Abstract
Antimony (Sb), a non-essential heavy metal, exerts severe toxic effects on the growth and development of plants. This study investigated the response of Brassica napus to Sb(III) stress under hydroponic conditions, focusing on Sb accumulation, physiological indexes, and transcriptome sequencing. Sb accumulation in different B. napus varieties showed consistent trends with physiological indicators (SOD, POD, CAT, MDA) in XZY512 root tissue. Both parameters increased with Sb concentration, reaching a peak at 75 mg/L before declining, suggesting that 75 mg/L Sb may be the optimal concentration for B. napus adaptation. Transcriptomic analysis identified 8,802 genes in root tissues and 13,612 genes in leaf tissues responsive to Sb stress, predominantly involved in oxidative stress responses, ABC transporters, glutathione metabolism, plant hormone signaling, and MAPK pathways. Physiological index changes were associated with upregulation of genes linked to antioxidants, including as CATs, GPXs, PERs, and GSTUs, in root tissues, whereas photosynthesis-related genes were mostly downregulated in leaf tissues. This work shows the potential of B. napus for phytoremediation efforts and offers important insights into its response mechanisms to Sb stress.
Similar content being viewed by others
Introduction
As urban industrialization progresses, the exploitation of mineral resources, along with the accumulation and smelting of heavy metals, has resulted in the widespread distribution of heavy metals and their compounds in the atmosphere, water bodies, and soil. Heavy metals such as cadmium (Cd), lead (Pb), chromium (Cr), antimony (Sb), among others, have caused significant environmental damage, resulting in severe pollution1,2. Sb is a crucial strategic resource with extensive applications in semiconductors, batteries, flame retardants, ceramics, weapons, and pharmaceutical materials3,4,5. Regrettably, improper handling has led to varying levels of Sb contamination in soil, adversely affecting the plant growth and development.
Sb exists in four oxidation states (-III, 0, III, and V) in the natural environment. Among these, inorganic Sb exhibits higher toxicity compared to its organic counterpart, and its toxicity varies depending on its oxidation states. Specifically, Sb(III) is ten times more toxic than that Sb(V)2,6,7. In recent years, research into the mechanisms of plant uptake and toxicity related to the heavy metal Sb has emerged a significant field. Studies on the sunflower and maize have revealed varying levels of tolerance to Sb toxicity among different crops, with maize showing greater susceptibility to Sb in soil8. Research demonstrates that Sb inhibits root elongation and germination in rice, significantly impacting rice productivity9. Further studies show that Sb(III) has a pronounced impact on rice root and stem length, as well as biomass. Exogenous Bio-SeNPs help mitigate Sb(III) toxicity, with genes such as OsCuZnSOD2, OsCATA, and OsGSH1 potentially involved in Sb detoxification10. Under heavy metal stresses, antioxidant enzymes like peroxidase (POD), superoxide dismutase (SOD), catalase (CAT), and glutathione (GSH), serves as critical indicators of plant’s antioxidative defense system. Studies on maize Sb tolerance have found that when soil Sb concentration exceeds 50 mg/kg, the activities of POD and SOD in maize plants are significantly inhibited11, indicating that the optimal Sb concentration for maize tolerance is below this threshold.
Brassica napus L., commonly known as rapeseed, is a globally significant oilseed crop extensively utilized in soil phytoremediation studies due to its rapid growth rate, substantial shoot biomass, and remarkable ability to adsorb heavy metals12,13,14,15. Additionally, RNA sequencing (RNA-seq) technology has been extensively employed to investigate the molecular mechanisms underlying plants responses to heavy metal stress. Researches have explored the effects of various metals, including Mn16, Cu17, Pb18, Cd19,20, and Sb21,22, in plant species such as Schima superba, Citrus grandis, Raphanus sativus L, Brassica oleracea L(Broccoli), Brassica juncea L, Festuca arundinacea. However, studies on the response of B. napus to Sb-induced stress are limited, and the molecular mechanisms underlying its response to Sb stress remain unclear.
In this study, we investigated Sb accumulation in several B. napus cultivars exposed to different Sb concentrations, evaluated physiological indicators in roots and leaves, and used RNA-seq to find differentially expressed genes (DEGs) associated with Sb toxicity., The results provide insights into the key regulatory networks and genes underlying B. napus tolerance to Sb, laying the groundwork for future research on Sb-tolerant variety selection and molecular mechanisms.
Results
Sb accumulation in different B. napus varieties
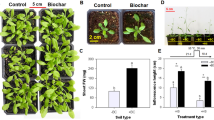
In this study, Sb accumulation in the entire plants of B. napus was used as a crucial indicator to evaluate its phytoremediation potential. The findings showed a progressive increase in Sb accumulation across six B. napus varieties as the Sb concentration increased, reaching a peak before declining (Fig. 1). Specifically, the highest levels were observed in the varieties XZY787, XZY512, ZYZ28, and QY18H at an Sb concentration of 75 mg/L, with values of 325.63, 561.42, 257.40, and 199.73 mg/kg, respectively. FY789 showed its maximum Sb accumulation of 222.29 mg/kg at 50 mg/L, while HYZ50 peaked at 204.35 mg/kg at a concentration of 25 mg/L. Among all the varieties, XZY512 exhibited the highest Sb content of 561.42 mg/kg at 75 mg/L, making it the focus of further investigation.
Variations in physiological indexes
To assess the impact of heavy metals on plants, toxicity to plant growth is frequently evaluated by measuring enzyme activities and metabolite levels, such as SOD, POD, CAT, and MDA23,24. This study measured the activities and contents of these enzymes or metabolites in both root and leaf tissues of B. napus XZY512 under varying Sb concentration treatments. Compared to the controls, root tissues showed increased activity or content of SOD (Fig. 2A), POD (Fig. 2B), CAT (Fig. 2C), and MDA (Fig. 2D) in response to rising Sb concentrations, particularly at low concentration. Peak values were observed around 75 mg/L Sb, followed by a decline. Additionally, SOD and POD activity changes in leaf and root tissues varied inconsistently under different Sb treatments (Fig. 2A-B). However, CAT activity in leaf tissues decreased under high-concentration Sb solutions (Fig. 2C), while MDA content notably declined (Fig. 2D). These results suggest that Sb stress significantly impacts B. napus growth, with physiological indices declining after reaching peak values, indicating a potential link to the plant’s adaptability to Sb stress or physiological damage caused by Sb toxicity.
Physiological indices analysis of root and leaf tissues of XZY512 under different Sb concentrations: SOD (A), POD (B), CAT (C), and MDA (D). The x-axis represents treatments with varying Sb concentrations. Different lowercase letters represent significant differences at p < 0.05 level using one-way ANOVA and Tukey’ s correction.
RNA sequencing data and identification of DEGs under Sb stress
To investigate the molecular mechanisms underlying Sb stress in B. napus, we conducted RNA-seq analysis on the roots and leaves treated with concentrations of 0 mg/L (control) and 75 mg/L Sb (treatment). The samples were designated as root control (RCK), root treatment (RTr), leaf control (LCK), and leaf treatment (LTr). High-throughput sequencing was performed on 12 samples, and the raw data were uploaded to the NCBI SRA database with accession numbers: SRR29906309 (RCK1), SRR29906308 (RCK2), SRR29906305 (RCK3), SRR29906304 (RTr1), SRR29906303 (RTr2), SRR29906302 (RTr3), SRR29906301 (LCK1), SRR29906300 (LCK2), SRR29906299 (LCK3), SRR29906298 (LTr1), SRR29906307 (LTr2), and SRR29906306 (LTr3). A total of 80.58 Gb of clean bases were obtained, with each sample yielding 5.97 Gb of clean data and a Q30 value exceeding 91.56%. All XZY512 leaf samples exhibited alignment rates above 93.25% when mapped to the ZS11 reference genome. However, root sample RTr3 showed a much lower alignment rate of 7.64% (Table S1). To maintain the integrity of subsequent analyses, the RTr3 sample was removed, and the analysis proceeded with the remaining 11 samples.
Using a filtering threshold of P-value < 0.05 and |log2(fold-change, FC)| > 1, we identified 8,802 DEGs in the RCK_vs_RTr comparison (4,274 upregulated, 4,528 downregulated) and 13,612 DEGs in the LCK_vs_LTr comparison (6,653 upregulated, 6,959 downregulated) (Fig. 3A). Notably, 2,218 DEGs were shared between root and leaf tissues, comprising 1,498 up-regulated genes and 720 down-regulated genes (Fig. 3B).
GO enrichment and KEGG pathway analysis of DEGs in root and leaf tissues
To deeper understand the function of DEGs between the control and treatment groups, we performed GO enrichment analysis on DEGs derived from RCK_vs_RTr, LCK_vs_LTr, and (RCK_vs_RTr)_vs_(LCK_vs_LTr) comparisons. We then classified the relevant GO terms statistically. The DEGs were primarily participate in functions such as cell, cell part, organelle, binding, catalytic activity, cellular process, metabolic process, and response to stimulus (Fig. S1). The comparison of upregulated and downregulated gene counts between RCK_vs_RTr and LCK_vs_LTr showed similar patterns. However, in the (RCK_vs_RTr)_vs_(LCK_vs_LTr) comparison, there was a notably higher count of upregulated genes among the shared DEGs pool.
GO enrichment and KEGG pathway analysis of DEGs between RCK_vs_RTr and LCK_vs_LTr revealed distinct patterns. Specifically, DEGs in RCK_vs_RTr were significantly enriched in GO terms related to response to oxidative stress (GO:0006979), response to toxic substance (GO:0009636), iron ion binding (GO:0005506), and glutathione transferase activity (GO:0004364) (Fig. 4A). In contrast, DEGs in LCK_vs_LTr were predominantly enriched in GO terms such as chloroplast thylakoid (GO:0009534), photosystem II (GO:0009523), response to abscisic acid (GO:0009737), and chlorophyll binding (GO:0016168) (Fig. 4C). KEGG pathway findings indicated that Sb stress treatment led to significant enrichment of DEGs in root tissues, particularly in pathways such as Glutathione metabolism (map00480), Plant hormone signal transduction (map04075), Starch and sucrose metabolism (map00500), ABC transporters (map02010), Brassinosteroid biosynthesis (map00905), and MAPK signaling pathway-plant (map04016) (Fig. 4B). Differently, DEGs in the leaves showed enrichment in pathways such as carbon fixation in photosynthetic organisms (map00710), photosynthesis (map00195), photosynthesis -antenna proteins (map00196), glutathione metabolism (map00480), and oxidative phosphorylation (map00190) (Fig. 4D). The top 25 enriched GO terms and KEGG pathways in the (RCK_vs_RTr)_vs_(LCK_vs_LTr) comparison included significant terms like response to toxic substance (GO:0009636), response to oxidative stress (GO:0006979), vacuolar membrane (GO:0005774), and glutathione binding (GO:0043295) (Fig. 4E). Key metabolic pathways comprised glutathione metabolism (map00480), protein processing in endoplasmic reticulum (map04141), ABC transporters (map02010), and Starch and sucrose metabolism (map00500) (Fig. 4F).
Based on the aforementioned findings, we propose that there are significant disparities in DEGs between the plant treated with 75 m g/L Sb and the control groups. Moreover, the observed DEGs exhibit a notable preference for root and leaf tissues. The toxic effects of Sb may primarily impact gene expression related to oxidative stress, glutathione metabolism, ABC transporters, and MAPK signaling pathway in root tissues. In leaves, the affected genes are mainly associated with Photosynthetic metabolism.
Top 25 GO terms and KEGG pathways enrichment analysis of DEGs in RCK_vs_RTr (A, B), LCK_vs_LTr (C, D), and (RCK_vs_RTr)_vs_(LCK_vs_LTr) (E, F). RCK, RTr, LCK, and LTr represent control and treatment samples of roots and leaves, respectively. The red font highlights the significant GO terms and KEGG pathways associated with Sb stress in the roots and leaves of B.napus.
DEGs analysis of antioxidant stress response by Sb-induced
During cellular metabolism, organelles produce a significant amount of reactive oxygen species (ROS), which are effectively regulated by the antioxidant system to maintain a dynamic equilibrium25. Nonetheless, excessive exposure to heavy metals triggers a substantial increase in ROS production, disrupting the homeostasis of the antioxidant system. As a result, there is a significant accumulation of ROS within the organism, causing enduring damage to cellular structures, proteins, and other essential components26. In this study, we observed that under Sb stress, key genes associated with oxidative stress response (58/64) (GO:0006979) were significantly upregulated in both root and leaf tissues of B. napus. This included a wide range of antioxidant genes, both enzyme and non-enzyme types, such as glutathione peroxidases (GPXs), ascorbate peroxidases (APXs), and peroxidases (PERs) (Table S2). Upon analyzing the DEGs regulating SOD, POD, and CAT activities, we discovered that genes related to SOD (SODC1), POD (PER), and CAT (CAT1) exhibited significantly higher expression in roots, aligned with their physiological activities under Sb treatment (Fig. 2, Table S2). Notably, only a small subset of SODC1, CAT, and PER genes showed no differential expression in leaf tissue, which corresponded with the physiologicalobservations in Sb-treated leaves (Fig. 2, Table S2). These findings suggest that the majority of antioxidant-related genes, including those regulating SOD and CAT activity, are upregulated in response to Sb-induced oxidative damage, thereby helping to mitigate the harmful effects of ROS, H2O2, and other toxic substances.
Analysis of DEGs in the glutathione metabolism and MAPK signaling pathways under Sb stress
Glutathione is a vital metabolite for plant survival, involved in detoxifying ROS and methylglyoxal (MG). It exists in two reversible states within cells: the monomeric reduced sulfhydryl form (GSH) and the oxidized disulfide dimer (GSSG)27. This study identified six groups of DEGs associated with the enrichment of Glutathione metabolism in B. napus root tissues. After Sb treatment, six glutathione peroxidase (GPX) genes in the GSH-GSSG conversion pathway were significantly upregulated. Similarly, three genes encoding 6-phosphogluconate dehydrogenase, decarboxylating genes (At3g02360) were also upregulated, while the Glucose-6-phosphate 1-dehydrogenase (G6PD3) was downregulated. Additionally, sixteen Glutathione S-transferase (GSTUs and GSTFs) genes involved in L-glutamate metabolism were detected, with GSTU genes predominantly upregulated, whereas GSTF genes showed a higher number of downregulated expressions. The L-amino acid synthesis-related genes Gamma-glutamyltranspeptidase 1 (GGT1) and Glutathione-specific gamma-glutamylcyclotransferase 2 (CHAC2) were upregulated in response to Sb stress, while CHAC1 exhibited the opposite trend (Fig. 5A, Table S3). As a major antioxidant and scavenger of free radicals, glutathione was found to upregulate the majority of the genes involved in its metabolism, suggesting its crucial role in regulating the metabolisms of GSH, L-glutamate, and L-amino acids.
Another important KEGG pathway enriched in this study is the MAPK signaling pathways. As shown in Fig. 5B, the Mitogen-activated protein kinase kinase kinase 1 (MEKK1) gene, through a series of phosphorylation events, may activate the expression of cell death defense response genes SUMM2 and RPS5 in the pathogen infection pathway. Additionally, genes involved in camalexin synthesis (WRKY33), ethylene synthesis (ACS6), and late defense responses to pathogens (WRKY29 and PR1) were also upregulated (Fig. 5B, Table S3). These results suggest that these genes play a positive regulatory role in the response to Sb-induced toxicity. We further analyzed the Salt/Drought/Osmotic stress regulatory pathways in the MAPK signaling pathway. Under Sb stress, the abscisic acid receptor genes PYL and PYR were downregulated, preventing the inhibition of the ABI (PP2C) gene, and leading to a significant upregulation of ABI1 (Fig. 5B, Table S3). The Serine/threonine-protein kinase genes (SRK2C) and (SRK2F), the Mitogen-activated protein kinase kinase kinase gene MAPKKK17, and the Mitogen-activated protein kinase 2 (MPK2) gene may collectively contribute to the plant’s adaptive response to Sb stress.
Glutathione metabolism (A) and MAPK signaling (B) pathway DEGs in B. napus roots following Sb treatment. Orange and blue squares indicate upregulated and downregulated DEGs, respectively, while gene IDs of the nodes in the KEGG pathway are highlighted in green font. The copyright license for image use is obtained.
DEGs associated with ABC transporters and photosynthesis pathway
ABC transporters are among the most common membrane protein families in plants, with subfamily members involved in the transport of anthocyanins, flavonoids, glutathione conjugates, and other compounds, crucial for plant detoxification28. In this study, a total of 15 ABC genes from the ABCB and ABCC subfamilies were discovered to be involved in the ABC transporter pathway in root tissues. Nine DEGs from the ABCB subfamily were upregulated under Sb stress, while in the ABCC subfamily, four genes (except ABCC9 and ABCC4) were downregulated in response to Sb toxicity (Fig. 6A, Table S3).
Under Sb stress, the photosynthesis pathway was the most significantly enriched among the DEGs in leaf tissues. Expression of five gene sets was notably downregulated, including core genes of the PSI reaction center (PSAE2, PSAG, PSAK, PSAO), PSII receptor-side (PSBB, PSBP, PSBR, PSBW), photosynthetic electron transport (DRT112, FD2, LFNR2, PETJ), Cytb6/f complex (PETC), and F-type ATPase (ATPC, ATPD) genes (Fig. 6B, Table S3). This compelling evidence indicates that the primary toxic effect of heavy metal Sb on the leaves of B. napus is a significant disruption of the molecular mechanisms of photosynthesis.
Expression patterns of ABC transporters and photosynthesis pathway-related genes. (A) Expression analysis of ABC transporter pathway-related genes in root tissues under Sb stress. (B) Expression analysis of photosynthesis pathway-related genes in leaf tissues under Sb stress. Orange and blue squares indicate upregulated and downregulated DEGs, respectively, while gene IDs of the nodes in the KEGG pathway are highlighted in green font.
Validation of RNA-Seq by qRT-PCR
To validate the RNA-seq data results, nine genes were selected for qRT-PCR verification, including DEGs related to antioxidant stress PER34 (BnaA01G0220100ZS) and GPX2 (BnaA03G0152400ZS), the Glutathione S-transferase GSTU10 (BnaC02G0283300ZS), transcription factor WRKY28 (BnaC07G0420700ZS) and WRKY75 (BnaC09G0529900ZS), the ABC transporter ABCB14 (BnaA09G0426500ZS), photosynthesis-related LHCA6 (BnaA08G0253100ZS), MAPK signaling pathway RBOHG (BnaA03G0308600ZS), and metal transport or binding protein HMA2 (BnaC07G0494200ZS) (Table S4). The findings indicated a strong positive correlation between the qRT-PCR and RNA-seq data, with correlation coefficients (R2) of 0.889 and 0.892 for DEGs in root and leaf tissues, respectively (Fig. 7A-B). This high correlation validates the reliability of the RNA-seq data.
Validation of transcript abundance from RNA-seq using qRT-PCR. (A) Transcript abundance of nine representative genes in root and leaf tissues validated by qRT-PCR. Green and orange bars represent Log2(Fold Change) from RNA-seq and qRT-PCR, respectively. (B) Co-linearity analysis of transcript abundance for nine representative genes in root tissue between RNA-seq and qRT-PCR data. C.Co-linearity analysis of transcript abundance for nine representative genes in leaf tissue between RNA-seq and qRT-PCR data. Blue circles represent the target genes being tested. The R2 value represents the level of linear correlation between transcript abundance and qRT-PCR data.
Discussion
Variations in Sb uptake and physiological indices in B. napus under Sb stress
Sb is a toxic heavy metal that can negatively affect plant growth and development. In this study, we conducted experiments with varying Sb concentrations to investigate the variations in Sb accumulation across different B. napus varieties. Our preliminary findings indicated that Sb accumulation in B. napus followed a dose-dependent pattern, with increased Sb concentration leading to higher uptake. A similar relationship has been observed in Brassica juncea, where arsenic accumulation demonstrated positively correlated with rising arsenic concentrations24. These results suggest that B. napus exhibits a toxicological response to Sb similar to that of arsenic, a clan element.
MDA, a product of lipid peroxidation, signifies reduced free radical scavenging capability in plants as its levels rise. The antioxidant enzyme system, including SOD, POD, CAT, and other enzymes, plays a crucial role in eliminating free radicals and ROS, thereby protecting cells from damage29. Previous studies have shown that the MDA content and SOD activity in rice root tissues significantly increase with rising Sb concentrations (0, 10, 20 mg/L), while POD activity decreases as Sb concentration increases27. In our study, the aforementioned physiological indices in B. napus root tissues exhibited an increased trend with rising Sb concentration, peaking at 75 mg/L, and subsequently declining (Fig. 2). This trend closely matched the Sb absorption results across six B. napus varieties (Fig. 1), suggesting that 75 mg/L may represent the threshold for optimal Sb adaptation in B. napus. In comparison to rice under Sb stress, the 20 mg/L treatment may not have reached the optimal Sb tolerance level for rice. In leaf tissues, which SOD and POD activities remained stable, CAT activity and MDA content were significantly affected by high-level Sb concentration (Fig. 2). In summary, Sb-induced stress significantly affects Sb absorption and the antioxidant system in B.napus, likely causing cellular damage and substantial physiological changes at around 75 mg/L (Fig. 8).
Effects of Sb stress on oxidative stress and ABC transporter genes
In response to Sb-induced oxidative stress, plants typically activate specific antioxidant enzymes and small molecule antioxidants to against the toxicity of Sb30,31,32. This investigation discovered that Sb stress significantly increased SOD, POD, and CAT activities in root tissues of B. napus. Moreover, the majority of antioxidant genes, including both enzymes and non-enzymes such as GPXs, APXs, and PERs, were significantly upregulated, actively contributing in mitigating Sb-induced oxidative stress. Our previous research revealed that 10 mg/L and 50 mg/L Sb stress activated the antioxidant enzyme system in rice, altered root structures, and induced toxicity33. Recent studies have also indicated that Sb(III) stress enhanced antioxidant enzyme activities and activates genes associated with oxidoreductase activity in tall fescue22. When considering the findings from rice and tall fescue, the upregulation of genes associated with oxidative enzyme, along with the corresponding increase in antioxidant enzyme activities (SOD, POD, and CAT) in this study, further confirm the pronounced toxicity of a 75 mg/L Sb concentration on B. napus root tissues (Fig. 7).
ABC transporters, the largest family of transporter proteins, are well known for their critical roles in metal detoxification and homeostasis34. In this study, we identified six DEGs in the ABCC transporter subfamily and nine in the ABCB subfamily during Sb stress (Fig. 6A, Table S3). Previous studies have shown that ABCB transporters are primarily involved in the transport of plant hormones, while ABCC transporters, located on both plasma and vacuolar membranes, play a key role in metal detoxification, transport, and vacuolar sequestration35. On the basis of DEG enrichment in the Plant Hormone Signal Transduction and Brassinosteroid Biosynthesis pathways and the upregulation of ABCB members under Sb stress, we hypothesize that ABCB transporters may facilitate the movement of plant hormones such as brassinolide, auxin and abscisic acid, thereby mitigating Sb toxicity. Moreover, ABCC1 and ABCC2 are involved in the vacuolar sequestration of mercury, arsenic, and cadmium, enhancing plant tolerance to these metals36. Similarly, studies on Bio-SeNPs treatment have demonstrated that OsABCC1 plays a role in Sb detoxification in rice10. Consequently, the DEGs identified in the ABCC subfamily in this study suggest that these genes may also be involved in Sb vacuolar sequestration.
Glutathione metabolism and MAPK signaling pathway respond to Sb stress
Glutathione metabolism is a crucial defense mechanism against stress in aerobic life supported by respiration and glycolysis. It not only effectively regulates ROS generated during aerobic metabolism (respiration and photosynthesis), but also plays a pivotal role in detoxifying the toxic by-product MG produced during glycolysis27. GPXs are essential enzymes in the antioxidant system, known to maintain the thiol/disulfide or NADPH/NADP+ balance within the reversible GSH-GSSG cycle, thereby regulating cellular redox homeostasis37. Transcriptomic analysis of B.napus roots under Sb stress identified several DEGs, including GPXs and ACG9, involved in the GSH-GSSG cycle. Previous studies have shown that GPXs are key players in oxidative stress responses to abiotic stresses such as cold, drought, heat, and salinity38 ,suggesting that GPX genes are also involved in maintaining cellular redox homeostasis under Sb toxicity stress in B.napus. In plants, the multifunctional enzyme glutathione S-transferase (GST) is classified into 14 distinct types, with tau (GSTU) and phi (GSTF) being the most abundant39. GSTs play a crucial role in responding to various abiotic stresses, including cold, salinity, and UV-B radiation40. In this study, differentially expressed GSTUs and GSTFs exhibited contrasting expression patterns in response to Sb stress, suggesting that GSTUs may positively regulate resistance to Sb-induced oxidative stress, while GSTFs may negatively modulate this response.
MAPK signaling cascades are well-established in plants for their involvement in stress adaptation, cell death, defense responses, ethylene signal transduction, and maintenance of ROS responses24,41. After Sb treatment, DEGs associated with the MAPK signaling pathway were significantly enriched in root tissues. In Arabidopsis, the expression of ACS2 and ACS6 are directly regulated by the downstream WRKY33 in the MPK3 and MPK6 cascade in response to pathogen invasion42. Additionally, WRKY29 has been shown to bind to the promoters of genes such as ACS5, ACS6, and ACS8, activating their expression and positively regulating ethylene production43. In this study, DEGs such as WRKY29, WRKY33, and ACS6 were responsive to Sb stress. Based on previous reports, it is hypothesized that ACS6 is upregulated by WRKY33, thereby regulating ethylene biosynthesis in response to Sb stress, while WRKY29 may negatively influence the Sb stress response through an as-yet undefined mechanism.
The ABA signaling module, another MAPK signaling cascade, consists of PYR/PYL/RCAR (Pyrabactin Resistance/Pyrabactin resistance-like/Regulatory Component of ABA Receptor, PP2C (Protein Phosphatase 2 C), and SnRK2s (SNF1-Related Protein Kinases type 2). This module is activated when drought signals are perceived by PYR/PYL/RCAR receptors, triggering conformational change that facilitates binding to the catalytic site of PP2C. This binding prevents PP2C from dephosphorylating SnRK2, leading to SnRK2 autophosphorylation44. In the present study, genes associated with the ABA signaling module, including PYR/PYL, PP2C, and SnRK2s, showed differential expression. These results suggest that Sb stress may be perceived by PYR/PYL receptors in a manner similar to drought signals, resulting in the downregulation of PYR/PYL genes, activation of PP2C’s catalytic site, and inhibition of SnRK2 autophosphorylation.
Effects of Sb stress on photosynthetic system
Photosynthesis is one of the most crucial physiological processes in plants, highly sensitive to environmental changes, and often serves as a key indicator of the impact of adverse environmental factors on plant growth and development45. Previous studies have shown that High concentrations of Sb(III) are found to significantly reduce PS II (Fv/Fm and PIABS) in maize leaf11 and cause a significant decrease in leaf pigment contents (chlorophyll a, b, carotenoid), net photosynthetic rate (Pn), stomatal conductance (Gs), evaporation rate (E), PSII maximum photochemical efficiency (Fv/Fm), and PSII electron transfer quantum yield rate (ΦPSII) of Acorus calamus46. This study used transcriptomics to evaluate changes in leaf DEGs under Sb stress, revealing significant downregulation of genes involved in the PSI reaction center, PSII receptor side, photosynthetic electron transport, Cytb6/f complex, and F-type ATPase. These results suggest that Sb stress causes substantial damage to B.napus leaves, impairing light absorption, transfer, and conversion, as well as photosynthetic electron transport, H+ transport, and ATP synthesis. Consequently, this leads to reduced energy production and slowed growth and development in plants.
In summary, the presence of the aforementioned DEGs in root and leaf tissues indicate that the response of B. napus to Sb stress involves a complex regulatory network, with these DEGs potentially playing a key role in the defense against Sb toxicity.
Conclusions
Sb accumulation in different B. napus varieties showed consistent trends with physiological indicators (SOD, POD, CAT, MDA) in XZY512 root tissue. Both parameters increased with Sb concentration, reaching a peak at 75 mg/L before declining, suggesting that 75 mg/L Sb may be the optimal concentration for B. napus adaptation. Transcriptomic analysis identified DEGs in B. napus root tissues under Sb stress, associated with oxidative stress response, ABC transporters, glutathione metabolism, plant hormone signaling, and MAPK signaling pathway. Notably, photosynthesis-related genes were significantly downregulated in leaf tissues. This study investigated the effects of heavy metal Sb on B. napus through Sb accumulation, physiological indices, and transcriptome sequencing, providing initial insights into the biological processes and gene regulatory networks in B. napus under Sb stress.
Materials and methods
Plant material and growth conditions
The commercial varieties used in this study, namely XZY787, FY789, XZY512, HYZ50, ZYZ28, and QY18H, were provided by the Oilseed Molecular Breeding Team at Hunan Agricultural University. The hydroponic system employed was adapted from Thakur et al.24. with specific modifications. Seeds were disinfected in a 50% sodium hypochlorite (v/v) solution for 2 min, followed by rinsed with distilled water. The seeds then were placed in perlite trays soaked with 1/2 Hoagland’s solution. Plants were cultivated in a phytotron for 35 to 40 days until they reached the 5-leaf stage, under a 12-hours light cycle, with a relative humidity of 50–60% and a temperature of 24 ± 2 °C. Afterward, the plants were transferred to hydroponic tanks containing a 1/2 Hoagland’s solutions. Following a 72-hours incubation, the plants were treated with Sb(III) solutions (C4H4KO7Sb·(1/2)H2O) at concentrations of 0, 10, 25, 50, 75, and 100 mg/L. Each variety and concentration were replicated three times. After the 72-hour treatment, samples were collected, rapidly frozen with liquid nitrogen, and stored in an ultra-low temperature freezer.
Determination of Sb accumulation and physiological indices
To assess Sb accumulation, both control and Sb-treated plants were dried. Initially, they were placed in a drying oven set at 105 °C for 30 min, followed by further drying at 75 °C until a constant weight was achieved. The sample processing procedure followed the method described by Liao et al.47. After sample digestion, the Sb accumulation was quantified using ICP-MS (PerkinElmer NexlON®2000, USA). To investigate the impact of varying Sb concentrations on the roots and leaves of B. napus, assay kits from Nanjing Jiancheng Bioengineering Institute were used to measure the activity of antioxidant enzymes. Specifically, the following assay kits were utilized: superoxide dismutase (SOD) assay kit (A001-3, WST-1), peroxidase (POD) assay kit (A084-3, colorimetric method), catalase (CAT) assay kit (A007-1, visible light method), and malondialdehyde (MDA) assay kit (A003-1, TBA method) to measure physiological indexes of the respective B. napus tissues. Measurements of Sb accumulation and physiological indices were conducted with at least three biological replicates.
RNA extract, library preparation and transcriptome sequencing
Based on the Sb accumulation results, a total of 12 samples (Three replicates each for roots and leaves) from XZY512, following a 72-hour treatment with 0 mg/L and 75 mg/L Sb concentrations, were selected for transcriptome sequencing. Total RNA was extracted using the RNAprep Pure Plant Plus Kit (Tiangen, Beijing, China), following the manufacturer’s protocol. The quality of the extracted RNA was evaluated using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA), Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA), and Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) prior to library preparation. Sequencing libraries were generated using NEB Next® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer’s instructions. The detailed instructions for library construction were followed according to the protocol described by Zheng et al.48. The libraries were sequenced with 2 × 150 bp paired-end reads using the Illumina Nova seq6000 platform (Jisi Huiyuan Biotechnology Co., Ltd., Nanjing, China). High-quality reads were then used to assemble unigenes following the method outlined by Haas et al.49. Finally, obtained sequence data were mapped to the reference genome of B. napus (ZS11) (https://www.ncbi.nlm.nih.gov/assembly/GCF_000686985.2/) utilizing the default parameters of the HISAT v2.1.050 software.
Comparative transcriptome analysis and gene functional annotation
To quantify the expression values of assembled transcripts, the fragments per kilobase of transcript per Million fragments mapped (FPKM) was employed as a quantification metric for transcripts or gene expression levels. DEGs among different treatment groups were identified using the DESeq R package51 with a significance threshold of P-value < 0.05 and |log2(fold-change, FC)| > 1. Subsequently, venn diagram and heatmap were generated to visualize the expression patterns of DEGs across various experimental conditions. Furthermore, Gene Ontology (GO) enrichment (http://www.geeontology.org) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (http://www.kegg.jp) analysis were conducted to screen DEGs significantly enriched in GO terms and KEGG pathways52, respectively, at a significance level of P < 0.05.
qRT-PCR validation
Total RNA (1 µg) was reverse-transcribed using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Japan). Quantitative real-time (qRT-PCR) was performed using SYBR Green II (SYBR® Premix Ex Taq™ Kit, Takara) on a CFX96 real-time system (BIO-RAD, USA). The qRT-PCR protocol included an initial denaturation step at 95 °C for 30 s, followed by 40 cycles of amplification at 95 °C for 5 s and extension at 60 °C for 30 s. To validate the RNA-Seq findings, nine DEGs showing significant differences among various treatments were selected for qRT-PCR analysis. Gene-specific qRT-PCR primers (Table 1) were designed using Primer-Premier 3.0 software. BnActin7, described by Chen et al.53, served as an internal control for normalizing gene expression levels. The relative expression of each DEGs was evaluated by the 2−ΔΔCt method. Each gene and sample were analyzed with three biological replicates and three technical replicates. The linear-regression function of the Microsoft Excel 2010 was used to calculate Pearson’s correlation coefficient between qRT–PCR (–ΔΔCT) and RNA-seq results (log2FC).
Data availability
The RNA-seq data from the 12 samples used in this study have been uploaded to the NCBI SRA database. The accession numbers are as follows: SRR29906309 (RCK1), SRR29906308 (RCK2), SRR29906305 (RCK3), SRR29906304 (RTr1), SRR29906303 (RTr2), SRR29906302 (RTr3), SRR29906301 (LCK1), SRR29906300 (LCK2), SRR29906299 (LCK3), SRR29906298 (LTr1), SRR29906307 (LTr2), and SRR29906306 (LTr3). RCK (1, 2, 3), RTr (1, 2, 3), LCK (1, 2, 3), and LTr (1, 2, 3) represent three biological replicates for the control and treated samples of roots and leaves, respectively.
References
Naja, G. M. & Volesky, B. Toxicity and Sources of Pb, Cd, Hg, Cr, as, and Radionuclides in the Environment (Kluwer Academic, 2017).
Zhang, Y., O’Loughlin, E. J. & Kwon, M. J. Antimony redox processes in the environment: a critical review of associated oxidants and reductants. J. Hazard. Mater. 431, 128607. https://doi.org/10.1016/j.jhazmat.2022.128607 (2022).
Ahmad, M. et al. Speciation and phytoavailability of lead and antimony in a small arms range soil amended with mussel shell, cow bone and biochar: EXAFS spectroscopy and chemical extractions. Chemosphere 95, 433–441. https://doi.org/10.1016/j.chemosphere.2013.09.077 (2014).
Okkenhaug, G. et al. Antimony (Sb) and lead (pb) in contaminated shooting range soils: sb and pb mobility and immobilization by iron based sorbents, a field study. J. Hazard. Mater. 307, 336–343. https://doi.org/10.1016/j.jhazmat.2016.01.005 (2016).
Wilson, S. C., Lockwood, P. V., Ashley, P. M. & Tighe, M. The chemistry and behaviour of antimony in the soil environment with comparisons to arsenic: a critical review. Environ. Pollut. 158, 1169–1181. https://doi.org/10.1016/j.envpol.2009.10.045 (2010).
He, M., Wang, N., Long, X., Zhang, C. & Shan, J. Antimony speciation in the environment: recent advances in understanding the biogeochemical processes and ecological effects. J. Environ. Sci. 75 https://doi.org/10.1016/j.jes.2018.05.023 (2018).
Salam, M. A. & Mohamed, R. M. Removal of antimony (III) by multi-walled carbon nanotubes from model solution and environmental samples. Chem. Eng. Res. Des. 91, 1352–1360. https://doi.org/10.1016/j.cherd.2013.02.007 (2013).
Tschan, M., Robinson, B., Johnson, C. A., Bürgi, A. & Schulin, R. Antimony uptake and toxicity in sunflower and maize growing in Sb III and Sb V contaminated soil. Plant. Soil. 334, 235–245. https://doi.org/10.1007/s11104-010-0378-2 (2010).
Cui, X. D., Wang, Y. J., Hockmann, K. & Zhou, D. M. Effect of iron plaque on antimony uptake by rice (Oryza sativa L). Environ. Pollut. 204, 133–140. https://doi.org/10.1016/j.envpol.2015.04.019 (2015).
Ran, M., Wu, J., Jiao, Y. & Li, J. Biosynthetic selenium nanoparticles (Bio-SeNPs) mitigate the toxicity of antimony (sb) in rice (Oryza sativa L.) by limiting sb uptake, improving antioxidant defense system and regulating stress-related gene expression. J. Hazard. Mater. 470, 134263. https://doi.org/10.1016/j.jhazmat.2024.134263 (2024).
Pan, X. et al. Sb uptake and photosynthesis of Zea mays growing in soil watered with sb mine drainage: an OJIP chlorophyll fluorescence study. Pol. J. Environ. Stud. 19, 981. https://doi.org/10.1017/S0032247409008626 (2010).
Angelova, V., Ivanova, R., Todorov, J. & Ivanov, K. Potential of rapeseed (Brassica napus L.) for phytoremediation of soils contaminated with heavy metals. J. Environ. Prot. Ecol. 18, 468–478 (2017).
Marchiol, L., Assolari, S., Sacco, P. & Zerbi, G. Phytoextraction of heavy metals by canola (Brassica napus) and radish (Raphanus sativus) grown on multicontaminated soil. Environ. Pollut. 132, 21–27. https://doi.org/10.1016/j.envpol.2004.04.001 (2004).
Nouairi, I. et al. Comparative study of cadmium effects on membrane lipid composition of Brassica juncea and Brassica napus leaves. Plant. Sci. 170, 511–519. https://doi.org/10.1016/j.plantsci.2005.10.003 (2006).
Zheng, Y. et al. Alleviation of metal stress in rape seedlings (Brassica napus L.) using the antimony-resistant plant growth-promoting rhizobacteria Cupriavidus sp. S-8-2. Sci. Total Environ. 858, 159955. https://doi.org/10.1016/j.scitotenv.2022.159955 (2023).
Liaquat, F. et al. Reprisal of Schima superba to mn stress and exploration of its defense mechanism through transcriptomic analysis. Front. Plant. Sci. 13, 1022686. https://doi.org/10.3389/fpls.2022.1022686 (2022).
Ren, Q. Q. et al. Physiological and molecular adaptations of Citrus grandis roots to long-term copper excess revealed by physiology, metabolome and transcriptome. Environ. Exp. Bot. 203, 105049. https://doi.org/10.1016/j.envexpbot.2022.105049 (2022).
Wang, Y. et al. Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (pb) stress with next generation sequencing. PLoS One. 8, e66539. https://doi.org/10.1371/journal.pone.0066539 (2013).
Han, M. et al. Transcriptome analysis reveals cotton (Gossypium hirsutum) genes that are differentially expressed in cadmium stress tolerance. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20061479 (2019).
Ma, X. et al. Comparative transcriptome analysis of broccoli seedlings under different cd exposure levels revealed possible pathways involved in hormesis. Sci. Hortic. 304, 111330. https://doi.org/10.1016/j.scienta.2022.111330 (2022).
You, L. et al. Molecular characterization and expression patterns of MTP genes under heavy metal stress in mustard (Brassica juncea L). Sci. Rep. 14, 17857. https://doi.org/10.1038/s41598-024-68877-8 (2024).
Fan, J. et al. Transcriptomic and metabolomic insights into the antimony stress response of tall fescue (Festuca arundinacea). Sci. Total Environ. 933, 172990. https://doi.org/10.1016/j.scitotenv.2024.172990 (2024).
He, Q. et al. Transcriptome profiles of leaves and roots of Goldenrain Tree (Koelreuteria paniculata Laxm.) In response to cadmium stress. Int. J. Environ. Res. Public. Health. 18 https://doi.org/10.3390/ijerph182212046 (2021).
Thakur, S., Choudhary, S., Dubey, P. & Bhardwaj, P. Comparative transcriptome profiling reveals the reprogramming of gene networks under arsenic stress in Indian mustard. Genome 62, 833–847. https://doi.org/10.1139/gen-2018-0152 (2019).
Toppi, L. S. et al. Response to cadmium in carrot in vitro plants and cell suspension cultures. Plant. Sci. 137, 119–129. https://doi.org/10.1016/S0168-9452(98)00099-5 (1998).
Heyno, E., Klose, C. & Krieger-Liszkay, A. Origin of cadmium-induced reactive oxygen species production: mitochondrial electron transfer versus plasma membrane NADPH oxidase. New. Phytol. 179, 687–699. https://doi.org/10.1111/j.1469-8137.2008.02512.x (2008).
Dorion, S., Ouellet, J. C. & Rivoal, J. Glutathione metabolism in plants under stress: beyond reactive oxygen species detoxification. Metabolites 11 https://doi.org/10.3390/metabo11090641 (2021).
Xiao, C. et al. Identification and functional characterization of ABC transporters for selenium accumulation and tolerance in soybean. Plant. Physiol. Biochem. 211, 108676. https://doi.org/10.1016/j.plaphy.2024.108676 (2024).
Vazirzadeh, A., Marhamati, A., Rabiee, R. & Faggio, C. Immunomodulation, antioxidant enhancement and immune genes up-regulation in rainbow trout (Oncorhynchus mykiss) fed on seaweeds included diets. Fish. Shellfish Immun. 106 https://doi.org/10.1016/j.fsi.2020.08.048 (2020).
Feng, R., Wei, C., Tu, S., Wu, F. & Yang, L. Antimony accumulation and antioxidative responses in four fern plants. Plant. Soil. 317, 93–101. https://doi.org/10.1007/s11104-008-9790-2 (2009).
Feng, R. et al. Toxicity of different forms of antimony to rice plant: effects on root exudates, cell wall components, endogenous hormones and antioxidant system. Sci. Total Environ. 711, 134589. https://doi.org/10.1016/j.scitotenv.2019.134589 (2020).
Ma, C. et al. Uptake, translocation and phytotoxicity of antimonite in wheat (Triticum aestivum). Sci. Total Environ. 669, 421–430. https://doi.org/10.1016/j.scitotenv.2019.03.145 (2019).
Duan, R. et al. Effects of antimony stress on growth, structure, enzyme activity and metabolism of Nipponbare rice (Oryza sativa L.) roots. Ecotoxicol. Environ. Saf. 249, 114409. https://doi.org/10.1016/j.ecoenv.2022.114409 (2023).
Ahad, A., Ahmad, N., Ilyas, M., Batool, T. S. & Gul, A. Plant Metal and Metalloid Transporters: The role of ABC Transporters in Metal Transport in Plants, 55–71 (Springer, 2022).
Dahuja, A. et al. Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant. 171, 785–801. https://doi.org/10.1111/ppl.13302 (2021).
Park, J. et al. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant. J. 69, 278–288. https://doi.org/10.1111/j.1365-313X.2011.04789.x (2012).
Madhu, Sharma, A., Kaur, A., Tyagi, S. & Upadhyay, S. K. Glutathione peroxidases in plants: innumerable role in abiotic stress tolerance and plant development. J. Plant. Growth Regul. 42, 598–613. https://doi.org/10.1007/s00344-022-10601-9 (2022).
He, L. et al. Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell. Physiol. Biochem. 44, 532–553. https://doi.org/10.1159/000485089 (2017).
Islam, S., Rahman, I. A., Islam, T. & Ghosh, A. Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: gaining an insight to their physiological and stress-specific roles. PLoS One. 12, e0187504. https://doi.org/10.1371/journal.pone.0187504 (2017).
Ding, F., Wang, C., Zhang, S. & Wang, M. A jasmonate-responsive glutathione S-transferase gene SlGSTU24 mitigates cold-induced oxidative stress in tomato plants. Sci. Hortic. 303, 111231. https://doi.org/10.1016/j.scienta.2022.111231 (2022).
Golldack, D., Li, C., Mohan, H. & Probst, N. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant. Sci. 5, 151. https://doi.org/10.3389/fpls.2014.00151 (2014).
Li, G. et al. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8, e1002767. https://doi.org/10.1371/journal.pgen.1002767 (2012).
Wang, Z. et al. WRKY29 transcription factor regulates ethylene biosynthesis and response in arabidopsis. Plant. Physiol. Biochem. 194, 134–145. https://doi.org/10.1016/j.plaphy.2022.11.012 (2023).
Manna, M., Rengasamy, B. & Sinha, A. K. Revisiting the role of MAPK signalling pathway in plants and its manipulation for crop improvement. Plant. Cell. Environ. 46, 2277–2295. https://doi.org/10.1111/pce.14606 (2023).
Dąbrowski, P. et al. Photosynthetic efficiency of perennial ryegrass (Lolium perenne L.) seedlings in response to Ni and cd stress. Sci. Rep. 13, 5357. https://doi.org/10.1038/s41598-023-32324-x (2023).
Zhou, X., Sun, C., Zhu, P. & Liu, F. Effects of antimony stress on photosynthesis and growth of Acorus calamus. Front. Plant. Sci. 9, 579. https://doi.org/10.3389/fpls.2018.00579 (2018).
Liao, G. et al. Efficiency evaluation for remediating paddy soil contaminated with cadmium and arsenic using water management, variety screening and foliage dressing technologies. J. Environ. Manage. 170, 116–122. https://doi.org/10.1016/j.jenvman.2016.01.008 (2016).
Zheng, X. et al. Unraveling the mechanism of potato (Solanum tuberosum L.) tuber sprouting using transcriptome and metabolome analyses. Front. Plant. Sci. 14, 1300067. https://doi.org/10.3389/fpls.2023.1300067 (2023).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. https://doi.org/10.1038/nprot.2013.084 (2013).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. https://doi.org/10.1038/s41587-019-0201-4 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–d592. https://doi.org/10.1093/nar/gkac963 (2023).
Chen, X., Truksa, M., Shah, S. & Weselake, R. J. A survey of quantitative real-time polymerase chain reaction internal reference genes for expression studies in Brassica napus. Anal. Biochem. 405, 138–140. https://doi.org/10.1016/j.ab.2010.05.032 (2010).
Acknowledgements
This research was supported by Hunan Provincial Natural Science Foundation of China (2023JJ50083, 2023JJ50475), the National Natural Science Foundation of China (32371589), Research Foundation of Education Bureau of Hunan Province, China (23B0809, 22B0844) and the construct program of plant protection applied characteristic discipline in Hunan Province.
Author information
Authors and Affiliations
Contributions
Xianjun Liu: Writing—original draft, Writing—review and editing, Resources. Liang You: Writing—review and editing, Methodology. Wencong Yu: Methodology, Data curation, Formal analysis. Yuhui Yuan: Data curation, Formal analysis. Wei Zhang: Material provision. Mingli Yan: Data curation. Yu Zheng: Data curation. Renyan Duan: Writing—review and editing. Guiyuan Meng: Writing—review and editing. Yong Chen: Writing—review and editing. Zhongsong Liu: Writing—review and editing, Fund support. Guohong Xiang: Writing—review and editing, Fund support.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, X., You, L., Yu, W. et al. Transcriptome profiles of leaves and roots of Brassica napus L. in response to antimony stress. Sci Rep 15, 9413 (2025). https://doi.org/10.1038/s41598-025-88521-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-88521-3