Abstract
To explore the potential effects and the corresponding mechanisms of brain and muscle arnt-like protein-1 (BMAL1) on the progression of intervertebral disc degeneration (IVDD) in vitro studies. The expression of BMAL1, SIRT1 and PINK1 were evaluated by the method of siRNA/pcDNA in the immortalized nucleus pulposus (NP) cells. The expression of SIRT1/PGC-1α pathway was assessed. The characteristics of NP cell, containing the activity and density, the level of apoptosis, inflammatory response, reactive oxygen species (ROS), senescence, and mitophagy were evaluated. The overexpression of BMAL1 was achieved with the pcDNA3.1, the expression of SIRT1 and PGC-1α were increased, the inflammatory response, the ROS, the level of apoptosis and senescence were decreased, however, the level of mitophagy, the activity and density of NP cell were enhanced. The BMAL1 inhibites the progression of IVDD by activating the SIRT1/PGC-1α pathway in the vitro studies.
Similar content being viewed by others
Introduction
Low back pain (LBP) has become a common human disease now. And it is closely related to intervertebral disc degeneration (IVDD)1,2. But the pathological mechanism of intervertebral disc degeneration (IVDD) is complicated and unclear now. Thus, exploring the pathogenesis and treatment of IVDD is of great significance to human health and social development.
Brain and muscle arnt-like protein-1 (BMAL1) is a core component of the circadian clock loop, which directs the sophisticated circadian expression of clock-controlled genes. And BMAL1 is indispensable for hard tissue development, including bone, cartilage and teeth3. In the context of IVDD, BMAL1 is closely involved in the maintenance of discs and vertebral health. It has been reported that knocking down BMAL1 induced a significantly decreased disc height index in mice. This study indicated a vital role for BMAL1 in maintenance of disc health4. But the exact role and potential mechanism of BMAL1 in the development of IVDD were unclear.
Some recent studies5,6found that BMAL1 could take some positive actions by promoting the silent information regulator factor 2 related enzyme 1 (SIRT1) in NP cells. It is well known that SIRT1 is a member of the class III HDAC family and is involved in regulating cell survival, metabolism, oxidative stress response, mitochondrial biogenesis and inflammation7. Besides, SIRT18,9 can enhance mitochondrial biogenesis by activating peroxisome-proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α).
Given the above findings, we hypothesize that BMAL1 inhibits the IVDD via regulating SIRT1 mediated cellular biological behaviors, including survival, metabolism, oxidative stress response, mitochondrial function, and inflammation. The present study might provide new insights of BMAL1 as a key regulator and potential treatment targets of IVDD.
Results
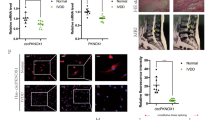
BMAL1, SIRT1 and PINK1 expression was downregulated in NP cells
To investigate the role of BMAL1, SIRT1, and PINK1 in NP cells, siRNA-mediated knockdown was performed. As shown in Fig. 1, the three siRNAs decreased the mRNA and protein levels of BMAL1, SIRT1 and PINK1 in NP cells. The siBMAL1#2, siSIRT1#1, and siPINK1#1 were selected for the subsequent experiments due to their high inhibition efficiency.
BMAL1 upregulated the SIRT1/PGC-1α pathway in NP cells
To determine whether BMAL1 overexpression or knockdown affected the SIRT1/PGC-1α pathway, NP cells were transfected with PCDNA3.1-BMAL1 and si-BMAL1. The expression of BMAL1, SIRT1 and PGC-1α was upregulated in the PCDNA3.1-BMAL1 group compared to the PCDNA3.1-NC group, confirming successful BMAL1 overexpression in NP cells. Conversely, BMAL1, SIRT1 and PGC-1α expression was downregulated in the si-BMAL1 group compared to the si-NC group, indicating effective BMAL1 knockdown. These results confirmed the successful establishment of the PCDNA3.1-BMAL1 and si-BMAL1 models in NP cells.
To determine whether SIRT1 was involved in BMAL1-mediated IVDD, SIRT1 was knocked down in the PCDNA3.1-BMAL1 group using siRNA. While BMAL1 expression remained unchanged, SIRT1 and PGC-1α expression were reduced in the PCDNA3.1-BMAL1 + si-SIRT1 group compared to the PCDNA3.1-BMAL1 group. Protein expression levels of BMAL1, SIRT1 and PGC-1α mirrored the RNA results, as shown in Fig. 2. These findings suggested that BMAL1/SIRT1/PGC-1α mediated IVDD.
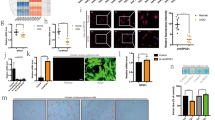
BMAL1 inhibited NP cell apoptosis and reduces ROS levels
To examine the effects of BMAL1 on apoptosis and ROS levels in NP cells, apoptosis and ROS were measured in different treatment groups. Apoptosis and ROS levels were reduced in the PCDNA3.1-BMAL1 group compared to the PCDNA3.1 group but increased in the si-BMAL1 group compared to the si-NC group. Additionally, apoptosis and ROS levels were higher in the PCDNA3.1-BMAL1 + si-SIRT1 group and PCDNA3.1-BMAL1 + si-PINK1 group compared to the PCDNA3.1-BMAL1 group. These results indicated that BMAL1 downregulated apoptosis and ROS levels in NP cells through SIRT1 and PINK1.
To assess the relationship between this signaling pathway and cell proliferation, OD values and cell density were measured. The BMAL1 group showed higher OD values and cell density at 96 h compared to the vector group, while the si-BMAL1 group exhibited lower values compared to the si-NC group. Furthermore, the BMAL1 + si-SIRT1 and BMAL1 + BMAL1 + si-PINK1 groups had reduced OD values and cell density compared to the BMAL1 group. These results demonstrated that BMAL1 upregulated NP cell proliferation through SIRT1 and PINK1. The results were shown at Fig. 3A–F.
BMAL1 suppressed inflammatory response in NP cells
To evaluate the role of BMAL1 in regulating inflammatory responses, levels of IL-1β, IL-6, and TNF-α were measured. IL-1β, IL-6, and TNF-α levels were increased in the si-BMAL1 group compared to the si-NC group but decreased in the BMAL1 group compared to the vector group. Moreover, IL-1β, IL-6, and TNF-α levels were higher in the BMAL1 + si-SIRT1 group and the BMAL1 + si-PINK1 group compared to the BMAL1 group, as shown in Fig. 4A–C. These findings indicated that BMAL1 downregulated inflammatory responses in NP cells through SIRT1 and PINK1.
BMAL1 reduced NP cell senescence
To investigate the effects of BMAL1 on cellular senescence, senescence markers were assessed in NP cells. Cellular senescence was reduced in the PCDNA3.1-BMAL1 group compared to the PCDNA3.1 group but increased in the si-BMAL1 group compared to the si-NC group. Additionally, senescence levels were higher in the BMAL1 + si-SIRT1 group and the BMAL1 + si-PINK1 group compared to the BMAL1 group. The mRNA and protein expression levels of senescence markers were consistent, as shown in Fig. 5A–D. These results suggested that BMAL1 inhibited NP cell senescence through SIRT1 and PINK1.
BMAL1 enhanced mitophagy in NP cells
To determine whether BMAL1 regulates mitophagy in NP cells, PINK1 and PARKIN expression was measured. PINK1 and PARKIN expression was increased in the PCDNA3.1-BMAL1 group compared to the PCDNA3.1 group but decreased in the si-BMAL1 group compared to the si-NC group. Furthermore, PINK1 and PARKIN expression was reduced in the BMAL1 + si-SIRT1 group and the BMAL1 + si-PINK1 group compared to the BMAL1 group, as shown in Fig. 6A–C. These findings indicated that BMAL1 upregulated mitophagy in NP cells through SIRT1 and PINK1.
Discussion
With the growth and natural aging, the intervertebral disc degeneration (IVDD) becomes inevitable. But it remains challenging to distinguish the differences between normal aging-related degeneration and pathological IVDD. Pathological IVDD can lead to the pain at low back and legs, which can affect quality of life. The metabolism of NP cells could play an important role at the degeneration of intervertebral disc. So it was important for all of us to know about energy metabolism of NP cell and explore strategies to inhibit pathological IVDD. The present study demonstrated that BMAL1 inhibited IVDD by activating the SIRT1/PGC-1α axis. The results highlight the role of BMAL1 in regulating IVDD through mechanisms involving the SIRT1/PGC-1α axis, NP cell activity and density, apoptosis, inflammatory response, reactive oxygen species(ROS), senescence, and mitophagy levels in NP cells.
BMAL1 upregulates the expression of SIRT1/PGC-1α axis in the NP cells. As a classic pathway regulating mitochondrial function, the SIRT1/PGC-1α pathway is involved in multiple pathological processes, including antiaging and oxidative stress10,11. IVDD is closely associated with aging and oxidative stress, and its incidence increases with age. SIRT1, a member of the NAD+-dependent Sir2 histone deacetylase family, regulates mitochondrial function and reduces oxidative stress12. It modulates celluar senescence and longevity through substrate acetylation and deacetylation, altering transcriptional and enzymatic activities13,14. SIRT1 enhances mitochondrial biogenesis and oxygen consumption by deacetylating PGC-1α15, a key regulator of mitochondrial biogenesis and function16. PGC-1α also enhances cellular antioxidant capacity, reducing inflammation-induced ROS17. So the SIRT1/PGC-1α pathway has played an important role in the IVDD, providing a potential therapeutic target. This study showed that BMAL1 upregulated the SIRT1/PGC-1α axis in NP cells, suggesting BMAL1 as a potential treatment for IVDD.
BMAL1 inhibites the inflammatory response by mediating SIRT1. SIRT1 exerts anti-inflammatory and antioxidative effects through pathways such as mitogen-activated protein and NF-kB18. It reduces NF-kB pathway activity and decreases inflammatory factor production19, partly through the deacetylation of P65 and C-JUN20. SIRT1 also promotes the deacetylation of PGC-α and forkhead box O(FOXO), enhancing mitochondrial activity and antioxidant capacity, thereby reducing inflammation-induced ROS17. Increased proinflammatory cytokines (e.g., IL-1, IL-6, IL-12, IL-17, TNF-α, TNF-γ) are characteristic of the IVDD microenvironment21. Inflammatory cytokines like IL-1β and TNF-α increase MMP13 expression, decrease collagen II and aggrecan expression, and induce NP cell apoptosis, contributing to IVDD22,23. This study demonstrated that BMAL1 inhibited inflammatory responses by mediating SIRT1, reducing IL-1β, IL-6 and TNF-a expression, thereby improving the NP cell survival microenvironment.
BMAL1 reduces the level of reactive oxygen species (ROS) in NP cells by mediating SIRT1. ROS, generated by external stimulus such as pro-inflammatory cytokines, nutrient deprivation, and mechanical loading, plays a critical role in IVDD progression24. Mitochondria are a major source of ROS, and mitochondrial-dependent ROS production has been observed in various disc cells, including NP cells25,26,27,28,29,30. Intracellular redox homeostasis depends on the balance between ROS generation and scavenging31,32. Excessive ROS disrupts this balance, causing oxidative stress and impairing cell function and viability31,33. This study showed that BMAL1 reduced ROS levels in NP cells by mediating SIRT1, promoting NP cell survival.
BMAL1 upregulates levels of mitophagy in NP cells by mediating SIRT1. Mitophagy, essential for mitochondrial turnover and maintenance, is triggered by pathological stresses such as mitochondrial membrane potential collapse, hypoxia, oxidative stress, and iron starvation34. It removes dysfunctional mitochondria, preserving cellular energy metabolism35. Mitophagy regulation involves molecules such as PINK-1 and parkin36. PINK-1 silencing increases NP cell senescence and ROS production, while parkin knockdown promotes apoptosis and mitochondrial impairment35,37. This study demonstrated that BMAL1 enhanced PINK-1 and parkin expression through the BMAL1/SIRT1 pathway, improving NP cell survival and delaying aging, thereby alleviating IVDD progression.
BMAL1 downregulates the apoptosis of NP cell by mediating the expression of the SIRT1. SIRT1, a nuclear protein, regulates apoptosis38, and mitochondria plays an important part in cellular apoptosis by activating mitochondrial outer membrane permeabilization39. In the present study, BMAL1 reduced NP cell apoptosis by activating the BMAL1/SIRT1 pathway, consistent with finding that SIRT1 overexpression confers resistance to oxidative stress and apoptosis40.
BMAL1 downregulates the senescence of NP cell by mediating SIRT1. Cellular senescence, characterized by irreversible cell cycle arrest and reduced proliferation, is driven by stress and injury41. Senescent disc cells secrete matrix proteases, cytokines, and chemokines, deteriorating the disc microenvironment42,43. The p53-p21-Rb pathway plays a key role in NP cell senescence44. This study showed that BMAL1 inhibited p53, p21 and Rb1 expression in NP cells through SIRT1 and PINK1, reducing cellular senescence.
BMAL1 upregulates NP cell activity and density by SIRT1 and PINK1. NP cell activity and density are critical for tissue function. This study demonstrated that BMAL1 enhanced NP cell activity and density, slowing IVDD progression.
BMAL1 regulates SIRT1 and PGC-1α downstream targets in mitochondrial function. BMAL1, as part of the BMAL1/CLOCK complex, binds to the E-box elements in the SIRT1 promoter, and BMAL1/CLOCK may directly regulate SIRT1 expression by binding to E-boxes in the SIRT1 promoter45. BMAL1 regulates circadian rhythms and ECM maintenance in annulus fibrosus cells46,47. PGC-1α is a master regulator of mitochondrial biogenesis and function, and it activates downstream targets such as NRF1/2 and TFAM, which are critical for mitochondrial DNA transcription and replication48.
Targeting BMAL1 through small-molecule agonists, gene therapy, or circadian rhythm modulation offers a promising approach to treating IVDD. These strategies aim to restore mitochondrial function, reduce inflammation, and mitigate cellular senescence, addressing the root causes of disc degeneration. Future animal experiments will provide critical insights into the safety, efficacy, and translational potential of these therapies, paving the way for clinical trials in humans.
Limitations
This study has some limitations. The use of immortalized NP cells in this study offers practical advantages, such as consistent proliferation and reduced experimental variability. However, immortalized cells may exhibit functional differences compared to primary NP cells, including altered metabolic activity, senescence profiles, and response to inflammatory stimuli. These differences could influence the interpretation of results related to apoptosis, ROS production, and inflammatory responses. Future studies should validate key findings using primary NP cells to ensure translational relevance. Additionally, the immortalization process may affect mitochondrial function and mitophagy, which are central to this study.
In vitro experiments often fail to replicate the mechanical load, nutrient supply, cell-matrix interactions, cellular heterogeneity, and inflammatory environment of the vivo NP microenvironment. Careful consideration of these limitations is essential when extrapolating results to in vivo conditions or clinical applications.
Conclusions
In conclusion, BMAL1 alleviates IVDD by activating the SIRT1/PGC-1α axis, regulating NP cell proliferation, apoptosis, inflammatory response, ROS levels, senescence, and mitophagy through vitro studies. These interconnected mechanisms collectively affect NP cell degeneration and provide potential therapeutic targets for IVDD. So the mechanism was described at Fig. 7.
Methods
Materials
Immortalized NP cells were purchased from iCell Bioscience Inc, Shanghai. Other materials contained BCA protein assay kit, ARNTL polyclonal antibody, CCK-8 cell proliferation assay kit, Human IL-1β ELISA kit, Human IL-6 ELISA kit, Human TNF-α ELISA kit, PARK2/Parkin polyclonal antibody, PGC-1α monoclonal antibody, PINK1 polyclonal antibody, RIPA buffer, SIRT1 polyclonal antibody, TRIzol reagent, Annexin-v-FITC/PI apoptosis detection kit, Reactive oxygen species (ROS) assay kit, NcmBlot rapid transfer buffer(20X), 7.5% ExpressCast PAGE gel, Primer sets for Reverse Transcription-Quantitative PCR (RT-qPCR), NovoStart SYBR qPCR SuperMix Plus and so on.
The siRNA required for the experiments was designed and synthesized by Qingke Biotechnology Co., Ltd. The sequences of siRNA are listed in Table 1.
SiRNA design and target specificity validation
Sequence sources: siRNA sequences targeting BMAL1, SIRT1, and PINK1 were designed using RNAi Designer and cross-validated using NCBI BLAST to minimize off-target effects.
Target validation: siSPOTR and siRNA scales were used to assess sequence specificity against the human transcriptome.
Control siRNA: A non-targeting scrambled siRNA served as a negative control, while GAPDH-targeting siRNA was used as a positive control for knockdown efficiency.
Knockdown efficiency verification: siRNA knockdown efficiency was confirmed 48 h post-transfection using RT-qPCR and Western blot.
Reverse transcription-quantitative PCR (RT-qPCR)
The total RNA was extracted from NP cells using TRIzol reagent. The RNA purity and concentration were measured using a spectrophotometer. The cDNA synthesis was performed using the ReverTra Ace qPCR RT Master Mix with gDNA Remove kit. The RT-qPCR was conducted using SYBR Premix ExTaq II on a Mx3005P real-time PCR System.
The results were normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. And the values were calculated using the 2−∆∆Ct method. Primers sequences for RT-qPCR were shown at Table 2.
Western blot (WB)
The total protein of the cells was extracted using RIPA buffer. The protein concentration was used with a BCA kit. The protein concentration was detected by the BCA method. The SDS-PAGE electrophoresis was achieved to transfer membrane and block. The Primary antibodies (BMAL1:Proteintech, 14268-1-AP, 1:5000; SIRT1: Proteintech, 13161-1-AP, 1:3000; PINK1: Proteintech, 23274-1-AP, 1:2000; PGC-1a: Proteintech, 66369-1-Ig, 1:10000; RB1: Proteintech, 10048-2-Ig, 1:8000; P53: Proteintech, 10442-1-AP, 1:6000; P21: Proteintech, 10355-1-AP, 1:2000; PARKIN: Proteintech, 14060-1-AP, 1:1500) and the anti-GAPDH antibodies (Proteintech, 10494-1-AP, 1:20000) were added, and it was incubated overnight at 4℃. After washing the membrane, the secondary antibody was added. After 1 h, the ECL was added for developing. The software Image J was used for image analysis. The GAPDH served as the loading control for normalizing protein expression levels.
Cell counting and proliferation assay
The cells were seeded in 96-well plates at a concentration of 5 × 103 per well at 0 h, 24 h, 48 h, 72 h and 96 h. 10 ml CCK-8 reagent was added to the each well after cell attachment. And the cells were incubated at 37 ℃ for 2 h. The cell growth rate was detected by measuring the optical density (OD) value at 450 nm.
Cell apoptosis assay
The cell apoptosis was tested on a FACS Calibur flow cytometer using an Annexin V-FITC/PI Apoptosis Detection Kit according to the manufacturer’s instructions. The apoptotic cells were detected by a high level of Annexin V-conjugated FITC fluorescence and a low level of PI fluorescence.
Reactive oxygen species (ROS) detection
ROS production in NP cells was measured by a ROS detection kit according to the manufacturer’s protocol. Cells were mixed with 1 ml of diluted DCFH-DA. The concentration of the DCFH-DA probe was 10 μm when diluted. The incubation time was 20 min at 37 ℃ in the dark. After removing excess DCFH-DA, cells were washed with PBS three times. The expression of green fluorescence was observed under a fluorescence microscope.
ELISA assay
The level of serum interleukin-6 (IL-6), interleukin-1β (IL-1β) and tumor necrosis factor-alpha (TNF-α) were measured by the ELISA kit according to the manufacturer’s protocol.
Statistical analysis
All the experiments were performed in biological triplicates. Each biological replicate was further analyzed with technical triplicates. The data were presented as the mean ± standard deviation.
The statistical analyses were achieved with GraphPad Prism 8 software. Analysis of differences were analyzed with one-way analysis of variance (ANOVA), Student’s test. Homogeneity of variance was assessed using Levene’s test, and normality was confirmed using the Shapiro–Wilk test. For all analyses, a P-value less than 0.05 from a two-tailed test was considered statistically significant.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- LBP:
-
Low back pain
- IVD:
-
Intervertebral disc
- NP:
-
Nucleus pulposus
- IVDD:
-
Intervertebral disc degeneration
- BMAL1:
-
Brain and muscle arnt-like protein-1
- SIRT1:
-
Silent information regulator factor 2 related enzyme 1
- PGC-1α:
-
Peroxisome-proliferator-activated receptor gamma coactivator 1 alpha
- ROS:
-
Reactive oxygen species
- FOXO:
-
Forkhead box O
References
Mirza, S. K. & Deyo, R. A. Systematic review of randomized trials comparing lumbar fusion surgery to nonoperative care for treatment of chronic back pain. Spine 32, 816–823 (2007).
Hanley, E. N. Jr et al. Debating the value of spine surgery. J. Bone Jt. Surg. Am. 92, 1293–1304 (2010).
Chen, G. & Tang, Q. Shaoling Yu, et al. The biological function of BMAL1 in skeleton development and disorders. Life Sci.. 1–7 (2020).
Suyama, K. et al. Circadian factors BMAL1 and RORαcontrol HIF-1αtranscriptional activity in nucleus pulposus cells: implications in maintenance of intervertebral disc health. Oncotarget 7 (17), 23056–23071 (2016).
Chen, Y. et al. Long noncoding RNA FLRL2 alleviated nonalcoholic fatty liver disease through Arntl-Sirt1 pathway. FASEB J. 33 (10), 11411–11419 (2019).
Zhang, J. et al. Effects of BMAL1-SIRT1-positive cycle on estrogen synthesis in human ovarian granulosa cells: an implicative role of BMAL1 in PCOS. Endocrine 53 (2), 574–584 (2016).
Meng, X. et al. Sirt1: role under the condition of ischemia/hypoxia. Cell. Mol. Neurobiol. 37 (1), 17–28 (2017).
Tian, L. et al. Pretreatment with Tilianin improves mitochondrial energy metabolism and oxidative stress in rats with myocardial ischemia/reperfusion injury via AMPK/SIRT1/PGC-1 alpha signaling pathway. J. Pharmacol. Sci. 139 (4), 352–360 (2019).
Waldman, M. et al. Regulation of diabetic cardiomyopathy by caloric restriction is mediated by intracellular signaling pathways involving ‘SIRT1 and PGC-1α‘. Cardiovasc. Diabetol. 17 (1), 111–123 (2018).
Li, J. et al. Radioprotective and antioxidant effect of Resveratrol in hippocampus by activating Sirt1. Int. J. Mol. Sci. 15, 5928–5939 (2014).
Liang, D. et al. SIRT1/PGC-1α pathway activation triggers autophagy/mitophagy and attenuates oxidative damage in intestinal epithelial cells. Biochimie 170, 10–20 (2020).
Ou, X., Lee, M. R., Huang, X., Messina-Graham, S. & Broxmeyer, H. E. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 32, 1183–1194 (2014).
Yao, H. & Rahman, I. Perspectives on translational and therapeutic aspects of SIRT1 in inflammaging and senescence. Biochem. Pharmacol.. 84, 1332–1339 (2012).
Ramis, M. R. et al. Caloric restriction, Resveratrol and melatonin: role of SIRT1 and implications for aging and related-diseases.Mech. Ageing Dev. 146–148 (2015).
Kitada, M. et al. Sirtuins and type 2 diabetes: role in inflammation, oxidative stress, and mitochondrial function. Front. Endocrinol. 10, 187 (2019).
Lagouge, M. et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1 alpha. Cell 127, 1109–1122 (2006).
Zhang, Y. et al. Recharge of chondrocyte mitochondria by sustained release of melatonin protects cartilage matrix homeostasis in osteoarthritis. J. Pineal. Res. 73, e12815 (2022).
Liu, Q. et al. Sparstolonin B suppresses rat vascular smooth muscle cell proliferation, migration, inflammatory response and lipid accumulation. Vasc. Pharmacol. 67–69, 59–66 (2015).
Deng, X. et al. The Chinese herb-derived Sparstolonin B surpresses HIV-1 transcription. Virol. J. 12, 108 (2015).
Yang, C. M. et al. Resveratrol inhibits BK-induced COX-2 transcription by suppressing acetylation of AP-1 and NF-kappa B in human rheumatoid arthritis synovial fibroblasts. Bilchem. Pharmacol. 132, 77–91 (2017).
Penolazzi, L. et al. Reciprocal regulation of TRPS1 and miR-221 in intervertebral disc cells. Cells 8 (10), 1170 (2019).
Hamamura, K., Lin, C. C. & Yokota, H. Salubrinal reduces expression and activity of MMP13 in chondrocytes. Osteoarthr. Cartil. 21 (5), 764–772 (2013).
Lian, C. et al. Collagen type II is downregulated in the degenerative nucleus pulplsus and contributes to the degeneration and apoptosis of human nucleus pulposus cells. Mol. Med. Rep. 16 (4), 4730–4736 (2017).
Feng, C. et al. ROS: Crucial Intermediators in the Pathogenesis of Intervertebral Disc Degeneration 5601593 (Oxidative Medicine and Cellular Longevity, 2017).
Gruber, H. E. et al. Mitochondrial bioenergetics, mass, and morphology are altered in cells of the degenerating human annulus. J. Orthopadic Res. 31 (8), 1270–1275 (2013).
Ding, F. et al. Role of mitochondrial pathway in compression-induced apoptosis of nucleus pulposus cells. Apoptosis 17 (6), 579–590 (2012).
Nasto, L. A. et al. Mitochondrial-derived reactive oxygen species (ROS) play a causal role in aging-related intervertebral disc degeneration. J. Orthopadic Res. 31 (7), 1150–1157 (2013).
Park, E. Y. & Park, J. B. High glucose-induced oxidative stress promotes autophagy through mitochondrial demage in rat notochordal cells. Int. Orthop. 37 (12), 2507–2514 (2013).
Park, J. S. et al. Accelerated premature stress-induced senescence of young annulus fibrosus cells of rats by high glucose-induced oxidative stress. Int. Orthop. 38 (6), 1311–1320 (2014).
Park, J. B., Byun, C. H. & Park, E. Y. Rat notochordal cells undergo premature stress-induced senescence by high glucose. Asian Spine J. 9 (4), 495–502 (2015).
Jones, D. P. Radical-free biology of oxidative stress. Am. J. Physiol.-Cell Physiol. 295 (4), C849–C868 (2008).
Trachootham, D. et al. Redox regulation of cell survival. Antioxid. Redox Signal. 10 (8), 1343–1374 (2008).
Betteridge, D. J. What is oxidative stress? Metab. Clin. Exp., 49(2, supplement1), 3–8 (2000).
Chen, G., Kroemer, G. & Kepp, O. Mitophagy: an emerging role in aging and age-associated diseases. Front. Cell. Dev. Biol. 1–15. (2020).
Saberi, M., Zhang, X. & Mobasheri, A. Targeting mitochondrial dysfunction with small molecules in intervertebral disc aging and degeneration. Gero Sci. 43, 517–537 (2021).
Chen, Y. et al. Mfn2 is involved in intervertebral disc degeneration through autophagy modulation. Osteoarthr. Cartil. 28 (3), 363–374 (2020).
Zhang, Z. et al. Parkin-mediated mitophagy as a potential therapeutic target for intervertebral disc degeneration. Cell. Death Dis. 9 (10), 1–16 (2018).
De Gregorio, E. et al. Relevance of SIRT1-NF-kB axis as therapeutic target to ameliorate inflammation in liver disease. Int. J. Mol. Sci. 21 (11), 3858 (2020).
Winter, J. M., Yadav, T., & Rutter, J. Stressed to death: mitochondrial stress responses connect respiration and apoptosis in cancer. Mol. Cell. 82, 3321–3332 (2022).
Xiang, Y. et al. Calorie restriction increases primordial follicle reserve in mature female chemotherapy-treated-rats. Gene 493 (1), 77–82 (2012).
Wu, Y. et al. Senolytics: eliminating senescent cells and alleviating intervertebral disc degeneration. Front. Bioeng. Biotechnol. 10, 1–15 (2022).
Feng, C. et al. Disc cell senescence in intervertebral disc degeneration: causes and molecular pathways. Cell. Cycle. 15 (13), 1674–1684 (2016).
Wang, F. et al. Aging and age related stresses: a senescence mechanism of intervertebral disc degeneration. Osteoarthr. Cartil. 24 (3), 398–408 (2016).
Calcinotto, A. et al. Cellular senescence: aging, cancer, and injury. Physiol. Rev. 99 (2), 1047–1078 (2019).
Yasukazu Nakahata, M. et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK mediated chromatin remodeling and circadian control. Cell 134 (2), 329–340 (2008).
Dudek, M., Swift, J., & Meng, Q. J. The circadian clock and extracellular matrix homeostasis in aging and age-related diseases. Am. J. Physiol. Cell. Physiol. 325 (1), C52–C59 (2023).
Zariel, I. et al. Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur. Cell. Mater. 30, 104–117 (2015).
Richard, C. & Scarpulla Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 1813 (7), 1269–1278 (2011).
Funding
This study was supported by Medical and Health Research Project of Zhejiang Province(NO.2022KY294, NO.2024KY301), Traditional Chinese Medicine Research Project of Zhejiang Province(NO.2024ZL951), Zhejiang Natural Fund Project (No. LQ21H090001), Natural Science Foundation of Ningbo (No. 2021J286, 202003N4235).
Author information
Authors and Affiliations
Contributions
P.S.: Review, edition, experiment, analysis, writing. Y.M.: Experiment, data collection, writing. X.Z.: Experiment, data collection, analysis. B.C.: Analysis, supervision. F.H.: Experiment, data collection. N.S.: Data collection. J.Z.: Data collection.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Guidelines
All methods were carried out in accordance with relevant guidelines and regulations.
Informed consent
This study did not involve human tissue samples or patient data. The research utilized commercially available cell lines obtained from iCell Bioscience Inc., which were ethically sourced and complied with relevant regulatory standards.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sang, P., Ma, Y., Zhang, X. et al. BMAL1 attenuates intervertebral disc degeneration by activating the SIRT1/PGC-1α pathway: evidence from vitro studies. Sci Rep 15, 9651 (2025). https://doi.org/10.1038/s41598-025-94029-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-94029-7