Abstract
WNT16 stands up as an essential gene for bone homeostasis. Here, we present new evidence of the functional role of a particular region within WNT16. Performing 4 C chromatin conformation analysis in three osteoblast-related cells (the human fetal osteoblast hFOB 1.19 cell line, Saos 2 osteosarcoma cell line and mesenchymal Stem Cells –MSC-), we identify physical interactions between the proximal part of WNT16 intron 2, shown here to be an active promoter in Saos 2 osteosarcoma cells, and several putative regulatory regions within CPED1. Analysis of previously published RNA-seq data from hFOB cells disclosed low expression of a region located downstream of this promoter. Our results suggest a novel regulatory mechanism of WNT16 in bone, mediated by physical interaction with various enhancer regions within CPED1.
Similar content being viewed by others
Introduction
In recent years, genome-wide association studies (GWAS) have been widely used for the identification of genes involved in complex diseases. In particular, up to 40 GWAS have been published identifying more than 500 genetic loci associated with bone parameters, reviewed in1,2,3. However, these studies are limited in several crucial ways. First, it is possible that the hit, which is identified by the lowest p-value, is not the causal SNP. In fact, all the regions in linkage disequilibrium with the hit SNP are candidates to bear the causal variant/s. Second, the vast majority of associated SNPs are found in intergenic or non-coding regions, which makes functional tests difficult. Third, the gene responsible for the association is not always the closest gene. For instance, if the SNP is in a regulatory region it is possible that it regulates the expression of a gene that is many kb away. For these reasons, an in-depth functional study of associated variants is necessary for the identification of the effector gene, which can be of great help to reveal new therapeutic targets for the treatment of common diseases, such as osteoporosis, the most common metabolic bone disorder. Surprisingly, despite the large number of genetic loci identified in bone-related GWAS, very few of them have been functionally characterized4,5,6.
One of the most consistent “bone-GWAS” signals is the genomic region of WNT16, which has been associated with skeletal phenotypes1,2,3,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,−22. In humans, the WNT16 protein has two alternative isoforms, WNT16a and WNT16b, and within bone cells it is predominantly expressed in early osteoblasts23,24. A regulatory role of WNT16 on bone has been confirmed by the analysis of knock-out (KO) and osteoblast-specific conditional KO mouse models, which showed spontaneous fractures as a result of low BMD and reduced bone strength8,9,24. On the other hand, the transgenic overexpression of Wnt16 in mouse osteoblasts results in higher BMD and bone resistance25,26,27.
Given the contribution of WNT16 to bone strength and its potential role as a therapeutic target for osteoporosis, it is essential to perform a comprehensive functional characterization and better understand its regulation. To this aim, we previously characterized the region corresponding to the WNT16 rs3801387 GWAS sentinel signal7, re-sequencing the WNT16 gene in individuals with extreme BMD phenotypes and genotyping some selected variants in a larger cohort28. As a result, we found two regulatory variants associated with lumbar spine BMD and in high linkage disequilibrium with the GWAS sentinel SNP (rs3801387)28. One of them affects the Kozak sequence and has already been functionally studied4, while the other (rs142005327) is located in intron 2, overlapping putative regulatory marks in primary osteoblasts (ENCODE). Here we set out to test the functionality of the WNT16 region containing the variant rs142005327. By performing functional studies of 4 C chromatin conformation and luciferase reporter assays, we have demonstrated a promoter activity of part of intron 2 and its physical interaction, in three osteoblast-related cells (the human fetal osteoblast hFOB 1.19 cell line, Saos 2 osteosarcoma cell line and mesenchymal Stem Cells –MSC-), with different putative regulatory regions within CPED1.
Materials and methods
Cell culture
The human osteosarcoma cell line Saos2 was used for luciferase reporter assays and 4 C-seq assays. It was obtained from the American Type Culture Collection (ATCC® HTB-85™) and grown in Dulbecco’s Modified Eagle Medium (DMEM; Sigma-Aldrich), supplemented with 10% Fetal Bovine Serum (FBS; Gibco, Life Technologies) and 1% penicillin/streptomycin (p/s; Gibco, Life Technologies), at 37 °C and 5% of CO2. Human foetal osteoblasts 1.19 (hFOB) and human medulla-derived mesenchymal stem cells (MSCs) were used for 4 C-seq assays. The hFOB 1.19 cell line was obtained from ATCC (ATCC® CRL-11372™) and grown in DMEM: F12 (1:1) medium without phenol red (Gibco, Life Technologies), supplemented with 10% FBS and 0.3 mg/ml Geneticin (Gibco, Life Technologies), at 34 °C and 5% of CO2. MSCs were kindly provided by Dr. José Manuel Quesada Gómez, from Instituto Maimónides de Investigación Biomédica, Hospital Universitario Reina Sofía, Córdoba, Spain29. These cells were grown in alpha-MEM medium (Gibco, Life Technologies), supplemented with 10% FBS, 1% p/s and 1x Glutamax (Gibco, Life Technologies), at 37 °C and 5% of CO2.
Luciferase reporter constructs
We cloned seven fragments (Full, Inv, 1, 2, A, B and C) of the WNT16-Intron 2 proximal region (depicted in Fig. 1) into the pGL3-promoter reporter vector (Promega) to create constructs EFull, EInv, E1, E2, EA, EB and EC, respectively, and into the pGL3-basic reporter vector (Promega) to create constructs PFull, PInv, P1, P2, PA, PB and PC, respectively. All fragments were cloned using the restriction enzymes XhoI and KpnI. The fragment named “Full” corresponds to the whole region with epigenetic marks. Fragment “Inv” is identical to fragment “Full” but in an inverted orientation (to check for enhancer function). “1” and “2” are partially overlapping fragments that correspond to the proximal and distal regions of fragment “Full”, respectively. The overlapping part of “1” and “2” corresponds to the region with high level of conservation in 100 vertebrates. Finally, fragments “A”, “B” and “C” are smaller fragments selected by specific reasons: fragment “A” bears the SNP found to be associated with BMD in our previous study28; fragment “B” contains the region conserved among 100 vertebrates; and fragment “C” corresponds to the region with more epigenetic marks. The size, genomic position, and primers used for cloning each region are specified in Supplementary Table S1. In all cases, the presence of the desired mutations and the absence of errors were verified by Sanger sequencing.
Luciferase gene reporter assays
We performed luciferase gene reporter assays to test the regulatory function of the WNT16-Intron 2 region (regulatory reporter assay). For the regulatory reporter assay, 1.1 × 105 Saos2 cells per well were cultured in 12-well plates, 24 h before the transfection. Each construct (empty, test fragment or positive control) was individually co-transfected, at a 10:1 proportion, with pRL-TK (total DNA = 1.1 µg). For enhancer assays, the empty vector was the pGL3-promoter backbone (Promega) and the SV40 enhancer was used as a positive control. For promoter assays, the empty vector was the pGL3-basic backbone (Promega) and the COL1A1-promoter (SP construct in30) was used as a positive control. Fugene HD was used following the manufacturer’s instructions. Forty-eight hours after transfection, cells were lysed, and the luciferase activity of Photinus pyrali and Renilla reniformis was measured using a Glomax Multi + luminometer (Promega), following the instructions of the Dual-luciferase reporter assay (Promega). Each experiment included three replicates and was repeated independently in minimum three separate experiments.
4 C-seq and AFourC software
4 C-seq was carried out at the Functional Genomics Service of the Centro Andaluz de Biología del Desarrollo (Sevilla, Spain). 4 C-seq libraries were generated from Saos2, hFOB 1.19, and MSC cell lines as described previously31,32. Four-bp cutters were used as the primary (DpnII) and secondary (Csp6I) restriction enzymes. For each cell line, a total of 1.6 µg of the library was PCR amplified for 6 different viewpoints (Primers and coordinates in supplementary Table S2). Samples were sequenced with Illumina Hi-Seq technology according to standard protocols at the Genomics Service of the Centro Nacional de Investigaciones Cardiovasculares (CNIC, Madrid, Spain). 4 C-seq data were analyzed as described previously33. Briefly, raw sequencing data were demultiplexed and mapped to the corresponding reference genome (GRCh37). Reads located in fragments flanked by two restriction sites of the same enzyme, in fragments smaller than 40 bp or within a window of 10 kb around the viewpoint, were filtered out. 4 C-seq data were normalized by the total weight of reads within ± 2 Mb around the viewpoint.
4 C-seq data were further analyzed following and adapting previously described pipelines34,35,36. Briefly, we assumed that the 4 C signal profile relative to the viewpoint v with coordinate xv on chromosome N is formed by three independent contributions. First, a constant background level common to all the chromosomes. Second, a negative exponential representing the monotonic decay of the 4 C signal with the genomic distance from the viewpoint34. Third, N gaussians with maximum values Pi, widths σi and centered around the coordinates xi, representing the peaks obtained from the genomic regions that are in physical contact with the viewpoint v:
To obtain the parameters of the 4 C profile for a given chromosome N and viewpoint v, we first estimated the background level B by computing the mean of the signal above the 99th percentile excluding the chromosome of interest. We then estimated the minimum detectable peak prominence as 5% of the maximum value of the 4 C profile on the chromosome of interest. Next, to estimate the genomic distance-dependent monotonic decay, we assumed a symmetric trend around the viewpoint and performed the exponential fit on the left-right averaged profile (Supplementary Fig. 1). To find the profile peaks, we smoothened the curve with a moving average of window size 30 reads and found all local maxima. To find only statistically significant targets, we rejected peaks based on three criteria. First, using the exponential fit and its error, we estimated the residual of each peak from the mean and set a p-value threshold of 0.001. Next, to remove read errors and negligible peaks, we rejected peaks belonging to gaussians with a width smaller than 16 reads and a maximum value smaller than the estimated minimum detectable peak prominence. Finally, to avoid interactions due to close contact, we rejected peaks closer than 35 Kb from the viewpoint. With this method, we obtain statistically significant interaction for every viewpoint and cell line in an automated fashion.
To obtain the mutual interactions in a given cell line, we filtered for mutual interactions that started at a viewpoint v1 and landed in a target within 20 Kb of a second viewpoint v2. Finally, we normalized the interaction strength to the maximum interaction strength in the cell line. Other (non-mutual) interactions in the highest quartile (more than 75% of the strongest interaction within the same cell line) are also shown in the plot.
All software used for the analysis of the 4 C data was implemented in Python and developed into a graphical user interface (GUI). The software package and the 4 C data files used to make the statistical analysis, are publicly available at https://github.com/Nikoula86/AFourC.githttps://github.com/Nikoula86/AFourC.git.
Statistical methods
The ratio of the Firefly and Renilla luciferase measurement was calculated (Relative luciferase units). All luciferase data were normalized using the empty vector activity (Pgl3-promoter, Pgl3-basic), and expressed as mean values ± SD. In the luciferase assay the differences between conditions were evaluated using the Welch Two Sample t-test, as homogeneity of variances was rejected using Bartlett test. The p-value was adjusted using Bonferroni’s correction. Statistical significance was set at p-value < 0.05. The statistical evaluation was carried out with Rstudio Version 3.6.1. The representations were performed using the Graphpad (Graphpad Prism 8).
Bioinformatic analysis
SRA data selection
In order to obtain as many reads as possible mapping to the second intron of WNT16, we selected 13 Sequence Read Archive (SRA) files from control RNA-seq experiments involving human osteoblast cells that had at least a blast hit against the intronic sequence. After mapping them, we restricted our focus to just the human foetal osteoblast (hFOB) cells and selected all the available SRA files from hFOB control experiments. The selected experiment IDs are:
SRR7939699, SRR7939693, SRR7939687, SRR7141491, SRR7141490, SRR7141489, ERR2603574, ERR2603576, SRR5087318, SRR4450223, SRR4450224, SRR4450225, SRR4450226, SRR1186922, SRR1186921, SRR1186916, SRR1186915, SRR1186910, SRR1186909, SRR3631725, SRR3631724, SRR3631723, SRR3631722, SRR3631721, SRR3631720, SRR3631719, SRR3631718, SRR3631717, SRR3631716, SRR3631714.
Due to the mixture of single and paired-end reads, we ran two independent mappings for each class of reads. To assess the levels of expression of the interrogated genes in the other cell types used, we also selected three sets of paired-end reads, SRR11819122 and SRR11819123 for the MSCs and SRR8616101 for the Saos-2 and mapped them separately.
SRA mapping and data visualization
The reads from the SRA files were extracted with fastq-dump (v2.8.1) and mapped against the human assembly GRCh3837 using STAR (v2.7.0d)38 with the default options for both programs. Using SAMtools (v1.9)39 we extracted the reads mapping to WNT16 (chr7:121319704–121346940 GRCh38) and used IGV (v2.8.2)40 to visualize the resulting output. The WNT16 gene models and conservation data were obtained from the UCSC tracks41 GENCODE v32 and Cons_30_Mammals, respectively.
Hypothetical isoform reconstruction
The intronic region where most of the reads mapped (chr7:121,330,337 − 121,330,889 GRCh38) was both translated and analysed for splicing donor sequences using Human Splicing Finder42 in order to recreate the most viable exon. This putative alternative reconstructed isoform was then aligned using blastp against the nr database, with Myotis davidii Wnt16 (ELK35449.1) being the best hit obtained.
Tissue specificity assessment
To assess the specificity of the expression we used available mapped reads from human tissues deposited in the EMBL-EBI ArrayExpress database (experiment E-MTAB-6814)43. The organs selected were: brain, cerebellum, heart, kidney, liver, ovary and testis, and the developmental stages were (when available) adult (29 years post birth) and 18–19 weeks post conception.
Results and discussion
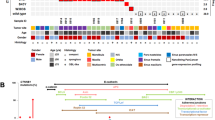
In the present work, we set out to perform a functional analysis of the WNT16 intron 2 region containing the variant rs142005327 (Fig. 1A, blue dot). To this aim, we focused on a 1 kb region in the proximal side of intron 2 (pale green background in Fig. 1) that shows epigenetic marks typical of regulatory elements. In particular, this region shows hypersensitivity to DNase I, enrichment of transcription factor binding sites and a CpG island (Fig. 1D). Moreover, analysis among vertebrates shows a conservation peak within intron 2, despite at lower levels compared to the exons (Fig. 1D). Interestingly, this region contains typical promoter histone modifications in osteoblasts, such as a low H3K4me1/H3K4me3 ratio, H3K4me2 or H2AFz, but also some contradictory signals on transcription activation such as a weak H3K27me3 or the lack of H3K27ac (Fig. 1E). Accordingly, this region is recognized by GeneHancer as part of a larger promoter element containing the WNT16b canonical promoter. Similarly, it is classified as several enhancer regions, distinct from the WNT16b promoter, by ENCODE Registry of candidate cis-Regulatory Elements (cCRES; Fig. 1F). To unambiguously determine the type of regulatory element of this region, we used a luciferase reporter assay and we tested seven fragments (colored bars in Fig. 1B and C) for promoter or enhancer activities in Saos2 cells. Two of the fragments showed both promoter and enhancer activities (Fragments B and C; Fig. 1B and C), one showed enhancer activity (Fragment A; Fig. 1B and C), and three only showed promoter activity (Fragments Full, 1 and 2; Fig. 1B and C). Fragment C showed the highest luciferase signal for both tests, being considerably higher in promoter activity (7.2-fold shares to empty vector in promoter activity, vs. 3-fold shares to empty vector in enhancer activity; Fig. 1B and C in purple). Fragment B also displayed luciferase activity as promoter (4.9-fold compared to the empty vector) and as enhancer (2-fold compared to the empty vector), again with higher promoter activity (Fig. 1B in dark blue). The remaining fragments showing promoter activity contain fragment B and/or C, indicating that these two fragments are the minimum with activity. This region contains the SNP rs73440215, associated with BMD8,44, and in high LD with the GWAS sentinel in7.
On the other hand, A, the smallest fragment containing rs142005327, failed to show promoter activity on its own and it only showed slight enhancer activity (1.5-fold compared to the empty vector; Fig. 1B and C). The COL1A1-promoter (SP construct in30) and the SV40 enhancer, used as a positive control in promoter and enhancer activities, showed 23.7-fold and 23.5-fold compared to the empty vector, respectively (data not shown). Consistent with our luciferase results, transcription factor ChIP-seq data from ENCODE (Fig. 1C) and TRANSFAC data show that many transcription factors are predicted to bind to fragments B and C (Supplementary Fig. S2).
Characterization of a regulatory element in WNT16 intron 2. (A) WNT16 isoforms from UCSC annotations of RefSeq RNAs; light blue dots indicate the position of rs142005327 (B) Luciferase promoter assay of different WNT16 intron 2 fragments. Normalized relative luciferase units (RLU; Ratio of Photinus pyrali and Renilla reniformis luciferase activities) of the different fragments cloned into the Pgl3-basic vector: Full, Inv 1, 2, A, B, C (see details in methods). Values are normalized to the activity of the empty vector, arbitrarily set at 1. Error bars indicate s.d. *p-value < 0.05; ****p-value < 0.0001. (C) Luciferase enhancer assay of different WNT16 intron 2 fragments. Normalized relative luciferase units (RLU; Ratio of Photinus pyrali and Renilla reniformis luciferase activities) of the different fragments cloned into the Pgl3-promoter vector: Full, Inv 1, 2, A, B, C (see details in methods). Values are normalized to the activity of the empty vector, arbitrarily set at 1. Error bars indicate s.d. *p-value < 0.05; ****p-value < 0.0001. (D) DNase: Hypersensitivity to DNAseI (ENCODE project); TF: Transcription Factor ChIP-seq Clusters (ENCODE project); CpG islands from UCSC genome browser; Cons 100 Verts: Vertebrate Multiz Alignment & Conservation (100 Species). (E) Post-translational modifications of different histones in osteoblasts (ENCODE project). (F) GeneHancer database of human regulatory elements included in GeneCards and ENCODE Registry of candidate cis-Regulatory Elements (cCRES). (G) Expresion levels of hFOB RNA-seq in SRA.
Knowing that the B-C region is a putative promoter in Saos 2 cells, we next decided to investigate whether the region immediately downstream is expressed in different cell types or tissues. To this aim, we used publicly available RNA-seq data from different tissues (Sequence Read Archive; SRA). With the help of this large dataset, we found that the few reads mapping to the WNT16 intron 2 are restricted to hFOB cell types exclusively (Fig. 1G and Supplementary Fig. 3A). None of these reads is connected with the exon 3, which is consistent with the hypothesis that they come from an enhancer/promoter RNA or other non-coding RNAs. Nevertheless, when translating the expressed region into protein and aligning it to all protein sequences with BLAST, we found a unique match with a Wnt16 protein in microbat (Myotis davidii). In contrast with the previous hypothesis, this result is consistent with the existence of an isoform of WNT16 that uses the B-C region of intron 2 as promoter. After further analysis, we unexpectedly but excitingly found that the bat protein is formed by a first exon strikingly similar to the first exon of the putative isoform (Supplementary Fig. 3B). The microbat protein is completed by 2 more exons that correspond to exons 3 and 4 of the human WNT16. These results confirm that, at least in one other species, a protein containing the amino acids coded by this previously undescribed human exon exists. In the future, experiments using different cell lines and developmental stages, will be crucial to elucidate whether such an isoform exists and, if appropriate, its role in humans.
To determine which regions might regulate the activity of this promoter, we performed a 4 C-seq assay on three relevant osteoblast-related cell types (Human fetal osteoblasts –hFOB-, an osteosarcoma cell line –Saos2-; and mesenchymal Stem Cells –MSC-) using the WNT16 intron 2 region (WNT16-In2; chr7:120970581–120971655, GRCh37/hg19) as a viewpoint (Fig. 2A, B and C and Supplementary Fig. 4). To confirm the interactions, we performed four additional 4 C-seq assays. Three of them were chosen using the regions with the strongest interaction with WNT16-intron 2 as viewpoints, namely the CPED1 main promoter (CPED1-MP), the region between CPED1 exons 11 and 12 (CPED1-R-11-12) and the region containing CPED1 exon 17 (CPED1-R-17). Additionally, due to the important function of FAM3C in bone45, we chose a fourth viewpoint at the FAM3C main promoter (FAM3C-MP). The reciprocal 4 C-seq assays confirmed the interactions with CPED1-R-11-12 and CPED1-R-17 in the three cell types used and with CPED1-MP for Saos2 and hFOB cells (black/grey arches in Fig. 2A, see Methods). Among them, WNT16 and CPED1-R-11-12 showed the strongest interaction (Fig. 2A, solid black arch). Interestingly, the CPED1-R-11-12 region was previously identified as an active promoter in the pre-osteoblast MC3T3-E1 cell line46. Unfortunately, we did not have sufficient MSC to test the CPED1-MP and FAM3C viewpoints, therefore we could not test these interactions for the MSC cell type. According to the GeneHancer database, both CPED1-R-11-12 and CPED1-R-17 are described as enhancers, while CPED1-MP and WNT16-In2 are described as promoters (Fig. 2C). In addition, for every cell line and viewpoint, we have identified several non-reciprocal interactions (blue arrows in Fig. 2A). Among them, we found interactions in the three cell types between the WNT16-In2 viewpoint and a genomic region within CPED1 (exon 22–23; CPED1-R-22-23) that is described as a regulatory element (vertical dashed line in Fig. 2A, B and C). Moreover, our data show that FAM3C does not strongly interact with either CPED1 or WNT16 (Fig. 2A and B). Our results are in contrast with those of the GeneHancer database, which only includes either interactions between the main promoter of CPED1 and some of the regulatory elements included in it, or between WNT16 and a region located between the two genes (Supplementary Fig. 5). The interaction between CPED1 and WNT16 that we report here is novel and may be bone cell type-specific. Despite these results, the resolution of the technique does not allow to discern whether the physical interaction with several regulatory elements of CPED1 occurs with the alternative promoter described here or through the canonical promoter of WNT16b. In the future, studies at higher resolution or in vivo will be necessary to precisely determine the interacting region.
(A) Summary of results obtained in 4 C-seq experiments using 5 viewpoints from the CPED1-WNT16-FAM3C region in human foetal osteoblasts (hFOB), Saos2 cells and mesenchymal stem cells (MSC). Observed contacts are depicted by arched lines. Grey scale arches represent reciprocal interactions and are color-coded according to the strength of the interaction (grey scale bar). Arrow-headed blue arches represent the non-reciprocal interactions with strength higher than 75% of the strongest interaction in the same cell line. Solid vertical lines represent the position of the viewpoints and the dashed line corresponds to the exon 22–23 CPED1 region. (B) Displayed Ensembl/Havana genes in this region are ING3_002 (ENST00000339121.5), ING3_001 (ENST00000315870.5), FAM3C (ENST00000359943.3), WNT16_2 (ENST00000361301.2), WNT16_1 (ENST00000222462.2), CPED1_1 (ENST00000310396.5) and CPED1_7 (ENST00000450913.2) from GRCh37/hg19. (C) GeneHancer track from the UCSC genome browser, enhancers are shown in grey and promoters in red.
Taken together, our results demonstrate that the alternative promoter region of WNT16 is physically interacting with CPED1. It is still an open question whether the WNT16 region is regulating CPED1 expression, or vice versa. However, previous studies have demonstrated that Wnt16-KO, including the alternative promoter region, does not affect CPED1 expression in mouse models24. Therefore, the interaction observed here is likely due to a regulatory role of CPED1 on WNT16 expression. To unambiguously demonstrate this regulation, it will be necessary to establish a CPED1-KO model and quantify the expression of WNT16 in bone cells.
The variants in the alternative promoter region of WNT16 may be partially responsible for the association of this locus with BMD in GWAS. However, we cannot exclude the involvement of other genes in this locus in this association47. For instance, Chesi et al.. (2019)6 performed a genome-wide promoter-focused Capture C and found that another gene in the region, ING3, plays an important role in the MSC cell fate decision to osteoblast lineage, which could synergize with WNT16 functions. Interestingly, although these authors did not find looping between the SNP rs142005327 and putative enhancers in regions of open chromatin, the interacting pattern that they described for this WNT16 promoter region is very much comparable with the one we present here, which essentially includes enhancers within CPED1. Chesi et al.6 used only primary human MSC-derived osteoblasts, while we have used cell types at different stages, from MSCs to Saos2. In the future it will be important to further dissect this regulatory region, for instance by studying different in vivo models.
Conclusion
In the present work we have performed a functional study of one region within WNT16, which harbours major consistent BMD/fracture GWAS signals. We have demonstrated the promoter activity of part of intron 2, and its physical interaction in osteoblast-related cells (the human fetal osteoblast hFOB 1.19 cell line, Saos 2 osteosarcoma cell line and mesenchymal Stem Cells –MSC-) with different putative regulatory regions within CPED1, including the main CPED1 promoter. Furthermore, using previously published RNA-seq data from hFOB cells, we observed low levels of expression of a region at 3’ of the intron 2 WNT16 promoter. Our results suggest a novel regulatory mechanism of WNT16 in bone cells that is mediated by physical interaction with various CPED1 regions although the functional implications of this interaction should be analyzed in future studies. Considering the important effect of WNT16 in BMD determination, the characterization and evaluation of regulatory elements could be translated into therapeutic strategies to treat bone related disorders, such as osteoporosis.
Data availability
All data and material will be available upon request. The software package and the 4 C data files used to make the statistical analysis are publicly available at https://github.com/Nikoula86/AFourC.git.The datasets generated and/or analysed during the current study are available in the ENA (European Nucleotide Archive), with accession number PRJEB84184.
References
Morris, J. A. et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 51 (2), 258–266 (2018).
Kemp, J. P. et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 49 (10), 1468–1475 (2017).
Kim, S. K. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS One. 13 (7), e0200785 (2018).
Hendrickx, G. et al. Variation in the Kozak sequence of WNT16 results in an increased translation and is associated with osteoporosis-related parameters. Bone 59, 57–65 (2014).
Roca-Ayats, N. et al. Functional characterization of the C7ORF76 genomic region, a prominent GWAS signal for osteoporosis in 7q21.3. Bone 123, 39–47 (2019).
Chesi, A. et al. Genome-scale capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat. Commun. 10 (1), 1260 (2019).
Estrada, K. et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44 (5), 491–501 (2012).
Zheng, H. F. et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. 8 (7), e1002745 (2012).
Medina-Gomez, C. et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 8(7), e1002718 (2012). (2012).
García-Ibarbia, C. et al. Missense polymorphisms of the WNT16 gene are associated with bone mass, hip geometry, and fractures. Osteoporos. Int. 24 (9), 2449–2454 (2013).
Koller, D. L. et al. Meta-analysis of genome-wide studies identifies WNT16 and ESR1 SNPs associated with bone mineral density in premenopausal women. J. Bone Min. Res. 28 (3), 547–558 (2013).
Zhang, L. et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum. Mol. Genet. 23 (7), 1923–1933 (2014).
Moayyeri, A. et al. Genetic determinants of heel bone properties: genome-wide association meta-analysis and replication in the GEFOS/GENOMOS consortium. Hum. Mol. Genet. 23 (11), 3054–3068 (2014).
Chesi, A. et al. A trans-ethnic genome-wide association study identifies gender-specific loci influencing pediatric aBMD and BMC at the distal radius. Hum. Mol. Genet. 24 (17), 5053–5059 (2015).
Zheng, H. F. et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 526 (7571), 112–117 (2015).
Mullin, B. H. et al. Genome-wide association study meta-analysis for quantitative ultrasound parameters of bone identifies five novel loci for broadband ultrasound Attenuation. Hum. Mol. Genet. 26 (14), 2791–2802 (2017).
Medina-Gomez, C. et al. Life-course genome-wide association study meta-analysis of total body BMD and assessment of age-specific effects. Am. J. Hum. Genet. 102 (1), 88–102 (2018).
Pei, Y. F. et al. Joint study of two genome-wide association meta-analyses identified 20p12.1 and 20q13.33 for bone mineral density. Bone 110, 378–385 (2018).
Kichaev, G. et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104 (1), 65–75 (2019).
Trajanoska, K. et al. Assessment of the genetic and clinical determinants of fracture risk: genome wide association and Mendelian randomisation study. BMJ 362, k3225 (2018).
Wang, H. et al. Genotype-by-environment interactions inferred from genetic effects on phenotypic variability in the UK biobank. Sci. Adv. 5 (8), eaaw3538 (2019).
Greenbaum, J. et al. A multiethnic whole genome sequencing study to identify novel loci for bone mineral density. Hum. Mol. Genet. 31 (7), 1067–1081 (2022).
Fear, M. W., Kelsell, D. P., Spurr, N. K. & Barnes, M. R. Wnt-16a, a novel Wnt-16 isoform, which shows differential expression in adult human tissues. Biochem. Biophys. Res. Commun. 278 (3), 814–820 (2000).
Movérare-Skrtic, S. et al. Osteoblast-derived WNT16 represses osteoclastogenesis and prevents cortical bone fragility fractures. Nat. Med. 20 (11), 1279–1288 (2014).
Movérare-Skrtic, S. et al. The bone-sparing effects of Estrogen and WNT16 are independent of each other. Proc. Natl. Acad. Sci. U S A. 112 (48), 14972–14977 (2015).
Alam, I. et al. Osteoblast-specific overexpression of human WNT16 increases both cortical and trabecular bone mass and structure in mice. Endocrinology 157 (2), 722–736 (2016).
Alam, I. et al. Bone mass and strength are significantly improved in mice overexpressing human WNT16 in osteocytes. Calcif Tissue Int. 100 (4), 361–373 (2017).
Martínez-Gil, N. et al. Common and rare variants of WNT16, DKK1 and SOST and their relationship with bone mineral density. Sci. Rep. 8 (1), 10951 (2018).
Casado-Díaz, A. et al. Cryopreserved human bone marrow mononuclear cells as a source of mesenchymal stromal cells: application in osteoporosis research. Cytotherapy 10 (5), 460–468 (2008).
García-Giralt, N. et al. In vitro functional assay of alleles and haplotypes of two COL1A1-promoter SNPs. Bone 36 (5), 902–908 (2005).
Fernández-Miñán, A., Bessa, J., Tena, J. J. & Gómez-Skarmeta, J. L. Assay for transposase-accessible chromatin and circularized chromosome conformation capture, two methods to explore the regulatory landscapes of genes in zebrafish. Methods Cell. Biol. 135, 413–430 (2016).
an de Werken, H. J. et al. 4 C technology: protocols and data analysis. Methods Enzymol. 513, 89–112 (2012).
Noordermeer, D. et al. The dynamic architecture of hox gene clusters. Science 334 (6053), 222–225 (2011).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507 (7492), 371–375 (2014).
Klein, F. A. et al. FourCSeq: analysis of 4 C sequencing data. Bioinformatics 31 (19), 3085–3091 (2015).
Irastorza-Azcarate, I. et al. 4Cin: A computational pipeline for 3D genome modeling and virtual Hi-C analyses from 4 C data. PLoS Comput. Biol. 14 (3), e1006030 (2018).
Schneider, V. A. et al. Evaluation of GRCh38 and de Novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 27 (5), 849–864 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21 (2013).
Li, H. et al. The sequence alignment/map format and samtools. Bioinformatics 25 (16), 2078–2079 (2009).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14 (2), 178–192 (2013).
Raney, B. J. et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC genome browser. Bioinformatics 30 (7), 1003–1005 (2014).
Desmet, F. O. et al. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37 (9), e67 (2009).
Cardoso-Moreira, M. et al. Gene expression across mammalian organ development. Nature 571 (7766), 505–509 (2019).
Surakka, I. et al. MEPE loss-of-function variant associates with decreased bone mineral density and increased fracture risk. Nat. Commun. 11 (1), 4093 (2020).
Määttä, J. A. et al. Fam3c modulates osteogenic cell differentiation and affects bone volume and cortical bone mineral density. Bonekey Rep. 5, 787 (2016).
Maynard, R. D., Godfrey, D. A., Medina-Gomez, C. & Ackert-Bicknell, C. L. Characterization of expression and alternative splicing of the gene cadherin-like and PC esterase domain containing 1 (Cped1). Gene 674, 127–133 (2018).
Gómez, A. E. et al. Multiple mechanisms explain genetic effects at the CPED1-WNT16 bone mineral density locus. Curr. Osteoporos. Rep. (2023).
Acknowledgements
We thank O. Monclús and M. Cozar for relevant technical assistance.
Funding
Funds for the study include grants PID2019-107188RB-C21, PID2020-117820GB-I00, PID2022-141461OB-I00, PID2023-146794NB-I00 and BFU2017-86152-P (Spanish MINECO), 2021SGR: 01093 (AGAUR, Catalan Government) and CIBERER (U720). NMG was a recipient of a FI predoctoral fellowship from Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR); NRA was a recipient of a FPU and CHU of a FPI predoctoral fellowship from the Spanish Ministerio de Educación Cultura y Deporte.
Author information
Authors and Affiliations
Contributions
NMG, DO, XN, DG and SB were involved in the Conceptualization. NMG was involved in the Data curation, Formal analysis. NMG, NRA, NU and NGG were involved in Investigation and Methodology. The bioinformatics study of the RNA-seq in hFOB cells was performed by CHU and JGF. The AFourC software was performed by NG. DG and SB were involved in the Funding acquisition. The first draft of the manuscript was written by NMG, DG, SB and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Martínez-Gil, N., Herrera-Ubeda, C., Gritti, N. et al. Regulation of WNT16 in bone may involve upstream enhancers within CPED1. Sci Rep 15, 9607 (2025). https://doi.org/10.1038/s41598-025-93259-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93259-z