Abstract
Dendrobium officinale is an edible medicinal herb. Its flowers contain various nutrients and active compounds, the abundance of which differs significantly across developmental stages. However, the molecular mechanisms underlying these variations remain poorly understood. This study employed combined transcriptomic and metabolomic analyses to investigate changes in gene expression and metabolite concentrations in Dendrobium flowers and buds. The analysis identified 2767 differentially expressed genes between the bud (group B) and flower (group F) stages, with 902 up-regulated and 1865 down-regulated in group B relative to group F. The number of differentially abundant secondary metabolites was 221, including 113 up-regulated and 108 down-regulated metabolites. The differential metabolites primarily consisted of lipids and lipid-like molecules, organic heterocyclic compounds, organic acids, and their derivatives, phenylacetones and polyketides, organic oxides, benzene ring-type compounds, hydrocarbons, and alkaloids with their derivatives. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the differentially expressed genes and metabolites revealed enrichment in pathways related to phytohormone signaling, phenylpropanoid biosynthesis, and zeaxanthin biosynthesis. Verification of 15 genes through quantitative real-time PCR (qRT-PCR) confirmed the accuracy of the transcriptomic analysis. Taken together, these findings contribute to an improved understanding of the nutritional and pharmacological characteristics of Dendrobium officinale flowers at different developmental stages.
Similar content being viewed by others
Introduction
Dendrobium officinale Kimura et Migo is a perennial herbaceous plant of the Orchidaceae family. It is mainly distributed in southwestern Anhui, eastern Zhejiang, western Fujian, northwestern Guangxi, Sichuan, and southeastern Yunnan in China. The plant thrives in semi-shaded rocky mountainous regions at elevations of up to 1,600 m above sea level. The flowering period spans from March to June, with racemes bearing 2–3 flowers emerging from the upper section of the old stem1. In the Chinese Pharmacopoeia, the strip of Dendrobium officinale represents the dried stem of the plant2. Dendrobium officinale is traditionally recognized for its effects in promoting saliva secretion, supporting gastric health, clearing heat, nourishing yin, and nourishing the skin3. In November 2023, the National Health Commission and the State Administration for Market Regulation included Dendrobium officinale in the official list of substances classified as both food and traditional Chinese medicinal materials4. The plant is rich in polysaccharides, flavonoids, polyphenols, alkaloids, various volatile components, amino acids, and essential macro- and micro-elements5,6,7,8,9. While the chemical compositions of stems, leaves, and flowers of Dendrobium officinale are similar, the concentrations of individual components vary. All parts of the plant show significant antioxidant activity, which correlates positively with the contents of total flavonoids, total polyphenols, and total alkaloids10. Ethanol extracts of Dendrobium officinale flowers demonstrate the highest levels of total phenolic and flavonoid contents and the strongest antioxidant activity11. Furthermore, studies have shown that Dendrobium officinale flowers exert a stable antihypertensive effect in spontaneously hypertensive rats12.
Dendrobium officinale has gained significant research interest due to its medicinal and edible properties13,14,15,16,17. Significant differences in the active components of Dendrobium officinale have been observed across different flowering periods and flower parts, particularly in the concentrations of polysaccharides, total flavonoids, and total phenols18. Studies have shown that the levels of polysaccharides, total flavonoids, and water-soluble extracts are generally higher in flowers harvested during the peak flowering period than those from hybrid and bulbous plants at the bud stage or micro-flowering stage19. However, limited knowledge exists regarding the growth, development, and metabolic regulation of Dendrobium officinale flowers, which hinders their further development and utilization. Transcriptomics and metabolomics are important techniques used to analyze differentially expressed genes (DEGs) and differentially abundant metabolites (DAMs), and both have been widely applied in analyses of animal, plant, and microbial tissues20.This study employed a combination of metabolomic and transcriptomic approaches to investigate differences in gene expression and metabolite content in Dendrobium officinale flowers at various developmental stages. The findings elucidate the regulatory mechanisms of active ingredient accumulation at different stages and provide a theoretical basis for resource utilization and the development of medicinal ingredients.
Materials and methods
Collection of samples
Samples of Dendrobium officinale flowers were collected from the Huqingyutang Dendrobium officinale planting base in Tonglu County, Zhejiang Province. Fresh flowers were harvested at the bud and peak flowering stages, specifically on the 10th day of the bud stage and the 5th day of the peak flowering stage (Fig. 1). The fresh flowers used for RNA and metabolites extraction were frozened in liquid nitrogen and stored at − 80 °C immediately. Samples used for bioactive ingredients measuments were dried in an oven at 50 °C to obtain dried materials. The dried samples were then crushed and passed through a 60-mesh sieve to prepare test samples for each treatment. Bud-stage samples were designated Group B, while peak flowering-stage samples were designated Group F for subsequent analyses.
(A) Buds (B) and flowers (F) of Dendrobium officinale. (B) The content of total flavonoids, total phenols, and polysaccharides in Dendrobium officinale flower at different developmental stages. (C) The content of total amino acid of Dendrobium officinale flower during development periods. (D) The content of different types of amino acids of Dendrobium officinale flower during development periods.
Determination of bioactive ingredients in Dendrobium officinale
The total flavonoid content was determined as described by Li et al.21. The total phenol content was determinedaccording to the method described by Liao et al.22. the polysaccharide content was measured using the method described by Chen et al.23 Determination of Free Amino Acids was performed accroding to GB/T 30,987 − 2020.
Extraction and analysis of metabolites
For metabolite extraction, 50 mg of dried Dendrobium officinale flower buds and flowers were weighed, and 1,000 µL of extraction solution containing an internal standard (1000:2; methanol: acetonitrile: water, volume ratio 2:2:1; internal standard concentration 2 mg/L) was added. The samples were vortexed for 30 s, ground with ceramic beads using a grinder at 45 Hz for 10 min, and sonicated for 10 min in an ice-water bath. Following incubation at -20 °C for 1 h, the samples were centrifuged at 12,000 rpm for 15 min at 4 °C. An aliquot of 500 µL of the supernatant was transferred to an EP tube and dried in a vacuum concentrator. The dried metabolites were reconstituted with 160 µL of extraction solution (acetonitrile: water, volume ratio 1:1), vortexed for 30 s, sonicated for 10 min in an ice-water bath, and centrifuged at 12,000 rpm for 15 min at 4 °C. A volume of 120 µL of the supernatant was transferred to a 2 mL vial, and 10 µL aliquots from each sample were combined to form a quality control (QC) sample. Metabolomic analysis was performed using a Waters Acquity I-Class PLUS ultra-high-performance liquid chromatography (UPLC) system coupled with a Waters Xevo G2-XS QTof high-resolution mass spectrometer (Waters Corporation, Milford, MA, USA). The chromatographic column used was the Acquity UPLC HSS T3 column (1.8 μm, 2.1 × 100 mm, Waters). The mobile phases for both positive and negative ion modes consisted of 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in acetonitrile (mobile phase B).
RNA extraction and transcriptomic sequencing analysis
Total RNA was extracted from the samples using the Tiangen DP441 RNAprep Pure Plant Kit, designed for polysaccharide- and polyphenol-rich plant tissues. High-quality RNA was selected for transcriptomic sequencing. Library quality was evaluated, and sequencing was performed in PE150 mode using the Illumina NovaSeq 6000 platform (San Diego, CA, USA). Bioinformatic analyses of sequencing data were conducted on the Baimaike Cloud platform (BMKCloud, www.biocloud.net). Offline data were filtered to obtain clean reads, which were then aligned to the reference genome to generate mapped data. Library quality was assessed, and structural, differential expression, functional annotation, and enrichment analyses were performed. The effective concentration of the cDNA library was confirmed to be greater than 2 nmol/L using qRT-PCR.
Joint analysis
DEGs and DAMs were jointly analyzed using the KEGG database to identify common pathways. Pearson correlation coefficients (PCCs) were calculated for DEGs and DAMs, with thresholds set at PCC ≥ 0.6 and P < 0.05. Significant correlations were indicated by PCC ≥ 0.6. Bidirectional OPLS-DA, PCA, and KEGG pathway comparative analyses were performed.
Verification of RNA sequencing (RNA-seq) data by quantitative real-time PCR (qRT-PCR)
Dried Dendrobium officinale flower buds and flower samples were used for RNA extraction with the RNeasy Plant Mini Kit (Qiagen, Germany) following the manufacturer’s protocol. cDNA was synthesized using the SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions. Real-time PCR was performed with PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, Waltham, MA, USA) under the following conditions: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The relative expression of target genes was normalized to GAPDH and calculated using the 2−∆∆Ct method. Results were expressed as fold changes relative to the control group. Primer sequences are provided in Table S1.
Statistical analysis
Principal component analysis (PCA) and Spearman correlation analysis were conducted to verify the repeatability of within-group and quantitative control samples. Compounds were annotated using the KEGG, HMDB, and LipidMaps databases. Differences were calculated based on grouping information, and t-tests were used to determine statistical significance (P < 0.05). Orthogonal partial least squares discriminant analysis (OPLS-DA) modeling was performed using the R package ropls, with 200 permutation tests to validate model reliability. The variable importance in projection (VIP) values was computed via multiple cross-validation. Differential metabolites were identified based on OPLS-DA model differentials, P-values, and VIP values using the criteria FC > 1, P < 0.05, and VIP > 1. DAMs enriched in KEGG pathway analysis were identified using the hypergeometric distribution test.
Results
Phenotypic and quality analyses of Dendrobium officinale flowers at different developmental stages
The flowering period of Dendrobium officinale is from March to June. The buds are light green and 1–2 cm long during the bud stage. The flowers during the flowering period are light yellow, 2–4 cm long, and curved in a folded shape with a diameter of 1.5–2.0 cm (Fig. 1A). By comparing and analyzing the total flavonoids, polysaccharides, and total phenols in the buds and flowers of Dendrobium officinale, it was found that there were significant differences in the contents of these three substances between the buds and flower samples. From Fig. 1B, it can be seen that the total flavonoids in Group B of Dendrobium officinale are slightly higher than those in Group F. The polysaccharide contents showed a trend opposite to that of the total flavonoids, with slightly lower concentrations in the flower buds compared to the flowers, while the trends in the total phenolic and polysaccharide contents were consistent, with group F significantly higher than group B.
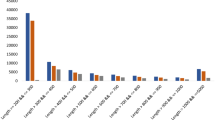
Measurement of the total amino acid contents and 24 types of amino acids, including phosphorus serine (P-Ser), taurine (Tau), phosphorus ethanolamine (PEA), aspartic acid (Asp), and threonine (Thr) in the flower buds and flower samples of Dendrobium officinale showed that there were no significant differences between the flower buds and flowers in terms of total amino acid content (Fig. 3A). Apart from non-significant differences in the contents of aspartic acid (Asp) and proline (Pro), the concentrations of all amino acids differed significantly (Fig. 1C and D), with methionine (Met) and histidine (His) showing the greatest differences. Amino acids with significantly higher contents in Group B relative to Group F included P-Ser, Tau, PEA, Gly, Ala, Met, Cysthi, b-Ala, g-ABA, and His, while those that were higher in Group F relative to Group B included threonine (Thr), serine (Ser), glutamic acid (Glu), valine (Val), cysteine (Cyr), isoleucine (Ile), leucine (Leu), tyrosine (Tyr), phenylalanine (Phe), ethanolamine (EOHNH2), lysine (Lys), and arginine (Arg).
Figure 1 Phenotype and quality of flower buds and flowers in Dendrobium officinale.
Transcriptomic analysis of Dendrobium officinale flowers at different developmental stages
Analysis of differentially expressed genes
To identify DEGs in Dendrobium officinale flowers at various developmental stages, RNA-seq analysis was performed on bud and flower samples. A total of 36.60 GB of clean data was obtained, with 5.89 GB of clean data per sample and Q30 base percentages exceeding 92.96%. Alignment of clean reads to the reference genome revealed sequence identities ranging from 88.67 to 89.68%. The results of the comparisons, prediction of alternative splicing, and analysis of gene structures identified 3545 genes, of which 1824 were functionally annotated.
DEGs were screened using the criteria Fold Change (FC) ≥ 2 and FDR < 0.01, as shown in Fig. 2A. A total of 2,767 DEGs were identified between Groups B and F, with 902 up-regulated and 1,865 down-regulated genes in Group B compared to Group F.
(A) Histogram of differentially expressed genes in Dendrobium officinale flower at different developmental stages. (B) GO classification of differentially expressed genes in Dendrobium officinale flower at different developmental stages. (C) Functional annotation and pathway classification of differentially expressed genes.
Gene ontology (GO) and KEGG analyses of differentially expressed genes
GO and KEGG enrichment analyses were conducted to explore the functions and pathways associated with DEGs. Among 18,806 annotated genes that were functionally annotated in the two groups of samples of Dendrobium officinale flower buds and flowers, 2,094 DEGs were categorized into biological process, cellular component, and molecular function GO domains. As shown in Figs. 2B and 816 DEGs were enriched in biological processes, 2,307 in molecular functions, and 1,632 in cellular components.
KEGG enrichment analysis identified 121 pathways associated with DEGs between developmental stages of Dendrobium officinale flowers (Fig. 2C). These pathways were grouped into five categories: cellular processes (3 pathways), environmental information processing (4 pathways), genetic information processing (9 pathways), metabolism (32 pathways), and organismal systems (2 pathways). A total of 421 DEGs were enriched in 10 key pathways, including 93 genes in plant-pathogen interactions, 83 genes in plant hormone signal transduction and MAPK signaling, 51 genes in plant biosynthesis, 47 in phenylpropanoid biosynthesis, and 42 in starch and sucrose metabolism. Other enriched pathways included endocytosis (22 genes), pentose and glucuronic acid interconversion (22 genes), RNA degradation (21 genes), RNA transport (20 genes), and ubiquitin-mediated proteolysis (20 genes).
The functional annotation and KEGG enrichment analysis of DEGs are shown in Fig. 2D. The top 20 pathways with the most significant enrichment are displayed, including biosynthesis of flavonoids and flavonols, brassinoles, flavonoids, keratin, cork and wax, and stilbenes, diarylheptanes and gingerols, as well as plant hormone signal transduction, monoterpene biosynthesis, biosynthesis of zeaxanthin, biosynthesis of phenylacetone, and interactions between plants and pathogens. The total flavonoid content of Dendrobium officinale flowers varied significantly at different developmental stages, which was related to the biosynthesis of flavonoids and flavonols, as well as the biosynthesis of flavonoids.
Figure 2 Analysis of differentially expressed genes between Dendrobium officinale flower buds and flowers.
Metabolomic analysis of Dendrobium officinale flowers at different developmental stages
Metabolomic analysis was conducted on bud and flower samples, followed by PCA and OPLS-DA, to compare the metabolic differences between Dendrobium officinale flowers at various developmental stages. As shown in Fig. 3A, the two sample groups are distributed in different regions, with PC1 and PC2 accounting for 26.91% and 23.95% of the variance, respectively. The cumulative variance contribution rate of these components is 50.86%, indicating a significant separation of metabolites between the flower buds and flowers at different developmental stages. The evaluation parameters of the OPLS-DA model were R²X = 0.435, R²Y = 0.999, and Q²Y = 0.592, all exceeding the threshold of 0.4. These values confirm the acceptability and reliability of the model. The results of the multivariate analysis demonstrated significant differences in metabolites between flower buds and flowers at different stages of development. Moreover, the differential metabolites identified based on VIP analysis showed strong representativeness.
(A) OPSL-DA of Metabolites of Dendrobium officinale flowers at different developmental. (B) Cluster heat map of differently expressed metabolites of Dendrobium officinale flowers at different developmental stages. (C) Cluster heat map of differently expressed metabolites of Dendrobium officinale flowers at different developmental stages. (D) Up-regulation and down-regulation fold change results of differential metabolites of Dendrobium officinale flowers at different developmental stages.
Figure 3 Analysis of differentially abundant metabolites in Dendrobium officinale flower buds and flowers.
A total of 3,860 metabolites were identified in Dendrobium officinale bud and flower samples, as shown in Fig. 3B. Based on OPLS-DA results, DAMs were identified using the criteria VIP ≥ 1, FC ≥ 2, or FC ≤ 0.5. This analysis revealed 221 DAMs, representing 0.57% of the total measured metabolites. Of these, 108 metabolites were up-regulated, and 113 were down-regulated in the flower group (Group F) compared to the bud group (Group B). The identified DAMs included 31 types of lipids and lipid molecules, such as terpenoid glycosides, diterpenes, fatty acids and conjugates, steroidal glycosides, fatty acids and conjugates, fatty alcohols, triterpenoids, bile acids, alcohols and derivatives, monoallyl glycerol, linear alcohol acids and their derivatives, steroid esters, glycerophosphoethanolamine, terpenoid glycosides, eicosanoids, hydroxysteroids, bile acids, alcohols and derivatives, sesquiterpenes, hydroxysteroids, and linear alcohol acids and their derivatives. Furthermore, 20 types of organic heterocyclic compounds were detected, including imidazoles, pyrrolo[2,3-d]pyrimidine, pyridines, hydroxyindoles, bilirubin, β-lactams, carbazoles, indoles, purine and purine derivatives, pyridine and its derivatives, pyridamine, and oxazoles. The analysis also identified 16 types of organic acids and their derivatives, including amino acids, peptides and analogs, organic sulfonic acids and their derivatives, medium chain ketone acids and their derivatives. Five types of nucleosides, nucleotides, and analogs were observed: purine ribonucleotides, purine 2’-deoxynucleotides, and pyrimidine 2’-deoxynucleotides. Furthermore, 14 types of phenylacetone and polyketide compounds were detected, including furan isoflavones, flavonoid glycosides, hydrolyzed tannins, isoflavone O-glycosides, linear diarylheptanes, ortho-methyl flavonoids, and hydroxycinnamic acid derivatives. Fourteen types of organic oxides were also observed, including carbohydrates and carbohydrate conjugates, alcohols and polyols, and carbonyl compounds. Seven types of benzene ring compounds were identified, such as benzoic acid and its derivatives, halogenated phenols, methoxyphenols, and benzophenones. Moreover, two types of hydrocarbons (cycloalkanes and branched unsaturated hydrocarbons) and two types of alkaloids and their derivatives were identified. Ten metabolites demonstrated significant up- or down-regulation based on their fold-change values (Fig. 3C).
KEGG enrichment analysis was performed to further understand the metabolic pathways associated with the DAMs. Of the total of 221 DAMs, 78 were found to be enriched in 48 KEGG pathways associated with metabolism. As shown in Figs. 3D and 20 significantly enriched metabolic pathways were identified based on the P-values, of which four were down-regulated, namely, linoleic acid metabolism (ko00591) and alpha-linolenic acid metabolism (ko00592), biosynthesis of benzoxazinoid compounds (ko00402), and degradation of valine, leucine, and isoleucine (ko00280). There were six up-regulated metabolic pathways, namely, biosynthesis of penicillin and cephalosporin (ko00331), taurine and low taurine metabolism (ko00430), pyruvate metabolism (ko00430), glycerol ester metabolism (ko00561), carbon fixation in photosynthetic organisms (ko00710), and caffeine metabolism (ko00030, pentose phosphate pathway caffeine metabolism). The three metabolic pathways with the most significant differences were vitamin B6 metabolism (ko00750), linoleic acid metabolism (ko00591), and folate biosynthesis (ko00790).
Figure 3 Analysis of differential metabolites in Dendrobium officinale flower buds and flowers.
Combined transcriptomic and metabolomics analysis
A comparison of pathways enriched with DEGs and DAMs revealed that 77 pathways were enriched with DEGs and 19 with DAMs, with 44 pathways shared between the two groups (Fig. 4A). The top 10 KEGG pathways with the highest number of co-enriched genes and metabolites, included the biosynthesis of ubiquinone and other terpenoid quinones, tyrosine metabolism, degradation of valine, leucine, and isoleucine, plant hormone signal transduction, tryptophan metabolism, cysteine and methionine metabolism, biosynthesis of carotenoids, pyruvate metabolism, biosynthesis of flavonoids and flavonols, and ascorbic acid and aldehyde metabolism (Fig. 4B).
(A) Wayne diagrams of differentially expressed genes and differential abundant metabolites. (B) Top 10 KEGG pathways of differentially expressed genes/differential metabolites. (C) Bubble diagram of KEGG enrichment pathway for differentially expressed genes/differential metabolites. (D) Kgml network diagram of differentially expressed genes/metabolites Note: ① Monoterpenoid biosynthesis; ② Cyanoamino acid metabolism; ③ Phenylpropanoid biosynthesis; ④ The biosynthesis of stilbenes, diarylheptanes, and gingerol; ⑤ The biosynthesis of flavonoids and flavonols; ⑥ Diterpenoid biosynthesis; ⑦ Cutin, suberine, and wax biosynthesis; ⑧ Zeatin biosynthesis; ⑨ Plant hormone signal transduction; ⑩ Flavonoid biosynthesis of flavonoids.
Analysis of the pathways enriched with both DEGs and DAMs between sample groups B and F identified plant hormone signal transduction pathways (ko04075) enriched with 83 DEGs and 1 DAM, including the down-regulated genes LOC110104313 and LOC110113419, the up-regulated gene LOC110103533, and up-regulated secondary metabolite dihydrozeatin (Fig. 4C). The LOC110092013 and LOC110108166 genes were up-regulated in gibberellin signaling, while the LOC110099634, LOC110096143, LOC110101984, and LOC110109011 genes were down-regulated. The monoterpenoid biosynthetic pathway (ko00902) was enriched with three DAMs, as well as the genes LOC110092385, LOC110099511, LOC110101309, LOC110102941, LOC110106061, LOC110111567, LOC110112888, LOC1145878344, and LOC114579673, all of which were up-regulated, while the genes LOC110096479 and LOC110114334 were down-regulated, with up-regulation of the secondary metabolite 7-Deoxyloganate and down-regulation of Loganate. The zeatin biosynthetic pathway (ko00908) was enriched with 16 DEGs and 2 DAMs, including the down-regulated genes LOC110113791, LOC110116586, LOC110105324, LOC110106158, LOC110107036, LOC110101928, and LOC110105063 and the down-regulated secondary metabolites adenosine-5’-monophosphate and up-regulated dihydrozeatin. The phenylpropanoid biosynthesis pathway (ko00940) was enriched with 47 DEGs and 1 DAM, with the up-regulated genes LOC110100008, LOC110098448, and LOC110103063 and down-regulated genes LOC110092440, LOC110109032, LOC110109033, LOC110110432, LOC110116261, and LOC110092517, and the up-regulated secondary metabolite 3,4-dihydroxystyrene. The carbon metabolism pathway (ko01200) was enriched with 20 DEGs and 2 DAMs, with the up-regulated genes LOC110114529, LOC110107472, LOC110096402, and LOC110114941 and down-regulated genes LOC110115773, LOC114578807, and LOC110115773, and the secondary metabolites glycerol and D-Erythrose-4-phosphate, both of which were up-regulated. Amino acid biosynthetic pathways (ko01230) were enriched with 19 DEGs and 3 DAMs, including the up-regulated genes LOC110114529, LOC110107472, LOC110108381, LOC110097663, LOC110105641, and LOC110104691 and down-regulated genes LOC110115773, LOC110110479, LOC114578503, LOC114578503, LOC110098602, LOC110095374, and LOC110110411. The enriched secondary metabolites were (S) -1-pyrroline-5-carboxylate, D-Erythrose-4-phosphate, and L-ornithine, of which L-Ornithine was up-regulated.
KEGG Markup Language (Kgml) stores detailed information about various components in the pathway, through which the relationship between DEGs and DAMs and their respective pathway networks can be plotted. A pathway may have multiple genes and metabolites, and a gene or metabolite may also belong to multiple pathways. The R package Graph can display the relationships between pathways, genes, and metabolites. This analysis, as shown in Fig. 4D, identified 10 pathways, namely, monoterpenoid biosynthesis, cyanoamino acid metabolism, plant hormone signal transduction, flavonoid biosynthesis, Cutin, cork and wax biosynthesis, suberine and wax biosynthesis, biosynthesis of flavonoids and flavonols, diterpenoid biosynthesis, zeatin biosynthesis, stilbenes, diarylheptanes, and gingerol (stilbeneid, diarylheptanoid, and gingerol biosynthesis), and phenylpropanoid biosynthesis. Compared to group F, the metabolites involved in the above pathways in group B were down-regulated by cyclohexene ether terpenoid glycoside (C16H22O10), up-regulated by ferulic acid salt (C16H22O10), and up-regulated by 7-deoxyglycinate (C16H24O9), while up-regulation of N-hydroxy-L-phenylalanine (C9H11NO3), Dihydrozeaxanthin (C10H15N5O), short leaf pine 3-acetate (C17H14O6) was observed, with down-regulation of 16 feruloyloxypalmitate (C26H40O6), up-regulation of quercetin 3- (2G xylose rutinoside) (C32H38O20), and downregulation of Kanfenol 3-O-beta-D-glucosyl-(1->2)-beta-D-glucoside (C27H30O16), up-regulation of cochineal glycoside (C27H30O15), gibberellin A36 (C20H26O6), and aconitine (C34H47NO11), and down-regulation of adenosine-5’-monophosphate (C10H14N5O7P), dihydrozeaxanthin (C10H15N5O), curcumin diglucoside (C33H40O16), and 3,4-dihydroxystyrene (C8H8O2).
From the KEGG pathway XML analysis of the transcriptomes and metabolomes, it was found that 10 genes were involved in both the cyanide amino acid metabolism pathway and the phenylpropanoid biosynthesis pathway, namely, LOC110095182, LOC110095272, LOC110100117, LOC110106639, LOC110109482, LOC110109642, LOC110113503, LOC110114035, LOC110115944, and LOC114578346. Nine genes were involved in the biosynthesis of phenylacetone, stilbene, diarylheptane, gingerol, and flavonoids, namely, LOC110092517, LOC110092996, LOC110093394, LOC110093422, LOC110093424, LOC110092334, LOC110103137, LOC110113109, and LOC110113968. Flavonoid and flavonol biosynthesis pathways were enriched with two genes, LOC110094145 and LOC110096779. The secondary metabolite involved in the biosynthetic pathway of zeaxanthin and the plant hormone signaling pathway was dihydrozeatin.
Figure 4 Combined analysis of differentially expressed genes/differential metabolites.
Verification of differentially expressed genes
To validate the Illumina sequencing data and the expression patterns of DEGs identified through RNA-Seq, qRT-PCR was performed on 15 DEGs involved in key pathways such as dihydrozeaxanthin synthesis, gibberellin signaling, phenylpropanoid biosynthesis, monoterpene biosynthesis, and zeaxanthin biosynthesis. Among these, genes including LOC110103533, LOC110092013, LOC110108166, LOC110100008, LOC110098448, LOC110092385, and LOC110099511 showed higher expression levels in Group B compared to Group F. On the other hand, genes LOC110104313, LOC110113419, LOC110099634, LOC110096143, LOC110113791, LOC110116586, LOC110096479, and LOC110114334 were expressed at higher levels in Group F relative to Group B (Fig. 5A). The qRT-PCR results were consistent with the RNA-Seq data for Groups B and F (Fig. 5B).
Figure 5 Verification of differentially expressed genes.
Discussion
Analysis of nutrient and metabolic pathways in Dendrobium officinale flowers at different developmental stages
Research on total flavonoids in Dendrobium officinale has primarily focused on comparing their contents across different organs. Stems, leaves, and flowers of Dendrobium officinale all contain polysaccharides, flavonoids, polyphenols, and alkaloids, with significant differences in concentrations. Among these, the total flavonoid content has been reported to be highest in the flowers10. In this study, significant differences in total flavonoid content were observed across developmental stages of Dendrobium officinale flowers. These differences were associated with flavonoid and flavonol biosynthesis, as well as the biosynthesis of flavonoid compounds. Metabolite analysis revealed 108 up-regulated and 113 down-regulated metabolites in flower samples (Group F) compared to flower buds (Group B). Among the down-regulated metabolites, aucubin and alternanthin were the most significantly reduced, with alternanthin, a type of flavonoid, corroborating the total flavonoid content results. The observed changes in polysaccharide contents during flower development were consistent with trends in total phenol content, with flower buds containing significantly lower levels of polysaccharides compared to flowers. Previous studies, such as those by Qiu et al., have shown that metabolism and transcription in white and purple Dendrobium officinale flowers at different flowering stages were primarily associated with pathways involved in flavonoid biosynthesis (ko00941), anthocyanin biosynthesis (ko00942), isoflavonoid biosynthesis (ko00943), and flavone and flavonol biosynthesis (ko00944). Specifically, changes in 19 flavonoids and 62 flavonol metabolites were identified as key factors driving the formation of white flowers24. Moreover, transient overexpression of CtFT genes in safflower leaves has been shown to increase total flavonoid content and up-regulate the expression of key enzyme genes involved in flavonoid biosynthesis pathways25.
From the perspective of the total amino acid content, this was found to be slightly higher in flower buds than in flowers, with differences in various types of amino acids. From the KEGG enrichment analysis of the DAMs, it was seen that the valine, leucine, and isoleucine degradation pathway was down-regulated in flower buds compared to flowers, which was consistent with the measured changes in the contents of valine, leucine, and isoleucine in flower buds and flowers. Further, D-Erythrose-4-phosphate was up-regulated in the amino acid biosynthesis pathway (ko01230) and is involved in the biosynthesis of aromatic amino acids such as tryptophan, tyrosine, and phenylalanine. In the tyrosine metabolism pathway (ko00350), the metabolite phenol was significantly up-regulated; L-tyrosine is an aromatic amino acid involved in the synthesis of polyphenolic compounds26, and this result was consistent with the higher content of tyrosine observed in flowers relative to flower buds. The three most significantly enriched KEGG pathways for DAMs in Dendrobium officinale flowers at different developmental stages were vitamin B6 metabolism, linoleic acid metabolism, and folate biosynthesis, with down-regulation of the folate biosynthesis pathway. Vitamin B6 is an effective antioxidant in plants, with antioxidant activity even higher than that of vitamin C or E. Vitamin B6 is also an important cofactor in amino acid biosynthesis. As a regulatory molecule for cell signaling, it regulates ion channels in the cell membrane, promoting stress resistance and protecting plants from environmental stresses, such as low temperature, ultraviolet radiation, oxidation, and osmotic pressure27. It plays an important role in plant growth and development and in coping with drought stress28. Alpha-linolenic acid metabolism and linoleic acid metabolism are pathways upstream of jasmonic acid (JA) hormone biosynthesis. Studies have shown that the JA content in Camellia oleifera flower buds was significantly increased during ovary and anther formation, which is consistent with the findings of this study29. Linoleic acid metabolism in plants has important physiological functions, including participating in inflammatory responses, regulating the immune system, protecting plants from environmental damage, and maintaining the structure and function of cell membranes. Folic acid provides the donor S-adenosylmethionine for methylation reactions, thereby directly or indirectly participating in all processes in plants and playing an important role in maintaining genomic stability30. Therefore, the differences in metabolic products during different developmental stages of Dendrobium officinale flowers result from one or more pathways that interact with each other, resulting in differences in active substances between flowers and flower buds.
Analysis of pathways in Dendrobium officinale flowers at different developmental stages
The joint analysis of transcriptomic and metabolomic data revealed significant changes in pathways related to plant hormone signal transduction (ko04075), phenylpropanoid biosynthesis (ko00940), and zeatin biosynthesis (ko00908).
Plant hormone signaling pathway
The biosynthesis of zeaxanthin and plant hormone signal transduction are interconnected pathways, highlighting the key role of plant hormones in light responses. In this study, dihydrozeatin was up-regulated in the auxin pathway of the plant hormone signal transduction pathway (ko04075). Gretchen Hagen 3 (GH3) was also up-regulated, while AUX/IAA and SAUR were down-regulated. The receptor GID1 and the gibberellin-regulated gene GAA36 were up-regulated in the gibberellin signaling pathway. Similarly, the receptor PYP/PYL in the abscisic acid signaling pathway and the receptor BKI1 in the brassinosteroid pathway were up-regulated. Gibberellin binds to its receptor GID1 to form a GA-GID1-DELLA complex with the DELLA protein. This complex is recognized by the E3 ubiquitin ligase complex SCFSLY1, which ubiquitinates the DELLA protein, promoting its degradation via the 26 S proteasome pathway. This process releases transcription factors bound to the DELLA protein, initiating the expression of gibberellin-responsive genes and promoting plant growth31. These findings are consistent with previous studies demonstrating the involvement of gibberellin signaling in flower bud differentiation and flowering32,33.
Various protein kinases and transcription factors (TFs) in plant hormone signaling pathways also play important roles in different tissues. Auxin treatment of plants results in the rapid induction of early auxin-responsive gene families, such as Aux/IAA, GH3, and SAUR, with up-regulation of gene expression34,35,36. Research by He Zishun et al. found that high levels of cytokinins in pear flower buds during the late stage of bud differentiation promoted flower differentiation37. A study by Wu Zhixiang et al. on endogenous hormones during lychee flowering showed that the content of endogenous GA3 decreased continuously throughout the entire lychee inflorescence differentiation process. The content of endogenous ABA decreased in the early stage, increased in the middle stage, and decreased in the later stage, while endogenous ZR contents decreased in the early stage and then increased, and the content of endogenous indole-3 acetic acid (IAA) decreased continuously in the early stage and then increased38.
In this study, the zeatin biosynthesis pathway (ko00908) was up-regulated by dihydrozeatin, while the associated enzymes isoprenyl transferase (ADP/ATP) EC 2.5.1.112, isoprenyl transferase (AMP) EC 2.5.1.27, and cytokinin dehydrogenase EC 1.5.99.12 were down-regulated. The initial and rate-limiting steps in cytokinin biosynthesis involve the conversion of AMP, ADP, or ATP and dimethylallyl diphosphate (DMAPP) into iP nucleotides catalyzed by isopentenyl transferase (IPT). This result aligns with findings by Duan et al. (1999), who observed that under field conditions, the dehulling of cotton delayed the peak appearance of IAA in corresponding main stem leaves and fruit branches, reduced zeaxanthin and its nucleosides, delayed the accumulation of isopentenyl adenine and its nucleosides (iP + iPA) in fruit branches and leaves, advanced the accumulation of dihydrozeaxanthin and its nucleosides (DHZ + DHZR), and reduced the abscisic acid (ABA) content39.
The phenylacetone biosynthetic pathway
The phenylpropanoid biosynthetic pathway (ko00940) is involved in key processes in plant seeds and plants under abiotic stresses such as salt and drought40. This study showed that secondary metabolites in the phenylpropanoid biosynthetic pathway of Dendrobium officinale flowers at different developmental stages were up-regulated by 3,4-dihydroxystyrene. Phenylpropanoid is metabolized through the shikimic acid, benzoate, and coumaric acid pathways, producing various phenylpropanoids and other compounds, including lignin, flavonoids, and plant antitoxin41. This is consistent with the finding of this study that the total flavonoid content in flowers was higher than that in flower buds. The application of hypophosphite in preventing and controlling rice phytophthora disease has shown that some DEGs were significantly enriched in the phenylpropanoid biosynthesis pathway42. Xiangxu Meng’s research showed that three TF families (MYB, C2H2, and NAC) could regulate all DEGs involved in phenylpropanoid biosynthesis. Studies have shown that up-regulated genes in the phenylpropanoid biosynthesis pathway may help enhance resistance against cucumber disease43. Studies indicate that phenylpropanoid biosynthesis is enhanced when plants are subjected to biotic or abiotic stress, and many of these compounds have been shown to contribute to plant resistance to pathogens and stress35. A study by Heng Wei et al. on different developmental stages of hazelnut ovary showed that the ChARF3 target gene was significantly enriched in the phenylpropanoid biosynthesis (ko 00940) and plant hormone signal transduction (ko 04075) pathways. ChARF3 is thought to regulate the auxin-induced genes and transcription factors MADS, AP 2/ERF, TCP, FT, and LFY. ChARF3 may regulate ovary initiation and ovule development by regulating genes related to auxin synthesis and transport, cell division and proliferation, and flower and fruit development44.
The zeaxanthin biosynthetic pathway
In this study, the secondary metabolite dihydrozeatin, part of the zeaxanthin biosynthetic pathway, was up-regulated. Dihydrozeaxanthin acts as an intermediate in zeaxanthin biosynthesis, converting from dihydrozeaxanthin nucleoside and then being converted into dihydrozeaxanthin-O-glucoside by glycosyltransferase. This cytokinin is known to stimulate flower bud formation. A study by Van et al. on tobacco flower bud formation in vitro demonstrated that BA, 9R-BA, and DHZ showed lower activity than IP and IPA, which were identified as the most active cytokinins44. Moreover, research on longan embryos at different developmental stages revealed that a significant number of DEGs were enriched in metabolic pathways, including phenylpropanoid biosynthesis, flavonoid biosynthesis, fatty acid biosynthesis, zeaxanthin biosynthesis, and tryptophan metabolism. During the early stages of longan embryo development, the expression levels of genes involved in the de novo synthesis of zeaxanthin were significantly up-regulated45. These findings, in conjunction with previous studies, confirm that cytokinins play a key role in flower bud formation.
Conclusion
This study revealed that Dendrobium officinale flower buds contain higher levels of total flavonoids and lower levels of total phenols and polysaccharides than flowers. Significant differences were observed in the contents of 22 amino acids, excluding aspartic acid and proline. Integrated transcriptomic and metabolomic analyses identified 2,767 DEGs between flower buds and flowers, with 902 genes up-regulated and 1,865 genes down-regulated in buds relative to flowers. Furthermore, 221 DAMs were identified, comprising 108 up-regulated and 113 down-regulated metabolites in flowers relative to buds. Further analysis highlighted the top 10 pathways with the highest numbers of co-enriched genes and metabolites, primarily associated with plant hormone signal transduction, including pathways related to gibberellin signaling and zeatin biosynthesis, which play critical roles in flower bud formation and flowering. Pathways involved in the biosynthesis of flavonoids and flavonols, as well as amino acid metabolism—such as tyrosine metabolism, degradation of valine, leucine, and isoleucine, tryptophan metabolism, and cysteine and methionine metabolism—were closely associated with the quality of flower buds and flowers. These findings provide a scientific basis for the development and utilization of Dendrobium officinale flowers, particularly in improving their nutritional and pharmacological qualities.
Data availability
The raw sequencing data are available in the National Center for Biotechnology Information Sequence Read Archive (SRA) under the BioProject accession number of PRJNA1191381.
References
Editorial Committee of Flora of China. Chinese Academy of Sciences. Flora of China, 117 (Science, 1999).
National Pharmacopoeia Committee Pharmacopoeia of the People’s Republic of China A. China Medical Science and Technology, 295 (202).
Ma, Q. et al. Progress in molecular biology research of dendrobium officinale. J. Mountain Agric. Biol. 42, 45–50 (2023).
Meng, Y. et al. Research on the production process and functionality of dendrobium officinale compound flower tea. Food Ferment. Ind. 47, 170–179 (2021).
Fan, J. K. et al. Analysis of nutritional components in dendrobium officinale flowers. J. Food Saf. Qual. Inspection. 12, 8334–8341 (2021).
Huang, B. et al. Determination of multiple mineral elements in stems and flowers of dendrobium officinale by microwave digestion inductively coupled plasma mass spectrometry. J. Food Saf. Qual. Test. 11, 8769–8774 (2020).
Huo, X., Zhou, J. H., Yang, X. J., Liu, W. W. & Huang, J. C. Study on volatile components of dendrobium officinale flowers. Chin. J. Traditional Chin. Med. 23, 735–737 (2008).
Xin, X. et al. Analysis of active components in different flowering periods and flower parts of dendrobium officinale. J. Zhejiang Agric. Forestry Univ. 36 (01), 200–205 (2019).
Huang, B., He, W., Wu, J. H., Wang, H. M. & Li, W. Simultaneous determination of phenolic components in stems, leaves, and flowers of dendrobium officinale by UPLC-MS/MS food science. 42, 262–268 (2021).
Tang, W. W., Xia, J. & Chen, Y. Functional components, antioxidant activity, and correlation of stems, leaves, and flowers of dendrobium officinale. Food Mach. 37, 45–50 (2021).
Ling, N. et al. Yu.Q.M. Study on the cytotoxicity and in vitro antioxidant activity of dendrobium officinale flower extract from Guangxi. Chin. Food Addit. 34, 141–148 (2023).
Long, H. Q. et al. Y. Experimental study on the hypotensive stability of dendrobium officinale flowers in spontaneously hypertensive rats. Zhejiang J. Traditional Chin. Med. 51, 726–729 (2016).
Xue, S. B., Zhu, T., Li, B. B. &Li, T. T. Application of transcriptome and metabolome combined analysis in plants. J. Shanxi Agricultural Univ. (Natural Sci. Edition). 42, 1–13 (2022).
Huang, Y. C. et al. Research progress of transcriptomics and metabolomics in plant abiotic stress. Jiangsu Agricultural Sci. 51,1–7. https://doi.org/10.15889/j.issn.1002-1302.2023.22.001(2023).
Song, J. L. et al. W.D. Comparative metabolomics analysis of the effects of mulberry twig Prunus cultivation on the metabolism of Varnishan yellow. J. Fungi. 42, 2231–2243 (2023).
Wei, G. L., Zhou, P. C., Liu, H., Yang, X. N. & Wei, K. H. Liao, J.H. Metabolomic analysis of different parts of acacia sinensis. Chin. Mod. Traditional Chin. Med. 25, 1575–1581 (2023).
Liang, H., Sun, H., Qian, J. & Zhang, , Y. Y. Progress in omics research on metabolic regulation of medicinal plants. Traditional Chin. Med. 8, 2085–2092 (2023).
Xin, X. X. et al. J.Active components of flowers in different flowering stages and floral structures of dendrobium officinale. J. Zhejiang Agric. Forestry Univ. 36, 200–205 (2019).
Huang, X. H., Wang, Z. H., Li, J., Miao, A. Q. & Ye, Q. S. Analysis of the main nutritional components and quality comparison of dendrobium flowers at different flowering periods. J. Trop. Crops. 38, 45–52 (2017).
Li, D. et al. S.Gapless genome assembly of ZH8015 and preliminary multi-omics analysis to investigate ZH8015’s responses against brown planthopper infestation. Rice Sci. 31, 317–327 (2024).
Li, Z. Study on the main medicinal components and hepatoprotective and anti-inflammatory effects of Dendrobium huoshanense with different growth years. Master’s Thesis (Jiangsu University, 2021).
Liao, X., Wu, Z., Yang, Y., Chen, G. & Zhan, Y. The total polyphenol and flavonoid content and antioxidant capacity of different solvent extracts of Gastrodia Elata. Food Ferment. Ind. 48, 157–164 (2022). (2022).
Chen, X. G. et al. Comparative analysis of chemical composition and antioxidant capacity of trifolium repens based on different sources: a case study of Fuyang district, Hangzhou City, Zhejiang Province. Zhejiang Agricultural Sci. 65, 320–324 (2024).
Yu, J. et al. Integrated analysis of the transcriptome and metabolome of brassica Rapa revealed regulatory mechanism under heat stress. Int. J. Mol. Sci. 24, 13993. https://doi.org/10.3390/ijms241813993( (2023).
Li, Z. Cloning of safflower FT gene and preliminary study on its involvement in flower formation and flavonoid synthesis. Master’s Thesis, Jilin Agricultural University, Changchun, China, (2023).
Duan, X. L., Zhu, J. F., Li, W., Wang, K. L. & Zhang, C. P. Metabolomic analysis of alfalfa during germination and seedling stages under salt stress. Chin. J. Grassland Sci. 46, 35–47 (2024).
Huang, L. Q. & Zhang, J. Y. Research progress on de Novo synthesis and metabolic conversion of plant vitamin B6. Northwest. Bot. J. 35, 2124–2131 (2015).
Li, L. et al. Metabolome analysis of alfalfa leaves under drought stress and screening of stomatal regulators. Grassland J. 31, 2671–2683 (2023).
Liu, B. et al. Y.P. Photoprotection contributes to freezing tolerance as revealed by RNA-seq profiling of rhododendron leaves during cold acclimation and deacclimation over time. Hortic. Res. 9, 2815–2829 (2022).
Gu, Y. Y. & Han, Y. Research progress on folate metabolism and biological enhancement in plants. Mol. Plant. Breed. 21, 3778–3784 (2023).
Claeys, H. & De Bodt, S. Gibberellins and Dellas: central nodes in growth regulatory networks. Trends Plant. Sci. 19, 231–239 (2024).
Jiang, X. D., & Zhao B.The effects of low temperature and Gibberellin on the regulation of flowering period of potted eight immortal flower. North. Hortic. 17, 67–75 (2024).
Zhang, S. et al. The role and possible mechanism of Gibberellin (ga) in the chilling-mediated blueberry dormancy release and germination. Sci. Hort. 336. https://doi.org/10.1016/j.scienta (2024).
Song, S. Q. et al. Research progress on the regulation of seed development, dormancy, and germination by cytokinins. Acta Bot. Sinica. 56, 218–231 (2021).
Wang, J. H., Zheng, Q. Y., Wang, C. X. & Zhou, A. Classification of soybeans from different habitats based on metabolomic-transcriptomic integration. Appl. Biol. Chem. 67, 30. https://doi.org/10.1186/s13765-024-00882-x (2024).
Wang, G., Chen, R., Han, X. & Xue, X. Effects and mechanism analysis of non-bagging and bagging cultivation on the growth and content change of specific substances of Fuji Apple fruit. Plants 12, 3309. https://doi.org/10.3390/plants12183309 (2023).
He, Z. S., Chen, D., Zhang, J., Zhang, S. L. & Zhang, H. P. The relationship between the number of flowers per inflorescence of Pear and plant hormones. Chin. Fruit Tree. 44, 21–23 (2019).
Wu, Z. X., Zhou, Z. D., Tao, Z. & Wang, L. X. Changes in endogenous hormones during flower bud differentiation of Feizixiao and Goose egg Litchi. J. Trop. Crops. 43, 42–45 (2005). 2005.
Duan, L. & He, Z. Effects of changes in source sink relationships on endogenous hormones in cotton leaves. Northwest. Bot. J. 6, 116–121 (1999).
Wang, Y., Cao, R., Yang, L. & Plants Transcriptome Analyses Revealed the Wax and Phenylpropanoid Biosynthesis Pathways Related to Disease Resistance in Rootstock-Grafted Cucumber. https://doi.org/10.3390/plants12162963 (2023).
Wei, H., Cheng, Y. & Sun, Y. Genome-Wide identification of the ARF gene family and ARF3 target genes regulating ovary initiation in Hazel via chip sequencing. Front. Plant Sci. https://doi.org/10.3389/FPLS.2021.715820 (2021).
Huang, Y., Cai, S. & Zhang, G. Transcriptome-Based analysis of Phosphite-Induced resistance against pathogens in rice. Plants 9 (10), 1334. https://doi.org/10.3390/plants9101334 (2020).
Meng, X., Wang, Y. & Li, J. RNA sequencing reveals phenylpropanoid biosynthesis genes and transcription factors for Hevea Brasiliensis reaction wood formation. Front. Genet. https://doi.org/10.3389/fgene.2021.763841 (2021). 12,763841.,.
Van der Krieken, W. M., Croes, A. F., Smulders, M. J. & Wullems, G. J. Cytokinins and flower bud formation in vitro in tobacco: role of the metabolites. Plant Physiol. 92(3), 565–569. https://doi.org/10.1104/pp.92.3.565 (1990).
Chen, Y. Transcriptome and Proteomic Analysis of Early Embryogenesis in Longan, as Well as Expression and Functional Analysis of Flowering time Related Genes (Fujian Agriculture and Forestry University, 2017).
Author information
Authors and Affiliations
Contributions
R.X. and R.S. initiated the concept of the article, thoroughly conducted the literature review, performed data analysis, crafted the illustrations, and penned the manuscript. H.Y. and W.X. lent invaluable experimental support and reviewed the manuscript for enhancement. Z.L., L.Q.and Y.W. contributed their expertise in assisting with the experiments. Co-authors finish the proofreading. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ren, X., Huang, Y., Zhang, L. et al. Integrated analysis of transcriptomics and metabolomics of Dendrobium officinale flowers at different developmental stages. Sci Rep 15, 10342 (2025). https://doi.org/10.1038/s41598-025-93889-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93889-3