free quantitative LC/MS proteomics: a tutorial with MSqRob. J. Proteom. 171, 23–36 (2018).
Cortes-Lopez, R. & Barjaktarevic, I. Alpha-1 antitrypsin deficiency: a rare disease? Curr. Allergy Asthma Rep. 20, 51 (2020).
Janciauskiene, S. M. et al. The discovery of alpha1-antitrypsin and its role in health and disease. Respir. Med. 105, 1129–1139 (2011).
Muley, M. M., Krustev, E., Reid, A. R. & McDougall, J. J. Prophylactic inhibition of neutrophil elastase prevents the development of chronic neuropathic pain in osteoarthritic mice. J. Neuroinflamm. 14, 168 (2017).
Backryd, E., Edstrom, S., Gerdle, B. & Ghafouri, B. Do fragments and glycosylated isoforms of alpha-1-antitrypsin in CSF mirror spinal pathophysiological mechanisms in chronic peripheral neuropathic pain? An exploratory, discovery phase study. BMC Neurol. 18, 116 (2018).
Wang, Z. et al. alpha1-Antitrypsin derived SP16 peptide demonstrates efficacy in rodent models of acute and neuropathic pain. FASEB J. 36, e22093 (2022).
Natale, C. A., Christie, M. J. & Aubrey, K. R. Spinal glycinergic currents are reduced in a rat model of neuropathic pain following partial nerve ligation but not chronic constriction injury. J. Neurophysiol. 129, 333–341 (2023).
Perez, J. et al. Dietary fat and protein interact in suppressing neuropathic pain-related disorders following a partial sciatic ligation injury in rats. Pain 111, 297–305 (2004).
Sharma, H. S. et al. Cerebrolysin attenuates exacerbation of Neuropathic Pain, blood-spinal Cord Barrier Breakdown and Cord Pathology Following Chronic Intoxication of Engineered Ag, Cu or Al (50–60 nm) nanoparticles. Neurochem. Res. 48, 1864–1888 (2023).
Sharma, H. S. & Olsson, Y. Edema formation and cellular alterations following spinal cord injury in the rat and their modification with p-chlorophenylalanine. Acta Neuropathol. 79, 604–610 (1990).
Liu, X. D. et al. DHX9/DNA-tandem repeat-dependent downregulation of ciRNA-Fmn1 in the dorsal horn is required for neuropathic pain. Acta Pharmacol. Sin. 44, 1748–1767 (2023).
Aktas, T. et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119 (2017).
Yang, L. et al. DExH-box helicase 9 modulates hippocampal synapses and regulates neuropathic pain. Iscience 27, 109016 (2024).
Mao, S. S. et al. A Schwann cell-enriched circular RNA circ-Ankib1 regulates Schwann cell proliferation following peripheral nerve injury. Faseb J. 33, 12409–12424 (2019).
He, J. T., Li, X. Y., Zhao, X. & Liu, X. Hyperpolarization-activated and cyclic nucleotide-gated channel proteins as emerging new targets in neuropathic pain. Rev. Neurosci. 30, 639–649 (2019).
Sommer, C., Leinders, M. & Uceyler, N. Inflammation in the pathophysiology of neuropathic pain. Pain 159, 595–602 (2018).
Dou, X. et al. The potential role of T-cell metabolism-related molecules in chronic neuropathic pain after nerve injury: a narrative review. Front. Immunol. 14, 1107298 (2023).
Lin, H. C. et al. Gabapentin reverses central hypersensitivity and suppresses medial prefrontal cortical glucose metabolism in rats with neuropathic pain. Mol. Pain. 10, 63 (2014).
Yoon, E. J. et al. Transcranial direct current stimulation to lessen neuropathic pain after spinal cord injury: a mechanistic PET study. Neurorehabil. Neural Repair. 28, 250–259 (2014).
Goulding, R. P., Burnley, M. & Wust, R. C. I. How priming exercise affects oxygen uptake kinetics: from underpinning mechanisms to endurance performance. Sports Med. 53, 959–976 (2023).
Franklin, B. A., Eijsvogels, T. M. H., Pandey, A., Quindry, J. & Toth, P. P. Physical activity, cardiorespiratory fitness, and cardiovascular health: a clinical practice statement of the American Society for Preventive Cardiology Part II: physical activity, cardiorespiratory fitness, minimum and goal intensities for exercise training, prescriptive methods, and special patient populations. Am. J. Prev. Cardiol. 12, 100425 (2022).
Tang, S. C. W. & Yiu, W. H. Innate immunity in diabetic kidney disease. Nat. Rev. Nephrol. 16, 206–222 (2020).
Rothschild-Rodriguez, D. et al. The effects of exercise on complement system proteins in humans: a systematic scoping review. Exerc. Immunol. Rev. 28, 1–35 (2022).
Khalafi, M. et al. Influence of different modes of exercise training on inflammatory markers in older adults with and without chronic diseases: a systematic review and meta-analysis. Cytokine 169, 156303 (2023).
Mahdirejei, H. A., Peeri, M., Azarbayjani, M. A. & Fattahi Masrour, F. Fluoxetine combined with swimming exercise synergistically reduces lipopolysaccharide-induced depressive-like behavior by normalizing the HPA axis and brain inflammation in mice. Pharmacol. Biochem. Behav. 232, 173640 (2023).
Ding, Z. & Du, L. Swimming exercise ameliorates depressive-like behavior by anti-inflammation activity, rebalancing gut Escherichia coli and Lactobacilli. Brain Res. 1797, 148113 (2022).
Xie, Y. et al. Swimming exercise reverses chronic unpredictable mild stress-induced depression-like behaviors and alleviates neuroinflammation and collapsing response mediator protein-2-mediated neuroplasticity injury in adult male mice. Neuroreport 33, 272–282 (2022).
Guo, J. B. et al. Comparative transcriptome profiling reveals changes of microRNAs response to exercise in rats with neuropathic pain. Neural Plast., 5597139 (2021).
Farzad, B. et al. Swimming training attenuates Allodynia and Hyperalgesia Induced by Peripheral nerve Injury in an adult male rat neuropathic model: effects on Irisin and GAD65. Pain Med. 19, 2236–2245 (2018).
Shen, J., Fox, L. E. & Cheng, J. Swim therapy reduces mechanical allodynia and thermal hyperalgesia induced by chronic constriction nerve injury in rats. Pain Med. 14, 516–525 (2013).
Chaplan, S. R., Bach, F. W., Pogrel, J. W., Chung, J. M. & Yaksh, T. L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods. 53, 55–63 (1994).
Ma, B. F. et al. Electroacupuncture alleviates Thalamic Pain in rats by suppressing ADCY1 protein upregulation. Pain Phys.. 25, E629–E640 (2022).
Dixon, W. J. Efficient analysis of experimental observations. Annu. Rev. Pharmacol. Toxicol. 20, 441–462 (1980).
Hargreaves, K., Dubner, R., Brown, F., Flores, C. & Joris, J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain 32, 77–88 (1988).
Bennett, G. J. & Xie, Y. K. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain 33, 87–107 (1988).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Yu, X. et al. Proteomics analysis of the spinal dorsal horn in diabetic painful neuropathy rats with electroacupuncture treatment. Front. Endocrinol. (Lausanne). 12, 608183 (2021).
Braga, C. L. et al. Proteomics profile of mesenchymal stromal cells and extracellular vesicles in normoxic and hypoxic conditions. Cytotherapy 24, 1211–1224 (2022).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Tang, D. et al. SRplot: a free online platform for data visualization and graphing. PLoS One. 18, e0294236 (2023).
Bader, G. D. & Hogue, C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 4, 2 (2003).
Acknowledgements
We thank the School of Medical Technology, Xuzhou Key Laboratory of Laboratory Diagnostics, Xuzhou Medical University for providing the experimental venue. We thank Biorender software, and Fig. 1 A was created with BioRender.com. And we also thank Mingjie Chen (Shanghai NewCore Biotechnology Co., Ltd.) for providing data analysis and visualization support.
Funding
This work was supported by Youth Fund of Jiangsu Basic Research Program in 2021 (BK20210907), Medical Research Project of Jiangsu Commission of Health (Z2022004).
Author information
Authors and Affiliations
Contributions
H.X.L. and J.B.G. jointly conceived the study. B.L.C. wrote the main manuscript text, and created the table and figure. Y.L.Z. contributed to the completion of the table and figuer. C.G. and C.C.Z. performed experiments and data analysis. Y.W. and S.B.W. assist the performance of experiments and data analysis. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics and consent to participate
The study was carried out in compliance with the ARRIVE guidelines (https://arriveguidelines.org) and approved by the Scientific Research Ethics Committee of Xuzhou Medical University (No: 202207S014).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, B., Guo, J., Gong, C. et al. Proteomic analysis of spinal dorsal horn in prior exercise protection against neuropathic pain. Sci Rep 15, 2391 (2025). https://doi.org/10.1038/s41598-025-86661-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-86661-0