Abstract
Target of rapamycin (TOR), discovered in Saccharomyces cerevisiae, is a highly conserved serine/threonine kinase acting as a regulatory hub between the cell and its environment. Like mammals, in fungi, the TOR complex 1 (TORC1) pathway is essential for coordinating cell growth in response to nutrient availability. The activation of TORC1 is similar in yeast and mammals, while its inhibition is more complex in mammals. This divergence of TORC1 regulation opens the question of how common are the yeast and mammalian variants in the fungal kingdom. In this work, we trace the evolutionary history of TORC1 components throughout the fungal kingdom. Our findings show that these fungi contain the mammalian-specific KICSTOR complex for TORC1 inhibition. They also possess orthologs of serine, arginine and methionine sensors of TORC1 pathway that orchestrate the response to nutrient starvation in mammals. The Rheb-TSC mediated activation of mammalian TORC1 that was lost in Saccharomycotina was also conserved in non-Dikarya. These findings indicate that the TORC1 pathway in non-Dikarya fungi resembles mammalian TORC1. Saccharomycotina lost many of the inhibitory components and evolved alternate regulatory mechanisms. Furthermore, our work highlights the limitations of using S. cerevisiae as a fungal model while putting forward other fungi as possible research models.
Similar content being viewed by others
Introduction
Target of rapamycin (TOR) protein is a member of the phosphoinositide-3-kinase related protein kinase family. It was first discovered in Saccharomyces cerevisiae through genetic screening of mutants resistant to the inhibitory effects of rapamycin1. Later on, it was also documented in filamentous fungi and described in other non-pathogenic fungi2 and in animals3. TOR signaling pathway is conserved in diverse eukaryotes, from yeast to humans and serves as a central regulatory hub for nutrient-sensing and maintaining cellular homeostasis4. In the presence of nutrients, the TOR pathway promotes cell growth by stimulating anabolic processes such as transcription, translation and ribosome biogenesis, while repressing catabolic processes such as autophagy5. Here, we introduce two best-known TORC1 pathway versions: the Saccharomyces cerevisiae (referred to as yeast) and Homo sapiens. For convenience, we introduce the gene names of both systems in Table 1.
mTOR
Mammals possess one TOR protein kinase, mTOR, which acts as a catalytic subunit for two complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). The former regulates cell growth while the latter is responsive to insulin-like growth factor 1 (IGF-1) signaling3. mTORC1 and mTORC2 share regulatory subunits mLST8 and Deptor. Both mTOR complexes have specific substrate recognition and regulatory components (PRAS40 for mTORC1 and mSin1 for mTORC2).
ScTOR
Budding yeasts on the other hand, possess two TOR proteins, Tor1 and Tor21 each of them forming a distinct complex, TORC1 and TORC2 respectively. Both complexes include a common regulatory protein Lst8 that functions as their primary interactor. TORC1 includes a novel component found in yeast, Tco89, which maintains cellular integrity during rapamycin treatment and stress6. Similarly, TORC2 has a yeast-specific component Avo2 that downregulates TOR signaling7.
Upstream TORC1 signaling
Under nutrient-rich conditions, presence of amino-acid activates the Rag GTPases (Gtr1/2 in yeast) that interact with LAMTOR/RAGULATOR scaffolds via roadblock domains. The RAGULATOR complex then regulates their translocation from cytosol to the surface of lysosomes in mammals (and vacuoles in yeast)14 (Fig. 1). While mammals have five LAMTOR proteins, the RAGULATOR in S. cerevisiae is reduced to a three-protein EGO complex. The EGO complex similarly interacts with the Gtr1-2 GTPases as LAMTOR does with Rag GTPases9. Ego1 is a homolog of Ltor1, while Ego2 and Ego3 are analogs with a similar structure to human Ltor2 and Ltor39.
In parallel, mTORC1 is also regulated by a GTP-bound active Rheb, which binds to mTORC1 65 Å away from the kinase domain, thereby increasing its effective substrate concentration and adopting the active conformation15. During nutrient starvation, the TSC complex causes hydrolysis of GTP bound to Rheb , keeping Rheb in a GDP-bound state and thereby, preventing it from activating mTORC116 (Fig. 1). Yeasts, in particular S. cerevisiae, contain Rheb but do not have TSC orthologs and hence, lack the Rheb-dependent regulation of TORC13.
Downstream TORC1 signaling
TOR signaling pathway is also governed by the GATOR/SEA-dependent regulation that helps maintain the localization of TORC1 at the lysosomal/vacuolar membrane during nitrogen starvation (Fig. 1). It comprises GATOR1 and GATOR2 subcomplexes (SEACIT and SEACAT in yeast)3. GATOR2 inactivates TORC1 by binding to GATOR1 (a complex that has a GTPase-activating protein (GAP) activity towards Rag A/B) which converts the GTP-bound active Rag A/B into GDP-bound inactive state1. The KICSTOR complex acts as a scaffold that recruits GATOR1 to the lysosomal surface13. In the absence of nutrient availability, the arginine sensing protein Castor interacts with GATOR2 by forming a dimer of Castor1 and Castor217. At the same time, the leucine sensor Sestrin1 binds to GATOR2. This binding induces GATOR1 activity by releasing its attachment to GATOR218. The starvation of amino acid methionine involves direct sensing in mammals using Samtor, which acts as the SAM (S-adenosylmethionine) binding effector19. Samtor contains a class I Rossman fold methyltransferase domain that binds S-adenosylmethionine (SAM), facilitating its interaction with KICSTOR bound GATOR1 in TORC1 inhibition20. The tumor suppressor gene FLCN (Lst7 in yeast) forms a complex with FNIP1/2 (Lst4 in yeast), and acts as a GAP towards Rag C/D. This promotes the binding of mTORC1 to the Rag heterodimers21. This coordination by the FLCN/FNIP1 complex contributes to metabolic homeostasis in mammals22.
S. cerevisiae do not have Samtor, and employ an indirect methionine sensing mechanism through protein phosphatase 2 A (PP2A) methylation. Decreased methionine levels lead to demethylation of the PP2A catalytic subunit PP2Ac, resulting in an inactivated phosphatase. This in turn, dephosphorylates Npr2 (mammalian Nprl2) and activates the SEACIT (GATOR1) mediated TORC1 inhibition19.
Shertz and co-workers in 2010 annotated the TORC1 pathway components in Zygomycetes (now Mucoro- and Zoopagomycota) and Chytridiomycota, a set of Dikarya members along with non-fungal Opisthokonta. They reported conservation of the Tor1 (mTor) kinase protein architecture among eukaryotes, along with duplications in most fungal groups2. Upstream regulators Tsc1 and Tsc2 comprising the TSC complex were identified in most fungal lineages except S. cerevisiae. A study by Manning and Cantley pointed out that loss of function of Rheb resulted in growth arrest phenotype in Schizosaccharomyces pombe23. A recent study on conservation of TORC1 nutrient signaling pathway in arbuscular mycorrhizal (AM) fungi pointed to the absence of yeast proteins Ego1, Ego3 and Tco89 in AM fungi24. This opened the question of how universal the Saccharomyces cerevisiae TORC1 pathway is to the fungal tree of life.
To address this question, we used mammalian and S. cerevisiae TORC1 pathway models as reference. We report the presence of different amino acid-specific sensor proteins, animal-specific regulatory complexes typical of mTOR across non-Dikarya fungi. Our findings suggest a mammalian mode of TORC1 pathway architecture in non-Dikarya fungal lineages. The possible implications of having an mTORC1-like mode of regulation starts from nutrient-sensing specificity in the environment, the balance of cell energetics via possible growth limitation strategies to pathogenicity regulation along with metabolite production.
Results
Distribution of TORC1 pathway components
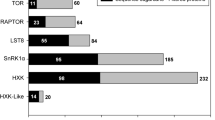
As the constituents of TORC1 regulation differ between mammals and yeasts, we looked for their distribution across the fungal kingdom, along with Metazoa and early Opisthokonts as outgroups. 32 out of 36 mammalian TORC1 components were identified in fungal proteomes (Fig. 2). It was interesting to find components of the human RAGULATOR complex (Ltor1-5) distributed not only across Early Diverging Fungi (EDF) but also in a few Dikarya members (Fig. 2). The homologs of Ltor2 found in Ascomycota were truncated, containing an N-terminal roadblock domain of 60 amino acids (Fig. 2).
The GATOR/SEA complex (combination of SEACIT and SEACAT complexes) of TORC1 inhibition pathway showed conservation across eukaryotes. Homologs of the mammalian KICSTOR, TSC complex and Sestrin were conserved across the phyla Mucoromycota (Mucoro-, Mortierello- and Glomeromycota), Entomophthoro- and Neocallimastigomycota, and scattered in the remaining EDF (Fig. 2). Loss of Castor and FLCN-FNIP1 proteins were observed in Rozello-, Chytridio- and Zoopago-, Kickexello- and Glomeromycota. It is worth noting that Dikarya members Ustilago- and Taphrinomycota possessed a complete TSC complex along with Rheb, pointing to the possible function of Rheb-dependent TORC1 activation (Fig. 2). The yeast-specific Tco89 protein was found in Mucoro- and Blastocladiomycota, along with duplications in Mucoromycota (Supplementary File S1).
Distribution of 36 TORC1 pathway components among model eukaryotes with at least two representatives from each fungal lineage. The nomenclature of proteins is in accordance with TORC1 protein names in Homo sapiens as listed in Table 1. The phylogenomic tree was built in Orthofinder and drawn in iTOL. The taxonomic ranks are in accordance with Voigt and coworkers 25.
The LAMTOR equivalent EGO complex is known only in S. cerevisiae. In order to assess its occurrence in other Saccharomycotina members, we based our search on proteomes from the y1000 + project26 encompassing a total of 1154 Saccharomycotina species.
The analysis of EGO complex proteins from 1154 Saccharomycotina members identified 575, 165 and 317 homologs for Ego1, Ego2 and Ego3, respectively. All 3 EGO complex subunits were present only in 6 out of 17 families (Saccharomycetaceae, Debaryomycetaceae, Saccharomycodaceae, Dipodascaceae, Wickerhaomycetaceae and Phaffomycetaceae). We observed that the oldest lineages of Saccharomycotina retained both EGO and LAMTOR components with a co-existence of both almost complete complexes in Trichomonascaceae. Two families Sporopachydermiaceae and Saccharomycopsidaceae have neither EGO nor LAMTOR components. This observation suggests a gradual exchange of the LAMTOR with EGO across the Saccharomycotina families and common losses of EGO components in Saccharomycotina (Supplementary file S1).
Transcriptomics of TORC1 pathway proteins
The presence of mRNA in transcriptomic data provides evidence of the gene being functional and active in a given organism. To examine the functionality of the candidate TORC1 genes in EDF, we analyzed the expression profiles of available transcriptomic data in EDF from various environmental conditions. A total of 28 out of 36 predicted genes encoding TORC1 pathway components were expressed (Fig. 3). For instance, the transition from aerobic vs. anaerobic growth of M. lusitanicus27 (Fig. 3, M1) changed the expression of 22 out of 28 predicted TORC1 pathway genes. The upregulation of genes was observed for a few of the TORC1 activation (FLCN, Rag GTPases, Ltor2, Raptor, Sin1, Rheb) and inactivation (GATOR2 complex, Sestrin, Tco89, and the KICSTOR complex protein SZT2) components, while GATOR1 complex genes DEPDC5, NPRL2 and NPRL3 were downregulated. Genes for KICSTOR complex proteins were both upregulated (SZT2) and downregulated (KICS2). In another study, R. microsporus exposed to murine macrophages28 (Fig. 3, M2) showed upregulation of Rag GTPases, Tsc1 and KICSTOR components ITFG2 and SZT2, and downregulation of Rheb and RAGULATOR components Ltor2 and Ltor3, while a similar study on R. delemar (Fig. 3, M3) found only Tsc1 upregulated. A dataset on R. delemar infection on A549 human airway epithelial cells29 for 6 h (Fig. 3, M4(a)) and 16 h (Fig. 3, M4(b)) showed downregulation of Castor1 in the former and upregulation in the latter stage of infection. The gene encoding Tor1 kinase protein was also downregulated in the 16 h infection phase, pointing to the inhibition of TORC1 pathway as a response to infection. In another dataset of mouse bone-marrow derived macrophages (BMDMs) infected with R. delemar for 1 h, 4 h and 18 h30, we found the expression of TORC1 pathway genes from samples infected for 18 h (Fig. 3, M5). Both TOR kinase and Rag GTPases were downregulated, along with KICSTOR complex proteins KPTN and SZT2, and GATOR components Nprl3 (Npr2) and Wdr24 (Sea2). Castor1 and Sestrin along with Rheb were upregulated. The study involving G. rosea treatment with G24, a synthetic analog of strigolactone, in its response to plant signals during the switch from asymbiotic to presymbiotic growth31 (Fig. 3, G1), observed a downregulation of 6 out of 8 (Tor1, FNIP1, Rag A/B, Rheb, Seh1, ITFG2) TORC1 components. The gene expression profiling of R. irregularis roots exposed to higher phosphate concentrations (300 µM, 500 µM)32 (Fig. 3, G2) identified a downregulated Tor1 kinase and an upregulated Nprl2. Another study on R. irregularis focused on differential expression of strigolactone-treated spores a day after inoculation33 (Fig. 3, G3) identified an upregulated Tor1, Raptor, Tsc2, Depdc5, SZT2, and downregulated RAGULATOR (Ltor2, Ltor3) and GATOR (Sect. 13, Seh1/Seh1l) proteins.
We also looked at the transcriptomic profiles of experiments obtained from Dikarya members. We observed a downregulation of Rag GTPases alongside upregulation of Rheb, Tsc1 and Nprl2 in T. reesei strains grown in carbon-deficit source (1% D-mannitol) (Fig. 3, A1). In another study of stress response against oxidative stress (treatment with 20mM H2O2) in N. crassa34 wild-type and upf1 knockout strains, TORC1 complex, Rag GTPases and GATOR1 components were upregulated in wild-type strains (Fig. 3, A2(a)). The upf1 knockout strains exposed to H2O2 showed an upregulated GATOR2 and a downregulated TSC complex (Fig. 3, A2(b)). The expression profiling of C. immitis35 in young and mature spherules displayed an upregulated Tor1 along with its activation components in mature spherules (Fig. 3, A3(b)) and downregulation of its inhibitory counterparts. The young spherules however, displayed low signals of upregulation of TORC1 pathway activation and higher signals in its downregulation counterparts (Fig. 3, A3(a)). Expression profiling of the cAMP/PKA signaling pathway in O. oligospora36 wild-type (Fig. 3, A4(a)) and ΔAoPkaC1 (Fig. 3, A4(b)) strains displayed low signals of upregulation of TORC1 pathway components in the former and downregulation in the latter.
Another study involving gene expression profiling of the basidiomycete C. cinerea upon deletion of cre1 gene (regulating carbon catabolite repression)37 (Fig. 3, B1) displayed upregulation of Tor1 and GATOR1 (Nprl3, Sect. 13) proteins, along with a downregulation of Tsc2. The R. solani AG1-IA strain upon infection of rice leaves (Fig. 3, B2) showed only a downregulated Lst8. The gene expression profiling of U. maydis38 wild-type (Fig. 3, B3(a)) and Pcrg1:grx4 (Fig. 3, B3(b)) strains grown in minimal medium for 24 h displayed downregulation of TORC1 pathway activation components.
Taken together, we see that in EDF, activation components RAGULATOR, Rag GTPases and Rheb-TSC complex most often respond to changes in fungal growth environments, alongside amino acid sensing proteins Castor and Sestrin, and KICSTOR complex for TORC1 inhibition (Fig. 3). On the other hand, much of the expression in Dikarya is seen for the GATOR/SEA complexes of TORC1 inhibition, with Rag GTPases and Rheb responding for activating TORC1 (Fig. 3).
Differential expression profiling of 28 TORC1 pathway genes expressed in different environmental conditions in members of Mucoromycota (M), Glomeromycota (G), Ascomycota (A) and Basidiomycota (B). The empty cells signify the absence of gene expression from the organism in the given environmental condition. Note that columns A1–B3.b lack the homologs of genes marked with *. M1—Mucor lusitanicus growth in anaerobic vs. aerobic conditions; M2—Rhizopus microsporus growth in presence vs. absence of murine macrophage; M3—Rhizopus delemar growth in presence vs. absence of murine macrophage; M4(a)—Rhizopus delemar host-pathogen interaction (6 h) using human airway epithelial cells (A549); M4(b)—Rhizopus delemar host-pathogen interaction (16 h) using human airway epithelial cells (A549); M5—Rhizopus delemar host-pathogen interaction (18 h) using mouse bone marrow-derived macrophages (BM); G1—Gigaspora rosea response to plant signals in the switch from asymbiotic to presymbiotic growth; G2—Rhizophagus irregularis growth in Lotus japonicus roots upon exposure to high vs. low concentrations of phosphate (500µM, 300 µM vs. 100 µM, 20 µM); G3—Rhizophagus irregularis association with Medicago truncatula and treatment with strigolactone for 24 h; A1—Trichoderma reesei QM6a and Δace1 strains grown in Mandels-Andreotti medium containing 1% D-mannitol as carbon source; A2(a)—Neurospora crassa wild-type strain treated with 20mM H2O2; A2(b)—Neurospora crassa upf1 knockout strain treated with 20mM H2O2; A3(a)—Coccidioides immitis gene expression profiling in young spherules; A3(b)—Coccidioides immitis gene expression profiling in mature spherules; A4(a)—Orbilia oligospora wild-type gene expression profiling of cAMP/PKA signaling pathway; A4(b)—Orbilia oligospora ΔAoPkaC1 gene expression profiling of cAMP/PKA signaling pathway; B1—Coprinopsis cinerea gene expression profiling upon deletion of cre1 regulating carbon catabolite repression; B2—Rhizoctonia solani AG1-IA transcriptome profiling upon infecting rice leaves; B3(a)—Ustilago maydis wild-type gene expression profiling in minimal medium (2% arabinose/glucose) after 24 h of growth; B3(b)—Ustilago maydis Pcrg1:grx4 strain gene expression profiling in minimal medium (2% arabinose/glucose) after 24 h of growth.
Phylogenetics of selected TORC1 pathway components
In order to understand whether duplications, losses, horizontal gene transfers impacted TORC1 pathway evolution, we performed phylogenetic analyses for all of the studied protein sequence sets (Supplementary file S2). We observed that in general, TORC1 pathway components were inherited vertically. Moreover, most of the TORC1 components occur as single copies and have a conserved domain architecture (an example of the Ltor3 protein phylogeny is shown in Fig. 4) with a few exceptions. For instance, the Rheb gene duplicated at least twice since we found paralogs in EDF and Basidiomycota. The KICSTOR complex protein SZT2 possesses more than two copies in selected Chytridiomycota (Chytriomyces confervae; Rhizoclosmatium globosum) and all members of Mucoromycota. Latter taxon also showed duplications of Castor2 and Sestrin (Supplementary file S1). The yeast-specific EGO complex proteins were present in most of the Saccharomycotina genera and their phylogenetic trees recapitulated the species phylogeny of the subphylum (Supplementary file S2).
Speculative TORC1 pathway model in non-dikarya fungi
Taking into account all of the homologs of TORC1 pathway components found in EDF, we propose a model of TORC1 regulation in EDF, taking two phyla as examples (Mortierellomycota: Lobosporangium transverasale and Zoopagomycota: Syncephalis psuedoplumigaleata respectively, Fig. 5). The presence of upto 2 out of 5 LAMTOR components might facilitate their interaction with Rag GTPases (Gtr1/Gtr2), followed by localization to the vacuolar membrane, thereby promoting TORC1 activation (Fig. 5). The TSC complex would also enable Rheb-TSC mediated TORC1 activation similar to mammalian TORC1. It is unsure how the localization of Rag heterodimers (Gtr1/Gtr2) to the vacuolar membrane and their subsequent binding to TORC1 occurs in the absence of the FLCN/FNIP1 (Lst4/Lst7) complex in Zoopagomycota (Fig. 5). With regards to TORC1 inhibition, the presence of Castor and Sestrin in Mortierellomycota and Sestrin in Zoopagomycota would promote specificity in amino acid sensing like mammalian TORC1 (Fig. 5). The presence of 3-protein KICSTOR instead of the complete 4-protein complex in Mortierellomycota might enable the recruitment of GATOR1 to the vacuolar surface and provide an additional layer of complexity on the TORC1 inhibitory dynamics of this fungal group. Taken together, most lineages possess an intermediate set of TORC1 components distinct from both mammalian and S. cerevisiae. Moreover, diverse fungal lineages retained TORC1 subunits typically associated with animals.
A tentative model of the TORC1 pathway for Mortierello- and Zoopagomycota. Proteins that share homology at the same position are depicted with the same color. The protein nomenclature is in accordance with Table 1.
Discussion
The study analyzes the evolutionary dynamics of TORC1 metabolism in the fungal tree of life. We show that EDF not only possesses more TORC1 pathway components than Dikarya, but they also share more similarities with the mammalian TORC1 pathway. The conservation of the ancestral TORC1 pathway varies among fungal lineages, with Mortierellomycota possessing the highest number of mTORC1 pathway components among fungal lineages, followed by Mucoro- and Glomeromycota.
The study by Petit and coworkers revealed that FLCN is specifically required for amino acid stimulated recruitment of mTORC1 to lysosomes by Rag GTPases, while FNIP1 promotes this recruitment and Rag interactions of FLCN39. Similarly in the case of S. cerevisiae, Lst7/Lst4 complex (FLCN/FNIP1 in humans, Table 1) was involved in trafficking of amino acid permease to the cell surface in response to nitrogen scarcity40. The conservation of the FLCN/FNIP1 complex in Mucoro-, Entomophthoro- and Neocallimastigomycota coupled with their upregulation in stress experiments of Mucorales, suggests a potential involvement of this complex in EDF, in line with Saccharomycotina (Fig. 3). The absence of this complex in Basidiomycota puts an open question on whether this phylum evolved an alternative TORC1 activation machinery.
The pentameric LAMTOR/RAGULATOR complex in humans requires the formation of Ltor2-3 and Ltor4-5 roadblock domain dimers, the former assisted by one alpha helix from Ltor1 (Table 1)9. The five-protein LAMTOR complex is reduced to four and three protein complexes in S. pombe41 and S. cerevisiae42 respectively. In S. pombe, Lam1 (Ltor1 in humans) forms a β-strand and wraps around Lam4 (Ltor5 in humans) and Lam2-3 roadblock heterodimer41. The Ego2 and Ego3 of S. cerevisiae do not dimerize but are held together by Ego19. The occurrence of Ltor2 and Ltor3 in Mucoro-, Neocalli-, Chytridio-, and Zoopagomycota (Fig. 1) coupled with the differential expression of Ltor2 and Ltor3 in stress environments (Mucoromycota, Fig. 3) points to the conservation of Ltor2-3 canonical roadblock heterodimer in EDF. The absence of Ltor4 in fungi, with the exception of a Chytridiomycota representative (Spizellomyces punctatus DAOM BR117: KND00403.1), alongside the occurrence of Ltor5 in Mucoro-, Glomero-, and few Chytridiomycota defers the possibility of formation of Ltor4-5 dimer in EDF. Similarly, lack of Ltor1 in EDF (except two species: Bifiguratus adelaidae and Anaeromyces robustus) points at a loss of N-terminal fold of Ltor1 that completes the complex structure and enables interaction with Rag GTPases. The occurrence and transcription expression of Ltor2, Ltor3 and Ltor5 in EDF points to a reduction in complexity of the RAGULATOR complex in EDF.
The occurrence of Rheb in all fungal groups except Kickxellomycota and Blastocladiomycota displays the conservation of this protein in the TORC1 pathway. The function of Rheb in S. cerevisiae was observed to regulate arginine and lysine uptake43. This finding supports the absence of arginine and lysine sensor proteins Castor and Sestrin from Saccharomycotina and possibly other Dikarya. It also indicates the function of Rheb beyond Rheb-TSC mode of TORC1 activation.
The conservation of both proteins of the TSC complex in Mucoro- and Neocallimastigomycota, along with Ustilago- and Taphrinomycota extends the possibility for the presence of a functional Rheb-TSC mode of TORC1 activation. The downregulation of Rheb and TSC complex proteins against oxidative stress response in N. crassa wild-type strain suggests a potential involvement of the Rheb-TSC mediated TORC1 activation in members of Ascomycota possessing both Tsc1 and Tsc2 (Fig. 3). Tsc2 contains the GAP domain responsible for hydrolysis of GTP-bound Rheb into a GDP-bound inactive form during nutrient starvation. Tsc1 is crucial in stabilization of the Tsc2 dimer and formation of a functional heterodimeric complex that localizes in the lysosomal membranes16. Hence, it is to be speculated whether the TSC complex is functionally active in the presence of only Tsc2, as observed in Basidio- and Zoopagomycota.
Studies in Drosophila melanogaster found that GATOR1 proteins Nprl2 and Nprl3 interact with each other and Depdc5 for lysosomal localization and inhibit TORC1 in response to nitrogen starvation44. The GATOR1 equivalent SEACIT proteins in yeast control nitrogen catabolite repression that downregulates proteins utilizing poor nitrogen sources in presence of preferred sources11. The GATOR2/SEACAT complex protein Seh1 (Seh1l) forms a dimer with Sea4 (Mios), inhibits activity of the GATOR1/SEACIT complex and prevents TORC1 inhibition11. The absence of Seh1 from selected EDF groups (Mucoro-, Kickxello-, Blastocladio- and Neocallimastigomycota) and Basidiomycota leaves an incomplete view on the mechanism of TORC1 inhibition by GATOR1/SEACIT (Fig. 2). The complete absence of the SEACIT/GATOR1 complex and a majority of SEACAT/GATOR2 complexes from Wallemiomycota (Basidiomycota) points to a loss of SEA/GATOR-dependent regulation of TORC1 in this fungal group (Fig. 2).
The conservation of KICSTOR components in Mucoromycota, along with their up- and downregulation in transcriptomic experiments opens the possibility of similarity between EDF and mammalian mode of TORC1 inhibition by KICSTOR (Fig. 3). Previously, the study by Wolfson et al. in 2017 stated that the KICSTOR is conserved only in vertebrates and not in fungi13. Our results expand the conservation of this complex in fungi. While KPTN and ITFG2 form a heterodimer, the SZT2 component of KICSTOR plays a central role in interaction with GATOR1. This localizes the KICSTOR complex to the lysosomal surface, and induces the GAP function of GATOR145. It is difficult to speculate whether the absence of one or more KICSTOR components could keep the complex active in fungal phyla Kicxello-, Zoopago-, Blastocladio- and Chytridiomycota.
Structural studies on Castor proteins revealed that Castor1 possesses arginine binding pockets and recognises arginine deprivation. Castor2 shares 63% similarity with Castor1 but lacks arginine recognizing sites17. Castor1 forms a homodimer and a heterodimer with Castor2, and together they bind to GATOR2 to induce TORC1 inhibition. The existence of Castor1 in EDF (Mucoro-, Mortierello-, Neocalli- and Entomophthoromycota) along with their up- and downregulation in transcriptomic stress experiments of Mucoromycota provides potential for mTORC1-like amino acid sensing (Fig. 3). However, the presence of only Castor2 in selected EDF and Dikarya does not confirm the extent of arginine sensitivity in fungi (Fig. 2). Structural studies discovered that this mechanism of binding of Castor1 to GATOR2 is similar to the one found in E. coli and cyanobacteria17. However, Chantranupong and colleagues (2016) found only one Castor-like protein in prokaryotes. This points at a duplication event of an ancestral Castor protein in the common ancestor of animals and fungi, that produced Castor1 and Castor2 in vertebrates and in parallel, loss of Castor1 in selected fungal lineages, including loss of both copies in Dikarya12.
Sestrin functions as an intracellular leucine sensor that negatively regulates the TORC1 signaling pathway through the GATOR complex. It is known that Sestrin evolved in the last common ancestor of animals, fungi and amoebozoa and was subsequently lost in the ancestor of S. cerevisiae46. The yeasts exapted leucyl tRNA synthetase (LeuRS) as the leucine sensor for TORC1 regulation. The conserved repertoire of Sestrin in members of phyla Mucoromycota points to their similarity to leucine sensitivity in mTORC1. While yeasts have adopted LeuRS as the leucine sensor, EDF have the ancestral Sestrin as leucine sensor (Fig. 3).
The yeast-specific novel TORC1 component Tco89 was also found in selected EDF groups (Mucoro- and Blastocladiomycota) (Fig. 2). Tco89 interacts with GTP-bound Gtr1 (RagA/B in humans, Table 1) and helps in maintaining cellular integrity of the cell during nutrient starvation6. The upregulation of Tco89 homologue in Mucor lusitanicus alongside Tor1 during growth in anaerobic conditions suggests its possible role in non-Dikarya fungi (Fig. 3).
TORC1 pathway is a critical metabolic step involved in regulating cellular life - development, survival, senescence, tumorigenesis, and inflammation. Rapamycin is a natural product with a broad range of biological activities such as immunosuppressive, antitumor, neuroprotective/neuroregenerative, and lifespan extension. It inhibits the TORC1 pathway, influencing the cascade of reactions having major roles in cell survival. Rapamycin exhibits inhibitory activity against opportunistic pathogenic Dikarya (Cryptococcus neoformans)47,48, and non-Dikarya (Mucor circinelloides)49. Our study showed that TORC1 genes respond in different ways to infection related conditions in M. lusitanicus, R. delmar and R. microsporus pointing at differentiation of the pathway regulation within Mucorales. Perhaps the background conditions play a subtle role in TORC1 pathway regulation. For instance, in the rice pathogen, Fusarium fujikuroi, rapamycin inhibits TOR pathway depending on nitrogen source conditions and concentrations50. In our study we found that all of the TORC1 genes are expressed at the same level in R. solani AG1-IA transcriptome profiling upon infecting rice leaves (Fig. 3).
The TSC complex is indispensable for pathogenicity in fungi, as reported from studies in Fusarium oxysporum highlighting the inappropriate activation of TORC1 upon deletion of Tsc251. The effect of TSC complex deletion was also observed in Trichoderma atroviride in the form of reduced mycoparasitic overgrowth and diminished production of secondary metabolites52. The TSC complex in EDF and selected Dikarya may add in regulation of the energetics of the cell with a possible role in pathogenicity. However, the role of this regulation is not the same for all infection models and may depend on the experimental conditions.
TORC1 pathway is a key energy regulator in the cell and as such, is of interest for the optimization of biotechnological procedures. It has been proven that in microalgae, triacylglycerols (TAGs) and starch accumulation is induced by TORC1 inactivation53. In the fungal kingdom, white-rot fungus Phanerochaete chrysosporium (Basidiomycota) was proved to secrete different extracellular proteins (copper radical oxidase and GMC oxidoreductases, glycoside hydrolases as well as amylase) in response to rapamycin TOR pathway inactivation54. But the effect of modulating the TORC1 pathway using rapamycin can be even more promising. It was shown that aflatoxin production in Aspergillus flavus is inhibited with rapamycin via TORC1 inactivation55. Information about the composition of TORC1 in different fungal phyla can impact future biotechnological applications. For instance, oil producing fungi such as Mortierello- along with Mucoromycota, may accumulate more oils in response to mTORC1-like inhibition (Fig. 5)56. According to our findings biotechnological settings should take into account differences in TORC1 architecture and regulation among individual organisms to achieve optimal bioproduction.
Having a mTORC1-like regulation with additional inhibitory complexes and amino acid sensors gives EDF a higher chance of phenotypic and regulatory plasticity in response to environmental conditions. Zhang et al. (2017) reported that yeast strains with reduced TOR signaling exhibited enhanced mitochondrial ROS during growth phase57. Taken together, the complexity of TORC1 regulation in fungi could pave the way for the flexibility of metabolic reprogramming along with balancing energetics and stress resistance (Fig. 5).
Methods
Homologous sequence search
The TORC1 pathway reference dataset was created using whole proteomes of model organisms: Homo sapiens (GCF_000001405.40), Mus musculus (GCF_000001635.27), Saccharomyces cerevisiae (GCF_000146045.2) and Schizosaccharomyces pombe (GCF_000002945.1). Individual records of the proteins were acquired from UniProt58, SGD59 and PomBase60 databases respectively (Supplementary file S1). They served as BLASTp queries (e-value < = 1e-5) against 183 proteomes of diverse fungi. For each set of proteins, CLANS clustering was performed (p-value threshold of 1e-10, expulsive force exponent set to 2) to remove potential nonspecific hits and group together the fungal homologs. Sequence groups were inspected for protein domain conservation and domain architecture using NCBI’s conserved domain database61 and PfamScan assessed by pfam_scan.pl62. For proteins displaying sequence divergence (Tco89, EGO complex), protein profiles were created using hmmpress63. They were scanned against the proteomes using hmmscan and the true positives were extracted by clustering of the resulting hits in CLANS. The EGO complex homologs were identified in a set of 1154 Saccharomycotina proteomes downloaded from the Y1000 + project64. To confirm the loss of genes, we searched the genomes with tBLASTn using proteins from the closest relative as queries.
Multiple sequence alignment and phylogenetic tree construction
Presence of TORC1 proteins were also searched in five basal Opisthokonts [Capsaspora owczarzaki (GCF_000151315.2)65, Sphaeroforma arctica (GCF_001186125.1)66, Fonticula alba (GCF_000388065.1)66, Salpingoeca rosetta (GCF_000188695.1)67, Monosiga brevicollis (GCF_000002865.3)68) as outgroups during phylogenetic tree construction. Multiple sequence alignments were performed for the proteins using MAFFT (v7.407)69 local alignment method with a maximum number of iterative refinements set to 100. Phylogenetic trees were constructed with IQ-TREE (v1.6.9)70 using maximum likelihood method with automated model selection and ultrafast bootstrap. A phylogenomic species tree was built with OrthoFinder71 (with mmseqs) for a set of 39 fungal taxa, 5 basal Opisthokonts, Amphimedon queenslandica (GCF_000090795.2), Homo sapiens (GCF_000001405.40), Mus musculus (GCF_000001635.27), Caenorhabditis elegans (GCF_000002985.6), Drosophila melanogaster (GCF_000001215.4), Pomacea canaliculata (GCF_003073045.1) with Dictyostelium discoideum (GCF_000004695.1) as outgroup. The trees were visualized using the iTOL online tool72. Lam1 from S. pombe was used as an outgroup for Ego1 tree. In the case of Ego2 and Ego3, the trees were rooted with the most ancestral Saccharomycotina representative having each of the proteins.
Transcriptomic data analysis of TORC1 homologs
The expression of TORC1 pathway proteins was checked in public transcriptomic datasets of EDF and Dikarya. Data was available in the form of transcriptomes from diverse cultural conditions for Mucor lusitanicus MS12, Rhizopus microsporus FP469, Rhizopus delemar 99–880, Gigaspora rosea DAOM 194,757, Rhizophagus irregularis DAOM 197198, Trichoderma reesei QM6a, Neurospora crassa OR74A, Coccidioides immitis RS, Orbilia oligospora ATCC 24,927, Coprinopsis cinerea okayama 7-130, Rhizoctonia solani and Ustilago maydis 521. The data was downloaded as fastq files from ENA server and were quality checked using FASTQC (v0.11.8)73. They were adapter-trimmed using fastp (v0.19.6) using default parameters74. The fastq reads were then aligned with the reference genome (downloaded from NCBI Datasets) using Hisat2 tool (v2.1.0)75. The SAM files obtained from the alignment were compressed into binary file format (BAM) using samtools (v1.10)76, and the aligned reads were counted using featureCounts (v1.6.3)77. The differential expression analysis was carried out using the DESeq2 R package for protein-coding gene expression from condition-specific transcriptomic datasets78. The Padj value was set to ≤ 0.05, and RNA-Seq reads were mapped on the protein-coding gene sequences. The log2fold change criteria [downregulation < 0 > upregulation] was used to determine the gene expression profiles. The heatmap of the expression profiles was generated using the ggplot2 R package79.
Data availability
All metadata processed in this study are deposited in zenodo: https://doi.org/10.5281/zenodo.1405166010.5281/zenodo.14051660. All protein identifiers, genomic assemblies, transcriptomic datasets are listed in Supplementary file S1. The figures of phylogenetic trees for TORC1 proteins are depicted in Supplementary file S2.
References
Morozumi, Y. & Shiozaki, K. Conserved and divergent mechanisms that control TORC1 in yeasts and mammals. Genes 12, 1 (2021).
Shertz, C. A., Bastidas, R. J., Li, W., Heitman, J. & Cardenas, M. E. Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom. BMC Genom. 11, 510 (2010).
Tafur, L., Kefauver, J. & Loewith, R. Structural insights into TOR signaling. Genes 11 (2020).
Dobrenel, T. et al. TOR signaling and nutrient sensing. Annu. Rev. Plant. Biol. 67, 261–285 (2016).
González, A. & Hall, M. N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36, 397–408 (2017).
Reinke, A. et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752–14762 (2004).
Thorner, J. TOR complex 2 is a master regulator of plasma membrane homeostasis. Biochem. J. 479, 1917–1940 (2022).
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Titze, S. & Kümmel, D. The difference is in the details: structural and mechanistic variations in the LAMTOR-Gtr/Rag module. Structure 31, 1010–1012 (2023).
Dai, X. et al. AMPK-dependent phosphorylation of the GATOR2 component WDR24 suppresses glucose-mediated mTORC1 activation. Nat. Metab. 5, 265–276 (2023).
Dokudovskaya, S. & Rout, M. P. SEA you later alli-GATOR—A dynamic regulator of the TORC1 stress response pathway. J. Cell. Sci. 128, 2219–2228 (2015).
Chantranupong, L. et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164 (2016).
Wolfson, R. L. et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017).
Lama-Sherpa, T. D., Jeong, M. H. & Jewell, J. L. Regulation of mTORC1 by the rag GTPases. Biochem. Soc. Trans. 51, 655–664 (2023).
Yang, H. et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373 (2017).
Yang, H. et al. Structural insights into TSC complex assembly and GAP activity on Rheb. Nat. Commun. 12, 339 (2021).
Gai, Z. et al. Structural mechanism for the arginine sensing and regulation of CASTOR1 in the mTORC1 signaling pathway. Cell. Discov. 2, 16051 (2016).
Kim, J. S. et al. Corrigendum: Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 5, 14029 (2015).
Lauinger, L. & Kaiser, P. Sensing and signaling of methionine metabolism. Metabolites 11 (2021).
Gu, X. et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 (2017).
Tsun, Z. Y. et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell. 52, 495–505 (2013).
Ramírez, J. A. et al. Folliculin interacting protein 1 maintains metabolic homeostasis during B cell development by modulating AMPK, mTORC1, and TFE3. J. Immunol. 203, 2899–2908 (2019).
Manning, B. D. & Cantley, L. C. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28, 573–576 (2003).
Zhou, X. et al. Genome-wide analysis of nutrient signaling pathways conserved in arbuscular mycorrhizal fungi. Microorganisms 9, 1 (2021).
Voigt, K. et al. Early-diverging fungal phyla: taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Divers. 109, 59–98 (2021).
Homa, M. et al. Differential Gene expression of Mucor lusitanicus under aerobic and anaerobic conditions. J. Fungi (Basel) 8 (2022).
Sephton-Clark, P. et al. Bacterial endosymbionts influence fungal transcriptional profiles with implications for host response in the human fungal pathogens Rhizopus microsporus and Rhizopus delemar. BioRxiv 1, 580746. https://doi.org/10.1101/580746 (2020).
Chibucos, M. C. et al. An integrated genomic and transcriptomic survey of mucormycosis-causing fungi. Nat. Commun. 7, 12218 (2016).
Andrianaki, A. M. et al. Iron restriction inside macrophages regulates pulmonary host defense against Rhizopus species. Nat. Commun. 9, 3333 (2018).
Tang, N. et al. A survey of the gene repertoire of Gigaspora Rosea unravels conserved features among glomeromycota for obligate biotrophy. Front. Microbiol. 7, 233 (2016).
Sugimura, Y. & Saito, K. Transcriptional profiling of arbuscular mycorrhizal roots exposed to high levels of phosphate reveals the repression of cell cycle-related genes and secreted protein genes in Rhizophagus Irregularis. Mycorrhiza 27, 139–146 (2017).
Tsuzuki, S., Handa, Y., Takeda, N. & Kawaguchi, M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus Irregularis. Mol. Plant. Microbe Interact. 29, 277–286 (2016).
Shen, S. et al. Sensing of H2O2-induced oxidative stress by the UPF factor complex is crucial for activation of catalase-3 expression in Neurospora. PLoS Genet. 19, e1010985 (2023).
Carlin, A. F. et al. Transcriptional analysis of Coccidioides Immitis mycelia and spherules by RNA sequencing. J. Fungi (Basel). 7, 1 (2021).
Zhu, M. C. et al. The cAMP-PKA signalling pathway regulates hyphal growth, conidiation, trap morphogenesis, stress tolerance, and autophagy in Arthrobotrys oligospora. Environ. Microbiol. 24, 6524–6538 (2022).
Pareek, M. et al. Preassembled Cas9 ribonucleoprotein-mediated gene deletion identifies the carbon catabolite repressor and its target genes in Coprinopsis Cinerea. Appl. Environ. Microbiol. 88, e0094022 (2022).
McCotter, S. W., Kretschmer, M., Lee, C. W. J., Heimel, K. & Kronstad, J. W. The monothiol glutaredoxin Grx4 influences iron homeostasis and virulence in Ustilago maydis. J. Fungi (Basel) 9 (2023).
Petit, C. S., Roczniak-Ferguson, A. & Ferguson, S. M. Recruitment of folliculin to lysosomes supports the amino acid–dependent activation of rag GTPases. J. Cell. Biol. 202, 1107–1122 (2013).
Pacitto, A. et al. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open. Biol. 5, 150174 (2015).
Tettoni, S. D. et al. Structure of the Schizosaccharomyces Pombe Gtr-Lam complex reveals evolutionary divergence of mTORC1-dependent amino acid sensing. Structure 31, 1065–1076e5 (2023).
Powis, K. et al. Crystal structure of the EGO1-EGO2-EGO3 complex and its role in promoting rag GTPase-dependent TORC1 signaling. Cell. Res. 25, 1043–1059 (2015).
Urano, J., Tabancay, A. P., Yang, W. & Tamanoi, F. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J. Biol. Chem. 275, 11198–11206 (2000).
Loissell-Baltazar, Y. A. & Dokudovskaya, S. SEA and GATOR 10 years later. Cells 10, 1 (2021).
Peng, M., Yin, N. & Li, M. O. SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437 (2017).
Brunkard, J. O. Exaptive evolution of target of rapamycin signaling in multicellular eukaryotes. Dev. Cell. 54, 142–155 (2020).
Cruz, M. C. et al. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell. Biol. 19, 4101–4112 (1999).
Wong, G. K., Griffith, S., Kojima, I. & Demain, A. L. Antifungal activities of rapamycin and its derivatives, prolylrapamycin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J. Antibiot. 51, 487–491 (1998).
Bastidas, R. J., Shertz, C. A., Lee, S. C., Heitman, J. & Cardenas, M. E. Rapamycin exerts antifungal activity in vitro and in vivo against Mucor circinelloides via FKBP12-dependent inhibition of Tor. Eukaryot. Cell. 11, 270–281 (2012).
Teichert, S., Wottawa, M., Schönig, B. & Tudzynski, B. Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot. Cell. 5, 1807–1819 (2006).
Navarro-Velasco, G. Y., Di Pietro, A. & López-Berges, M. S. Constitutive activation of TORC1 signalling attenuates virulence in the cross-kingdom fungal pathogen Fusarium oxysporum. Mol. Plant. Pathol. 24, 289–301 (2023).
Segreto, R. et al. The TOR kinase pathway is relevant for nitrogen signaling and antagonism of the mycoparasite Trichoderma Atroviride. PLoS ONE. 16, e0262180 (2021).
Pancha, I., Chokshi, K., Tanaka, K. & Imamura, S. Microalgal target of rapamycin (TOR): a central regulatory hub for growth, stress response and biomass production. Plant. Cell. Physiol. 61, 675–684 (2020).
Nguyen, D. V. et al. Target of rapamycin pathway in the white-rot fungus phanerochaete chrysosporium. PLoS ONE. 15, e0224776 (2020).
Li, G., Cao, X., Tumukunde, E., Zeng, Q. & Wang, S. The target of rapamycin signaling pathway regulates vegetative development, aflatoxin biosynthesis, and pathogenicity in. Elife 12 (2024).
Zhao, H. et al. Comparative genomics of provides insights into lipid metabolism: two novel types of fatty acid synthase. J. Fungi (Basel). 8, 1 (2022).
Zhang, N. & Cao, L. Starvation signals in yeast are integrated to coordinate metabolic reprogramming and stress response to ensure longevity. Curr. Genet. 63, 839–843 (2017).
UniProt. The universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Wong, E. D. et al. Saccharomyces genome database update: server architecture, pan-genome nomenclature, and external resources. Genetics 224, 1 (2023).
Rutherford, K. M., Lera-Ramírez, M. & Wood, V. PomBase: a global core biodata resource—growth, collaboration, and sustainability. Genetics. https://doi.org/10.1093/genetics/iyae007 (2024).
Marchler-Bauer, A. et al. NCBI’s conserved domain database. Nucleic Acids Res.. 43, D222–D226 (2015).
Mistry, J. et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Groenewald, M. et al. A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina. Stud. Mycol. 105, 1–22 (2023).
Suga, H. et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4, 2325 (2013).
Ruiz-Trillo, I. et al. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 23, 113–118 (2007).
Fairclough, S. R. et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca Rosetta. Genome Biol. 14, R15 (2013).
King, N. et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788 (2008).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Minh, B. Q. et al. Corrigendum to: IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 2461 (2020).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Andrews, S. et al. FastQC: A Quality Control Tool for High Throughput Sequence Data (2010).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360 (2015).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Hadley, W. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Acknowledgements
We are grateful for insightful comments and suggestions by Eugenio Mancera Ramos and Joanna Kaminska.
Funding
This work was supported by National Science Centre grants (#2021/41/B/NZ2/02426 to A.M.)
Author information
Authors and Affiliations
Contributions
D.B. and A.M. designed the study and performed analyses. D.B., M.P. and A.M. drafted the manuscript. D.B. prepared the figures, A.M. conceptualized the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barua, D., Płecha, M. & Muszewska, A. Non-dikarya fungi share the TORC1 pathway with animals, not with Saccharomyces cerevisiae. Sci Rep 15, 5926 (2025). https://doi.org/10.1038/s41598-025-89635-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89635-4