Abstract
Infertility, recognized by the World Health Organization (WHO) as a disease affecting the male or female reproductive system, presents a global challenge due to its impact on one in six individuals worldwide. Given the high prevalence of infertility and the limited available resources in fertility care, infertility creates substantial obstacles to reproductive autonomy and places a considerable burden on fertility care providers. While existing research are exploring to use artificial intelligence (AI) methods to assist fertility care providers in managing in vitro fertilization (IVF) cycles, these attempts fail in accurately predicting specific aspects such as medication dosage and intermediate ovarian responses during controlled ovarian stimulation (COS) within IVF cycles. Our current work developed Edwards, a deep learning model based on the Transformer-Encoder architecture to improve the prediction outcomes. Edwards is designed to capture the temporal features within the sequential process of IVF cycles, It could provide the options of treatment plans as well as predict hormone profiles, and ovarian responses at any stage upon both current and historical data. By considering the full context of the process, Edwards demonstrates improved accuracy in predicting the final outcomes of the IVF process compared to previous approaches based on traditional machine learning. The strength of our current deep learning model stems from its ability to learn the intricate endocrinological mechanisms of the female reproductive system, especially for the context of COS in IVF cycles.
Similar content being viewed by others
Introduction
Infertility is defined by the World Health Organization (WHO) as a disease of the male or female reproductive system characterized by the failure to achieve pregnancy after 12 months or more of regular unprotected sexual intercourse1. Ranked as the eighth most severe disability globally, infertility affects one in every six individuals at some point in their lives. It could be even more challenging in developing countries2. According to the World Report on Disability (2011), 32.5 million people of reproductive age in low- and middle-income countries suffer from infertility issues. However, despite its high prevalence, resources for fertility care remain limited. For example, in the United States, a leader in advanced artificial reproductive technology (ART), there were only 1,700 reproductive endocrinology and infertility (REI) specialists in 2020. This imbalance between high demand and limited resources restricts access to infertility care for low-income families3,4 and imposes an excessive workload on fertility care providers.
In vitro fertilization (IVF) is one of the most widely utilized ART methods. In 2016, IVF accounted for more than 99% of all ART cycles in the United States5. IVF treatments typically align with the woman’s menstrual cycle and proceed through four sequential phases: priming, controlled ovarian stimulation (COS), induction of final oocyte maturation (trigger phase), and oocyte retrieval (OR). REI specialists play a critical role in decision-making, including determining stimulation protocols, modifying medications, and scheduling follow-up visits and OR dates. These decisions are based on ovarian responses (e.g., follicle count and size) and hormone profiles, including estradiol (E2), progesterone (P4), follicle-stimulating hormone (FSH), and luteinizing hormone (LH). Clinical judgments also rely on data from current and prior cycles, creating a feedback loop where patient responses and outcomes in later stages of the cycle depend on earlier decisions. This dynamic and sequential process necessitates robust yet labor-intensive data and workflow management practices.
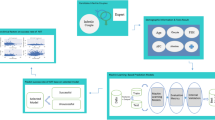
The architecture of Edwards (Edwards-Pro). We categorized the key elements of IVF process and patient demographic info into 123 subcategories, which are mapped into a high-dimension trainable embedding space with a lookup dictionary. The characteristics of each element, and the context of IVF process are thus captured and represented by the embedding space and the parameters of the Transformer Encoder. We named this deep learning approach as Edwards. We proposed a combined system with Edwards and a knowledge-based decision support system, called Edwards-Pro, to improve the accessibility of this approach, as well as to improve of the performance of predicting treatment plans.
Some pioneering studies leveraged artificial intelligence (AI) approaches to assist fertility care providers in managing IVF treatment cycles more effectively. For instance,6 introduces decision support systems based on knowledge-based algorithms for day-to-day management during controlled ovarian stimulation. This approach achieved high accuracies for four critical clinical decisions: 0.92 for deciding on continuing or stopping treatment, 0.96 for triggering and scheduling oocyte retrieval or canceling the cycle, 0.82 for adjusting medication dosage, and 0.87 for determining the number of days until follow-up. However, its performance in advanced predictions, such as determining exact medication dosages, was limited. We developed a more comprehensive decision support system using knowledge-based algorithms that incorporated a broad spectrum of clinical scenarios and IVF protocols7. Still, this system was not designed to predict ovarian responses, hormone profiles, and final COS outcomes, such as oocyte maturity (measured by metaphase II (MII) oocyte rate), 2 pronuclear (2PN) embryo rate, and blastulation rate. Loewke’s group8 employed machine learning techniques to determine the optimal date for triggering during ovarian stimulation. Their model was developed to forecast the number of collected metaphase II (MII) oocytes upon triggering on different dates and then determine the optimal trigger date. This study explored an uncharted area of predicting optimal trigger dates based on IVF cycle outcomes. However, its efficacy in forecasting the MII number was suboptimal, with a coefficient of determination (R2) value of 0.64 and 0.62 when triggering one day earlier or later, respectively. The aforementioned studies did not fully account for the temporal features of IVF cycles. Each phase within an IVF cycle is sequential and heavily influenced by preceding steps. We hypothesize that integrating a machine learning model capable of capturing these temporal features could significantly enhance predictive performance.
Since the breakthrough in 2012, deep learning algorithms have achieved continuous success in various fields, including games9, medicine and biology10, automotive technology11, and the IT industry12,13. As a key branch of deep learning, sequential learning algorithms aim to tackle tasks involving sequential data by extracting temporal features from the underlying patterns. In particular, the Transformer model14, a leading sequential learning architecture, has achieved remarkable success in natural language processing (NLP) and multi-modal tasks in recent years. One of the most recognized examples worldwide is ChatGPT12. Powered by a 1.5-billion-parameter multilayer Transformer Decoder, it can nearly pass the Turing test and compose comprehensive articles of up to 2,000 words on specific topics. The unprecedented performance of ChatGPT among pre-Transformer language models significantly highlights the Transformer’s ability to capture semantic contexts in human language. Because the IVF process resembles a causally correlated sequential process, akin to an article constructed with grammar and logic, we propose that a Transformer-based deep learning approach could effectively model female reproductive endocrinology within the IVF process. In this analogy, monitoring visits during IVF are akin to sentences, while key elements of these visits correspond to words.
In the current research, we propose a Transformer-Encoder-based deep learning model, called Edwards (dedicated to Sir Robert Geoffrey Edwards, who was awarded the Nobel Prize in Physiology or Medicine for the development of IVF), designed to capture temporal features in daily ovarian stimulation and response. This approach, unlike previous studies, is capable of predicting treatment plans, hormone profiles (e.g., serum estradiol (E2), progesterone (P4), follicle-stimulating hormone (FSH), luteinizing hormone (LH)), and ovarian response on any cycle day, based on current and prior treatment plans and assessments. Furthermore, it could achieve enhanced accuracy in predicting the final outcomes of the IVF cycle by considering the broader process context.
Results
In Edwards, we used a multi-layer Transformer Encoder to learn the representations of the key elements in IVF process. These key elements included demographic data (more details shown in Table 1), treatment plans, hormone profiles, and follicular measurements (more details shown in Table 2), categorized and mapped into a lookup dictionary. We applied a self-supervised training method, Masked LM15, as the pre-training strategy. During the pre-training process, these elements were projected into a high-dimension trainable vectorized embedding space through the aforementioned lookup dictionary, the characteristics of each element, and the context of IVF process were thus captured and represented by the vectored embedding space and the parameters of the Transformer Encoder. The downstream tasks (e.g., predicted treatment plans, final outcomes of IVF cycles, etc) are addressed by fine-tuning the pre-trained model. In addition, we developed Edwards-Pro by integrating the knowledge-based decision support system proposed by our previous study7 into Edwards, in order to improve the accessibility of this approach, as well as to improve the predictions of treatment plans.
We used historical clinical data collected over almost ten years from New Hope Fertility Center (NHFC) to train and verify our approach. The clinical data including the aforementioned key elements were collected from patients’ monitoring in every visit. The dataset for training the deep learning model contained 30,552 IVF cycles with 239,047 monitoring visits from January 2013 to December 2021. Another dataset of 1,804 cycles containing 8,364 visits from January 2022 to July 2022 was used as the validation dataset. More details about the data preprocessing, model architecture, and training strategies are addressed in Section 4 and Figure 1.
Evaluation strategy for two-phase predicting targets
Our approach provides predictions for two distinct phases in IVF COS cycles. Phase I focuses on key elements during monitoring visits, such as treatment plans, hormone profiles, and follicular measurements. Predictions for these elements in visit #n are based on all data from the previous #n-1 visits. Phase II targets the final outcomes of IVF cycles, such as MII rate, 2PN rate, and blastulation rate (more details shown in Table 3), predicted using data from the entire IVF cycle (Table 4).
Both phases were framed as classification tasks for two reasons: 1. Classification tasks align naturally with our approach, where key elements of the IVF process are categorized into data points for the training and validation datasets. 2. Clinically, REI specialists typically make decisions based on ranges of hormone profiles and follicular measurements rather than exact values. Additionally, the rates of MII, 2PN, and blastulation, defined as proportions of retrieved oocytes, are more accurate criteria for assessing IVF outcomes, as they correlate closely with patient factors such as age, ovarian reserve, and stimulation response.
We designed a targeted evaluation strategy for these two-phase predictions. For Phase I, which can be applied during any monitoring visit, we divided the 1,804 cycles in the validation dataset into 8,364 input sequences. In each sequence, data from visits beyond the predicted monitoring visit were excluded. For Phase II, we used the full dataset from each cycle, as final IVF outcomes depend on the entire ovarian stimulation process. To benchmark our deep learning model, we implemented traditional machine learning approaches referenced in prior studies6,8. Additionally, we developed a sequential learning baseline model-Sequence-to-Sequence (Seq2Seq)16, based on Long Short-Term Memory (LSTM) units17, to assess our model’s ability to capture temporal features effectively.
The results of phase I predictions
Tables 5 and 6 summarize the experimental outcomes for Phase I predictions. The main distinction between Edwards-Pro and Edwards lies in Edwards-Pro’s enhanced ability to predict treatment plans; both models performed identically for other prediction categories. In nearly all treatment plan categories (Table 5), sequential learning models, including Seq2Seq, Edwards, and Edwards-Pro-outperformed traditional machine learning approaches, achieving improvements of at least 10% in average precision (AP), 14% in the area under the receiver operating characteristic curve (AUROC), and 4% in top-2 accuracy. The exception was the Follitropin category, which had an imbalanced label set; while AdaBoost achieved the best AP (93.0%), this was due to predicting only the dominant class. For categories linked to clinical judgment, such as Day# (next visit date), Follitropin (COS dosage), and oral contraceptives, Edwards-Pro improved Edwards’s performance by 2.9% (AP), 5.8% (AUROC), and 11.6% (top-2 accuracy). In clinical assessment-related predictions (Table 6), sequential learning models excelled across all categories except FSH and follicular measurements, both of which had imbalanced datasets similar to Follitropin. Conversely, for E2 and LH-each with 9 and 8 classes, respectively-sequential learning methods achieved substantial gains in AP, exceeding traditional machine learning methods by at least 10.1%. While sequential learning methods demonstrated consistent performance, traditional machine learning models varied greatly between categories. For instance, AdaBoost ranked second for P4 but fell to the bottom three for LH. These findings confirm that sequential learning models leverage temporal features effectively to predict ovarian response and subsequent treatment plans in IVF cycles. Edwards-Pro and Edwards outperformed Seq2Seq in most categories due to the superior temporal feature extraction capabilities of Transformer models. Additional details are discussed in Section 3.
The results of phase II predictions
The experimental results of Phase II predictions were summarized in Table 7. Edwards|Edwards-pro surpassed all the baseline models in all the three categories of final outcomes. For the predictions of MII rate, Edwards|Edwards-pro beat Seq2Seq by 4.1%, and beat the traditional machine learning models by 12.1% at minimum; for 2PN rate, Edwards|Edwards-pro beat Seq2Seq and the traditional machine learning models by 4.5% and 23.0% at minimum, respectively; for blastulation rate, Edwards|Edwards-pro beat Seq2Seq by 3.9%, and beat the traditional machine learning models by 22.9% at minimum. We also assessed the statistical significance of our experiments. The statistical difference between Edwards|Edwards-pro and Seq2Seq, and between Edwards|Edwards-pro and traditional baseline models was p value \(<=0.05\) and p value \(<=0.01\), respectively (more details in Section 4.2). These huge improvements are a solid evidence to indicate that sequential learning models, especially by Edwards|Edwards-pro, attained the context and mutual relationships among the key elements of IVF process and thus boosted the performance of predicting the final outcomes. In contrast, the traditional machine learning approaches were not able to capture the correlation and causality within IVF process so that their performance, especially for 2PN rate and blastulation rate (3-class task), was only slightly better than random selection. The MII, 2PN, and blastulation rate trended down in turn, among the experimental results of all the approaches tested in our experiments. These results were reasonable because they represented the natural trend. In addition, the 2PN and blastulation rate might be affected by sperm quality, embryologists’ technical competency, and chemical and physical factors of culturing media18,19. In Section 3, we will discuss further about how to improve our approach on the above two tasks in the future study.
Discussion
In this paper, we proposed Edwards, a Transformer-based deep learning model to navigate IVF dynamics. To our best knowledge, Edwards is the first attempt to take advantage of sequential deep learning algorithms to explore the management and response of controlled ovarian stimulation (COS) and then accurately predict almost all key elements of IVF cycles, such as treatment plans, hormone profiles, and ovarian response on any visit cycle day. Our work novelly discovered that the inherent correlations among the essential elements of COS are well captured by Transformer-based deep learning approaches. As a result, Edwards is capable to provide accurate predictions for all kinds of essential elements of COS at any phase. This unique feature of Edwards may provide REI specialists useful information to make optimal clinical judgments. More essentially, our model achieved the best performance in the literature in predicting the final outcomes of IVF cycles by more than 12% improvement for the MII rate, 32% improvement for the 2PN rate and 30% for the blastulation rate. Edwards-Pro, the enhanced version of Edwards that integrates with the knowledge-based decision support system developed by our previous work, could provide more flexibility to healthcare providers for purposes in clinical practices.
Visualized Explanation for the Pre-training and Embedding Space
t-Distributed Stochastic Neighbor Embedding (t-SNE)20 is a popular tool to visualize high-dimension data. It is a nonlinear dimensionality reduction algorithm to embed high-dimension data in a two- or three-dimension space for visualization. More specifically, t-SNE models pairwise similarities between data points in both high- and low-dimensional spaces using conditional probabilities. By minimizing the Kullback-Leibler (KL) divergence of the distributions between the high- and low-dimensional space, t-SNE can capture complex nonlinear relationships within the data.
We used t-SNE to visualize the embedding space of the key elements in IVF process for both Edwards and Seq2Seq, as shown in Figure 2 and 3, respectively. The 2-D visualization by t-SNE of the embedding space of Edwards displays distinct clusters among the key elements in IVF process. In our study, Edwards clearly clustered each category in the dimension space. For example, although we used the same symbol to represent multiple categories (e.g., \(\triangleleft\) for ’ocp’ and ’est’, \(\triangledown\) for ’folli’, ’clomid’, and ’fem’), the subcategories under a category were closer with each other and distinguished from different categories by gradual color changes. Moreover, our work showed that the distance among subcategories was able to represent the underlining relationships among them. We take the category ’cycle day:’ for instance: since we know that a regular menstruation cycle length is about 28 days in average for most of women, ’day #0–3’ is close to ’day #28-’ except for the patients with a long or short menstrual cycle. Without any extra information imported, Edwards captured the cyclical pattern of menstruation cycles of females. The 8 subcategories of menstruation cycle day# place in turn as a circle, with a small gap between ’day #0–3’ and ’day #28-’ (Figure 2). In contrast, the subcategory of ’Estradiol’ (E2) almost fell into a curve pattern in t-SNE 2-D visualization because E2 levels rise proportionally with follicular growth during COS.
However, the embedding space of Seq2Seq did not have a comparable t-SNE visualized graph to show the clear clusters as Edwards (Figure 3). For example, the categories ’cycle day:’ and ’Estradiol’ lacks clear geometric patterns. This discrepancy highlights why Edwards outperforms most of the Phase I and II prediction tasks comparing to Seq2Seq. There are two plausible explanations for these findings: i) As a sequential learning model based on recurrent neural networks (RNNs), Seq2Seq is hindered by inherent limitations of RNNs, including challenges with long-range temporal dependencies21, and the vanishing gradient problem22,23. Both the intrinsic shortcomings make Seq2Seq very difficult to be trained. In contrast, Transformer is naturally free from the intrinsic problems of Seq2Seq since Transformer uses dot-product to learn temporal features in parallel. ii) MLM, the pre-training strategy of Edwards, is another key technology in our study: for each mask token in pre-training process, the loss was calculated based on the whole input sequence including both the parts before and after the mask token. As a result, the training process for each mask token was based on the whole sequence that apparently contains richer and more accurate context information. Seq2Seq, on the other hand, discards parts of the input sequence following each training token due to its iterative nature. This results in most training iterations failing to utilize the complete sequence context.
The distinct representation of key elements in the IVF process suggests that Edwards has a deeper understanding of the mechanisms underlying controlled ovarian stimulation and response. To validate this assumption, we conducted six groups of cross-validation experiments involving three key elements: E2, trigger, and cycle day. The validation strategy involved predicting two of the elements given a specific subcategory of the third. For instance, we tested the prediction accuracy for E2 and cycle day when trigger shots were taken. The results of these cross-validations are summarized in Tables 8,9,10,11 with heatmaps of confusion matrices provided as from Figure S5 to S32 in Supplementary Information. Edwards consistently outperformed Seq2Seq across all cross-validation groups. Notably, Edwards demonstrated a significantly greater advantage over Seq2Seq in predicting triggers, a task requiring comprehensive domain knowledge and understanding of intricate relationships.
Future work
Sperm quality is a major factor influencing 2PN and blastulation rates in IVF cycles. According to24, approximately 40% of infertility cases are attributed to male factors. Key procedures in embryology lab work, such as intracytoplasmic sperm injection (ICSI) and embryo culture, also play a critical role in determining these rates25. provided strong evidence that ICSI improves fertilization and implantation rates. The morphological evaluation of embryos at specific time points during culture, including multinucleation during early cleavage stages26,27 and cleavage timing28,29,30, offers valuable insights into embryo quality.
We hypothesize that integrating these datasets can enhance the accuracy of 2PN and blastulation rate predictions. In future work, we plan to develop a Transformer-based multi-modal model to learn temporal features from sperm parameters and time-lapsed video clips of embryo culturing. This model will include a vision encoder and a modality backbone. The vision encoder will extract high-dimensional visual features from the video clips of embryo culture. The modality backbone will process the abstract representation of the multi-modal input data to predict 2PN and blastulation rates. The multi-modal input will incorporate context from the stimulation cycle (output of the Transformer Encoder in Edwards), sperm quality data (e.g., male partner demographics, sperm motility, and progression rate), embryo culture preparation details (e.g., ICSI usage, culturing media information), and sequential data from video clips of embryo culture (e.g., multinucleation timestamps and cleavage timings).
Methods
Datasets
In this study, we used clinical data from the NHFC dataset, which includes 30,552 IVF cycles from 12,460 patients over nearly 10 years. This study was conducted in accordance with all relevant guidelines and regulations, as approved by the WCG Institutional Review Board (IRB) (Approval Number: 20230491). The requirement for informed consent was waived by the WCG IRB due to the use of fully de-identified retrospective data. All patient data used in this study were de-identified to ensure confidentiality. A detailed demographic summary of the dataset is provided in Table 4. We categorized the key elements of the IVF process and patient demographics into 123 subcategories, as shown in Tables 1 and 2. Inspired by studies in natural language processing (NLP)31,32,33and bioinformatics34, these subcategories were treated as the basic elements of the training corpus, forming a lookup dictionary that maps to a trainable high-dimensional embedding space. Additionally, five functional tokens were incorporated into the corpus. Specifically, ’[CLS]’ and ’[SEP]’ were used as the starting and ending anchors for a monitoring visit, ’[UNK]’ substituted rare elements not included in the corpus, ’[MASK]’ served as the mask token for self-supervised pre-training, and ’[PAD]’ was the padding token for input alignment. In contrast to standard NLP tasks, the input sequence in this work was organized as follows: [CLS], each element of monitoring visit #1, [SEP], [CLS], each element of monitoring visit #2, [SEP], ... [CLS], each element of monitoring visit #n, [SEP]. More formally, for a patient \(pt \in \{PT_{1}, PT_{2}, ..., PT_{n}\}\) in the dataset, the demographic info in cycle c is denoted as \(demo_{c}^{pt} = \{A_{c}^{pt}, B_{c}^{pt}, MC_{c}^{pt}\}\), where \(A_{c_{i}}^{pt}\), \(B_{c_{i}}^{pt}\), \(MC_{c_{i}}^{pt}\) represent the patient’s age, BMI, and Menstruation Cycle Length, respectively. An arbitrary monitor visit #i in cycle c is \(v_{c_{i}}^{pt} = \{D_{c_{i}}^{pt}, T_{c_{i}}^{pt}, H_{c_{i}}^{pt}, S_{c_{i}}^{pt}\}\), where \(D_{c_{i}}^{pt}\) represents the Menses Day#, \(T_{c_{i}}^{pt}\), \(H_{c_{i}}^{pt}\), and \(S_{c_{i}}^{pt}\) represent the set of all the corresponding categories in Treatment Plan, Hormone Profiles, and Follicular Measurements, respectively. The final outcomes in cycle c (if available) was \(o_{c}^{pt} \in \{MII_{c}^{pt}, 2PN_{c}^{pt}, BLST_{c}^{pt}\}\), \(MII_{c}^{pt}, 2PN_{c}^{pt}, BLST_{c}^{pt}\) denoted MII rate, 2PN rate, and blastulation rate, respectively. An example of the input sequences is provided in Supplementary Information.
For Phase I predictions, given the causality and correlation in the context of sequential learning tasks, we applied a strict validation strategy to avoid the data leakage problem. Specifically, for predictions of the treatment plan at monitoring visit #n, we fed the input sequence before monitor visit #n, along with the hormone profiles and follicular measurements at visit #n. For predictions of hormone profiles and follicular measurements at monitor visit #n, we only fed the input sequence before monitor visit #n. For Phase II predictions, a different validation strategy was applied. Since not all IVF cycles in our dataset had final outcomes-due to patient preferences and clinical scenarios-we ultimately collected 8,920 cycles with MII oocytes as outcomes from both oocyte cryopreservation and fertilization cycles, and 6,750 cycles with 2PN and blastocyst outcomes (blastocyst frozen cycles only). Additionally, the data for these three outcomes were excluded from the pre-training process, ensuring that data leakage was not a concern. We then fed the input sequence from monitor visit #1 through to the visit upon trigger and performed 10x10-fold validation sessions to assess the robustness of our model for Phase II predictions. In each fold, we randomly split the dataset into a training and validation set with a 9:1 ratio.
Model design
In the clinical management of IVF COS cycles, the health providers would like to accurately predict a range and trend of the key elements of IVF process rather than an exact value. For example, a generally acknowledged standard of LH surge is that the current LH level is 1.8 times higher than the previous level when the leading follicle is 15 millimeter (mm) or bigger. In reality, it is extremely technically difficult to accurately predict the exact levels of either the key elements or the final outcomes of IVF process due to the variants, such as the wide range of female endocrinological profile and demographic difference. In addition, the training dataset used by this approach was quite small (56 megabyte file size) comparing to the titanic scale of datasets used in current Large Language Models (LLM). For example, the dataset of GPT-4 was 1 petabyte file in size35, \(1.8*10^7\) times larger than our training dataset. Given the above unique features in IVF cycles and the limitation in data resource, our approach implemented the Phase I and II predictions as a series of multi-label classification tasks. For an arbitrary Phase I prediction \(PI_{c_{i}}^{pt} \in \{M_{c_{i}}^{pt}, T_{c_{i}}^{pt}, H_{c_{i}}^{pt}, S_{c_{i}}^{pt}\}\), our approach aimed to predict the following conditional probability:
for an arbitrary Phase II prediction \(PII_{c}^{pt} \in \{MII_{c}^{pt}, 2PN_{c}^{pt}, BLST_{c}^{pt}\}\), our approach aimed to predict the following conditional probability:
The efficacy of the above tasks depended on a hidden prerequisite of causalities or correlations among the key elements during IVF process. This prerequisite has been proven by IVF clinical practices for more than 40 years36. We thus hypothesized that the above tasks would be similar to other sequential learning tasks, such as NLP tasks. There are plenty of existing approaches to solve NLP tasks, such as tf-idf37, skip-gram31, recurrent neural networks (RNN)16,17, and attention mechanisms38,39. However, our current study had the following unique challenges: i) Unlike discrete and unique elements in other sequential learning tasks, most of the key elements in IVF process were continuous assessment. For example, estradiol (E2), one of the hormone profiles, varied in a range of from 20 pg/mL to more than 5,000 pg/mL during ovarian stimulation in IVF process. We used categorization to address this problem, as shown in Table 2. All the thresholds among the subcategories were defined by the experienced REI specialists from NHFC. ii) The key elements in IVF process have a naturally hierarchical structure after they are categorized into the subcategories, which brings more challenges to learn the relationships among the key elements. For example, it is obvious for human to understand that the subcategories of E2 are similar to each other much more than the ones of other categories. All the subcategories of the key elements, however, were mapped into a flattened lookup dictionary so that our model had no information about the relationship among these subcategories at all until it was pre-trained. iii) Not all the clinical information and inputs could be included in the datasets.
We hypothesized that Transformer14would be the proper fundamental architecture for our model based on the above analysis. Transformer is the state-of-the-art deep learning model for sequential learning tasks, most of recent breakthroughs in the field of NLP, such as ChatGPT, and CLIP13, are powered by this model. We followed the customization to the structure of Transformer by BERT15by using the encoder part of Transformer only and using masked language model (MLM) as the self-supervised pre-training strategy. After reviewing previous research12,15,40,41, Transformer Decoder based models would have more powerful performance under ultra-large scale of training dataset. For example, the 100-time larger training set made GPT-312achieved a huge performance growth comparing to GPT-241. Therefore, we selected Transformer Encoder based models as the fundamental architecture in our approach, given that the dataset in our study was exponentially smaller than the one in GPT-3. In addition, MLM was a naturally proper pre-training strategy for this study. When randomly masking one token of an input sequence, We could used this token as the label to calculate the loss value and then to do the back propagation process in MLM training process. This strategy was able to efficiently reuse the training dataset, because randomly masking a token among an input sequence could expand the scale of the training dataset.
We fine-tuned the pre-trained model to address the downstream multi-label classification tasks in Phase I and II predictions. For each task, we added a separate full-connected layer to output the desired predictions. Since most of the Phase I predictions had more than 5 classes, We used AP score, AUROC as the metrics. In addition, we used top-2 as an extra metrics in the phase I predictions with the following reasons: i) During COS in IVF treatments, multiple feasible options could be applied in each visit. The categorization for the key elements caused inevitable over-constraints. For example, the estradiol level 3,001 pg/mL is categorized into ’>3000 pg/mL’, however, it is also acceptable to predict it into another category ’2000–3000 pg/mL’. ii) In the field of sequential learning, top-2 is a common metric to assess models’ capability of capturing context. The labels for the Phase II predictions were balanced well so that AP score could perfectly assess the accuracies of them. Meanwhile, we could randomly split the dataset into the training dataset for fine-turning and the validation dataset in the Phase II predictions (more details in Section 4.1), we then evaluated the statistical significance of the Phase II predictions: we used Mcnemar’s test42and 10x10-fold cross-validation t paired test43 to assess the statistical significance of our experiments. Both the two tests for the three Phase II predictions demonstrate that the statistical difference between Edwards|Edwards-pro and Seq2Seq, and between Edwards|Edwards-pro and traditional baseline models is p value \(<=0.05\) and p value \(<=0.01\), respectively.
Training
We used Bayesian optimization44 to seek the optimal set of hyperparameters for pre-training. We applied a Transformer Encoder with 5 hidden layers and 6 attention headers, each of the hidden layers had a 228-dimension attention layer and a 1602-dimension fully-connected layer. For the trainable embedding space mentioned in Section 4.1, the vector dimension of the embedding space was 128. On the side of training settings, the max length of input sequences was 128 tokens, the learning rate was 4.36e-05, the batch size was 11, and the weight decay was 2.5e-04. We used MLM as the pre-training strategy, Adam45as the optimiser, and cross entropy as the loss function. The pre-training process ran 200 epochs. After each epoch, we randomly picked a subset of training set to calculate AP scores as the criterion of saving checkpoints of the model weights, The last checkpoint of the model was used as the pre-trained model. For the further downstream tasks, we applied a lower learning rate 1e-05 to fine-tune the pre-trained model. A Nvidia TITAN RTX 24GB GPU was used for pre-training, and an additional NVIDIA GeForce GTX 1070 8GB GPU was applied for downstream tasks in parallel. Our approach was implemented based on Pytorch framework46.
Integrating deep learning into a knowledge-based decision support system
Knowledge-based decision support systems are a type of expert systems, which are computer programs that use a knowledge base of facts and rules to simulate the problem-solving abilities of a human expert in a specific domain. In our previous work7, we developed a unique knowledge-based decision support system that was able to predict the following treatment plans based upon the current hormone profiles, follicular measurements, and current treatment plans. Although this knowledge-based decision support system could not predict hormone profiles and follicular measurements in next visits and the final outcomes of the IVF cycles, it gave medical providers some flexibility to modify the treatment plan based on the non-medical factors, such as bad weather, and rescheduled appointments. Here, we integrated this knowledge-based decision support system into Edwards in order to provide recommendations in the treatment plan based on the knowledge-based decision support system as well as other treatment plans generated by the deep learning model and self-defined treatment plans by users. Meanwhile, for each portfolio of treatment plans, the deep learning model generated the corresponding predictions of hormone profiles and follicular measurements in next visit for each option among treatment plans. The performance of the integrated system is shown in Table 5.
Data availability
The data that support the findings of this study are available from New Hope Fertility Center but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Requests for data access can be directed to the corresponding author at the email address provided in the author list.
References
Vander Borght, M. & Wyns, C. Fertility and infertility: Definition and epidemiology. Clinical biochemistry 62, 2–10 (2018).
Njagi, P. et al. Financial costs of assisted reproductive technology for patients in low-and middle-income countries: a systematic review. Human reproduction open 2023, hoad007 (2023).
Kelley, A. S., Qin, Y., Marsh, E. E. & Dupree, J. M. Disparities in accessing infertility care in the united states: results from the national health and nutrition examination survey, 2013–16. Fertility and Sterility 112, 562–568 (2019).
Brautsch, L. A. S., Voss, I., Schmidt, L. & Vassard, D. Social disparities in the use of art treatment: a national register-based cross-sectional study among women in denmark. Human Reproduction 38, 503–510 (2023).
Sunderam, S. et al. Assisted reproductive technology surveillance-united states, 2018. MMWR Surveillance Summaries 71, 1 (2022).
Letterie, G. & Mac Donald, A. Artificial intelligence in in vitro fertilization: a computer decision support system for day-to-day management of ovarian stimulation during in vitro fertilization. Fertility and Sterility 114, 1026–1031 (2020).
Wang, X. et al. A knowledge-based decision support system for in vitro fertilization treatment. In 2020 IEEE International Conference on E-health Networking, Application & Services (HEALTHCOM), 1–8 (IEEE, 2021).
Fanton, M. et al. An interpretable machine learning model for predicting the optimal day of trigger during ovarian stimulation. Fertility and Sterility 118, 101–108 (2022).
Silver, D. et al. Mastering the game of go with deep neural networks and tree search. nature 529, 484–489 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature 596, 583–589 (2021).
Hu, Y. et al. Planning-oriented autonomous driving. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 17853–17862 (2023).
Brown, T. et al. Language models are few-shot learners. Advances in neural information processing systems 33, 1877–1901 (2020).
Radford, A. et al. Learning transferable visual models from natural language supervision. In International conference on machine learning, 8748–8763 (PMLR, 2021).
Vaswani, A. et al. Attention is all you need. Advances in neural information processing systems 30 (2017).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 (2018).
Sutskever, I., Vinyals, O. & Le, Q. V. Sequence to sequence learning with neural networks. Advances in neural information processing systems 27 (2014).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural computation 9, 1735–1780 (1997).
Bormann, C. L. et al. Deep learning early warning system for embryo culture conditions and embryologist performance in the art laboratory. Journal of Assisted Reproduction and Genetics 38, 1641–1646 (2021).
Wale, P. L. & Gardner, D. K. The effects of chemical and physical factors on mammalian embryo culture and their importance for the practice of assisted human reproduction. Human reproduction update 22, 2–22 (2016).
Van der Maaten, L. & Hinton, G. Visualizing data using t-sne. Journal of machine learning research 9 (2008).
Sutskever, I. Training recurrent neural networks (University of Toronto Toronto, ON, Canada, 2013).
Hochreiter, S. Untersuchungen zu dynamischen neuronalen netzen. Diploma, Technische Universität München 91, 31 (1991).
Bengio, Y., Simard, P. & Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE transactions on neural networks 5, 157–166 (1994).
Kumar, N. & Singh, A. K. Trends of male factor infertility, an important cause of infertility: A review of literature. Journal of human reproductive sciences 8, 191 (2015).
Van Steirteghem, A. C. et al. High fertilization and implantation rates after intracytoplasmic sperm injection. Human reproduction 8, 1061–1066 (1993).
Hardy, K. Cell death in the mammalian blastocyst. Molecular human reproduction 3, 919–925 (1997).
Alikani, M. et al. Cleavage anomalies in early human embryos and survival after prolonged culture in-vitro. Human reproduction 15, 2634–2643 (2000).
Plachot, M. & Mandelbaum, J. Oocyte maturation, fertilization and embryonic growth in vitro. British medical bulletin 46, 675–694 (1990).
Munné, S. & Cohen, J. Chromosome abnormalities in human embryos. Human reproduction update 4, 842–855 (1998).
Alikani, M. et al. Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertility and sterility 71, 836–842 (1999).
Mikolov, T., Chen, K., Corrado, G. & Dean, J. Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781 (2013).
Le, Q. & Mikolov, T. Distributed representations of sentences and documents. In International conference on machine learning, 1188–1196 (PMLR, 2014).
Angelov, D. Top2vec: Distributed representations of topics. arXiv preprint arXiv:2008.09470 (2020).
Asgari, E. & Mofrad, M. R. Continuous distributed representation of biological sequences for deep proteomics and genomics. PloS one 10, e0141287 (2015).
OpenAI. Gpt-4 technical report (2023). arXiv: 2303.08774.
Johnson, M. H. Robert edwards: the path to ivf. Reproductive biomedicine online 23, 245–262 (2011).
Sparck Jones, K. A statistical interpretation of term specificity and its application in retrieval. Journal of documentation 28, 11–21 (1972).
Bahdanau, D., Cho, K. & Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473 (2014).
Cheng, J., Dong, L. & Lapata, M. Long short-term memory-networks for machine reading. arXiv preprint arXiv:1601.06733 (2016).
Radford, A., Narasimhan, K., Salimans, T., Sutskever, I. et al. Improving language understanding by generative pre-training. OpenAI blog (2018).
Radford, A. et al. Language models are unsupervised multitask learners. OpenAI blog 1, 9 (2019).
McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 12, 153–157 (1947).
Dietterich, T. G. Approximate statistical tests for comparing supervised classification learning algorithms. Neural computation 10, 1895–1923 (1998).
Snoek, J., Larochelle, H. & Adams, R. P. Practical bayesian optimization of machine learning algorithms. Advances in neural information processing systems 25 (2012).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014).
Paszke, A. et al. Pytorch: An imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems 32, 8024–8035 (Curran Associates, Inc., 2019).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, J., Liu, Z., Zhang, C. et al. A deep learning approach to understanding controlled ovarian stimulation and in vitro fertilization dynamics. Sci Rep 15, 7821 (2025). https://doi.org/10.1038/s41598-025-92186-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-92186-3