Abstract
To investigate the differences in gene expression and functional enrichment associated with anaplasia and laterality in retinoblastoma patients. Two datasets were selected from the GEO database of the National Center for Biotechnology Information of the US, including chip data of gene expression in tumor tissue from 36 patients with retinoblastoma. After grouping by anaplasia or laterality, the data were reanalyzed with Student’s t-test in R and Bioconductor software. The functional enrichment of genes significantly related to anaplasia and laterality was explored via GSEA. The functions of synapse assembly and synaptic signaling were enhanced in clinical patients with severe anaplasia, while the function of photoreceptors was reduced; abnormal conjugate eye movement and lipid metabolic processes were upregulated in unilateral patients. Gene expression may be different in retinoblastoma patients with different anaplasias and lateralities. The consequential difference in gene functional pathways could be associated with clinical symptoms.
Similar content being viewed by others
Introduction
Retinoblastoma (RB), which predominantly occurs in infants and children, has a global incidence ranging from 1 in 20,000 to 1 in 15,000 and has a high mortality rate1,2,3. Both the degree of anaplasia and laterality (unilateral or bilateral) significantly influence the prognosis and survival rate of RB patients4,5. Unilateral RB showcases increased promise for long-term cause-specific and other-cause survival compared to cases of bilateral RB6. Increasing severity of cellular anaplasia is also associated with high-risk histologic features and worse clinical outcomes in RB5.
The genetic mechanisms underlying the occurrence and development of RB are complex and diverse7,8,9. In 2020, Alvarez Suarez et al. reported that there was a statistically significant difference in gene expression between tumor tissues with unilateral RB and those with bilateral RB. They found that genes related to embryonic development (e.g., FPM2; MAN2A1) were more highly expressed in unilateral RB, while genes associated with visual perception and sensory perception of the light stimulus (e.g., GUCA1C; GABRR2; MYO3B) were more highly expressed in bilateral RB10. In another study, researchers found that tumors of differing anaplastic grade manifest pronounced differences in gene expression profiles. Notably, the expression of photoreceptor and nucleoporin are highly dysregulated in severe anaplasia (e.g., PDE6C; RHO)11. However, we believe that the analysis of gene data in the aforementioned studies is incomplete. For instance, the former did not examine the gene expression among tumors of differing anaplastic grade, while the latter did not conduct an analysis of the gene expression in samples based on laterality.
Therefore, we re-grouped and conducted a secondary analysis of the aforementioned gene chip data. In conjunction with the findings of previous studies, we sought to explore the differential gene expression and functional enrichment in retinoblastoma patients with different anaplasias and lateralities to understand the gene expression features which may be associated with clinical development of RB.
Materials and methods
Public data
We sourced the gene chip data from the GEO database of the National Center for Biotechnology Information (NCBI) (study numbers GSE141209 and GSE110811)10,11. GSE141209 includes chip data for 8 RB patients generated from the Affymetrix Human Transcriptome Array 2.0 detection platform. GSE110811 includes chip data for 28 RB patients generated from the Affymetrix Human Gene 2.0 detection platform.
The GSE141209 dataset included 3 unilateral patients and 5 bilateral patients, and the basic sample information is shown in Table 1. The GSE110811 dataset included 19 unilateral patients and 9 bilateral patients, and the basic sample information is shown in Table 2. The grading of anaplasia was based on the pathological grading method proposed by Mendoza et al.5.
Methods and software
Shapiro–Wilk test was performed to check the data normality for each gene. P-value > 0.05 in Shapiro–Wilk suggested acceptable normality. Gene data failed in Shapiro–Wilk test was labeled with # sign. Then the samples were divided into four different groups based on anaplasia or laterality, and Student’s t-test was used to compare the differences in gene expression between groups. The four groups were as follows:
-
1.
All GSE141209 samples were divided into a severe anaplasia group (4 patients) and a negative anaplasia group (4 patients).
-
2.
Thirteen patients with moderate anaplasia in the GSE110811 dataset were divided into a unilateral group (8 patients) and a bilateral group (5 patients).
-
3.
Seventeen unilateral patients in the GSE110811 dataset were divided into a severe anaplasia group (9 patients) and a moderate anaplasia group (8 patients).
-
4.
Nine bilateral patients in GSE110811 were divided into a moderate anaplasia group (5 patients) and a mild anaplasia group (4 patients).
Statistical analysis of the gene chip data was performed with the statistical analysis software R (version 4.0.3) and Bioconductor (version 3.12)12,13. RMA (Bioconductor Version 3.1.2) was used to process the raw microarray files, and Limma to analyze the expression difference. Genes were first ranked by adjusted P-values (FDR), then P-values less than 0.05 and fold change (log2) greater than 1.5 were used to further filter genes. Gene functional enrichment analysis was performed with GSEA (version 4.1.0)14,15. Gene transcripts selected by adjusted P-values in expression difference were compiled into ranked lists by absolute fold changes. GSEA Preranked was run on the lists with MSigDB (Version 7.2) and 1000 permutations. P-values were used to present the extent to which the biological functions were enriched. The EKit software (version 1.0.1) of Hangzhou Daohe Tianfu Biotechnology Co., Ltd. was used for automatic data processing and graphic production.
Results
Differential gene expression and function analysis between severe anaplasia and negative anaplasia patients in the GSE141209 dataset
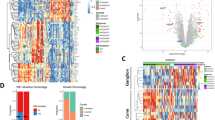
The GSE141209 dataset was regrouped based on anaplasia grade, and we compared gene expression between the severe anaplasia group and the negative anaplasia group. Among the 500 genes with the greatest changes in expression, 83 had P-values ≤ 0.05 and greater than 1.5-fold changes (log2). Functional enrichment analysis of these genes revealed that there was a statistically significant functional enrichment in the sensory perception (P = 0.033) of genes such as LRIT3, USH2A, RGS9, OR2L2, and OR2L5. These genes were significantly down-regulated in the tumor tissues of patients with severe anaplasia and are often closely related to eye diseases. The results are shown in Fig. 1 and Table 3.
The effect of laterality on gene expression in GSE110811 samples with moderate anaplasia
We divided 13 samples of patients with moderate anaplasia in the GSE110811 dataset into unilateral and bilateral disease groups, including 5 bilateral patients and 8 unilateral patients, respectively, and analyzed the gene expression and functional enrichment of different lateral samples. Among the 1000 genes with the greatest changes in expression, 95 had P-values ≤ 0.05 and a change greater than 1.5-fold (log2). Related functional enrichment included abnormal eye conjugation movement (P = 0.083) (Fig. 2a) and lipid metabolism function (P = 0.072) (Fig. 2b). The expression of related genes in unilateral samples, including SNRPN, TYRP1, NPC2, ARL13B, GK, and CBR4, was significantly greater than that in bilateral samples (P < 0.05). There was a statistically significant functional enrichment in inhibiting cell proliferation in bilateral samples (P = 0.025) (Fig. 2c), and the expression of related genes in bilateral samples, including NFIB, AKIRIN1, SOD2, MIR329-1, and MIR182#, was greater than that in unilateral samples (P < 0.05). The results are shown in Fig. 2.
The effect of anaplasia on gene expression in unilateral samples of GSE110811
We divided 17 unilateral samples from the GSE110811 dataset into a severe anaplasia group (9 samples) and a moderate anaplasia group (8 samples) and analyzed their gene expression. Among the 1000 genes with the greatest changes, 282 genes had P-values ≤ 0.05 and greater than 1.5-fold changes (log2). Table 4 shows the genes with lower expression and corresponding statistically significant vision-related functional enrichment in samples with severe anaplasia. Table 5 shows the genes with higher expression and corresponding statistically significant vision-related functional enrichment in severe anaplasia samples. Figure 3 shows the functional enrichment results related to moderate anaplasia. Figure 4 shows the functional enrichment results related to severe anaplasia.
The effect of anaplasia on gene expression in bilateral samples of GSE110811
There were 9 bilateral samples in GSE110811, including 5 from patients with moderate anaplasia and 4 from patients with mild anaplasia. Among the 1000 genes with the greatest changes, a total of 80 genes had P-values ≤ 0.05 and greater than 1.5-fold changes (log2). However, no vision-related functional enrichment was found.
Discussion
A large amount of publicly available gene chip research data related to diseases has accumulated, providing researchers with convenient research materials. Researchers can reanalyze public data from new scientific perspectives to quickly study new scientific problems16. One limitation often seen in such data, especially in clinical areas, is the limited sample size resulting in issues of statistical power. Though it is still worth to re-analyze the limited data carefully for rare diseases. In the study published recently, the authors found differences in the expression of cone and ganglion/neuronal markers of two retinoblastoma subtypes. Cone markers (such as GUCA1C, ARR3) were overexpressed in subtype 1 tumors, whereas neuronal/ganglion markers (such as PCDHB10) were overexpressed in subtype 2 tumors17. The genes and related functions were also found in our results (result III). Therefore, we believe that our research provided some real insight related to gene expression features in retinoblastoma.
As we know, heritable RB result from a germline mutation and an acquired somatic mutation, whereas non-heritable RB arise when two somatic mutations are present in the same transformation suppressor gene18. Germline pathogenic variants are more frequent in bilateral RB than in unilateral RB. Approximately, 45% of retinoblastoma patients are hereditary cases (15% unilateral and 30% bilateral) while the others are sporadic19. However, heritable RB has been reported in 5% to 15% of non-familial unilateral RB cases20,21. Previous study demonstrates the utility of grading for anaplasia in distinguishing RB patients at increased risk of developing metastasis, especially in patients without identifiable high-risk histologic features5. Given that clinical studies have shown that both anaplasia and laterality affect the clinical decision and prognosis of RB18, this study examined publicly available high-throughput gene expression data that were not fully analyzed, reorganized the samples, and compared gene expression related to anaplasia and laterality.
Analysis of the public GSE141209 dataset revealed that compared to those of unilateral patients, the tumor tissues of bilateral patients with RB were enriched in vision-related functions, and the expression of multiple related genes was increased. However, the impact of anaplasia on gene expression has not been explored10. This study compared the gene expression of negative and severe anaplasia samples in the GSE141209 dataset and revealed that the expression of functional genes related to sensory perception was inhibited in severe anaplasia samples, indicating that both anaplasia and laterality are important factors affecting clinical conditions. There were only 8 GSE141209 samples, which has certain limitations. Therefore, we further conducted research with the public dataset GSE110811.
GSE110811 revealed that different degrees of anaplasia were associated with various gene expression characteristics, and functional genes related to visual perception, such as light perception, exhibited significantly reduced expression in severe anaplasia patients. However, lateral effects have not been explored11. This study included 28 patients in the GSE110811 dataset for further analysis of the relationship between anaplasia/laterality and gene expression. Samples with the same laterality or grading of anaplasia were selected in the study to minimize the mutual interference between the two factors:
-
1.
We compared gene expression between the bilateral and unilateral groups of 13 moderate anaplasia patients and found that the expression of multiple genes affecting eye conjugation movement and lipid metabolism was enhanced in the unilateral group, while the expression of genes inhibiting cell proliferation was enhanced in the bilateral group. This finding is different from the lateral comparison results of the public dataset GSE141209, indicating an objective difference between the two public datasets.
-
2.
We compared the gene expression of 17 unilateral patients in the severe and moderate anaplasia groups and found a statistically significant decrease in the expression of genes related to sensory perception and light perception in the severe anaplasia group. This finding is similar to the results of the entire GSE110811 sample, as well as our analysis of anaplasia in the public dataset GSE141209. However, the related genes were not completely consistent. Notably, the expression of the RGS9 gene was significantly decreased in the severe anaplasia group according to both sets of public data, suggesting that RGS9 may have important functions in severe anaplasia patients.
-
3.
We compared the gene expression of 9 bilateral patients in the moderate and mild anaplasia groups and did not detect any vision-related genes or functional changes. This may be related to the small difference in anaplasia between the two groups.
The results above indicate that different anaplasias and lateralities are featured by different vision-related tumor genes in the pathogenesis of RB. This study suggested the possibility of unique functional gene features related to anaplasia and laterality, which may jointly affect the clinical development of RB. At current stage, we focused on the corresponding gene expression characteristics of different laterality and anaplasia. In future, we will explore how the gene expression is regulated and the roles of germline/somatic mutations in expression regulation. Also, the current sample sizes limited the robustness of the conclusion. Future research with larger sample size both in public data and in-house cohort is also essential to elucidate the full impact of these genetic differences on diagnosis and treatment strategies. In addition, although public data can provide research materials for new research ideas, they also have certain sample limitations. For example, in this study, we can only choose a fixed research factor to study another factor; otherwise, it is difficult to rule out the mutual influence between the two factors. However, this has important significance for the next step of research, suggesting that attention should be given to avoiding interference between anaplasia and lateral factors when collecting self-owned samples and designing experiments.
Data availability
The datasets analysed during the current study are available in the GEO database of the National Center for Biotechnology Information (NCBI) (study numbers GSE141209 and GSE110811).
References
Ortiz, M. V. & Dunkel, I. J. Retinoblastoma. J. Child Neurol. 31, 227–236. https://doi.org/10.1177/0883073815587943 (2016).
Dimaras, H. et al. Retinoblastoma. Lancet (Lond., Engl.) 379, 1436–1446. https://doi.org/10.1016/s0140-6736(11)61137-9 (2012).
Kivelä, T. The epidemiological challenge of the most frequent eye cancer: retinoblastoma, an issue of birth and death. Br. J. Ophthalmol. 93, 1129–1131. https://doi.org/10.1136/bjo.2008.150292 (2009).
MacCarthy, A. et al. Retinoblastoma in Great Britain 1963–2002. Br. J. Ophthalmol. 93, 33–37. https://doi.org/10.1136/bjo.2008.139618 (2009).
Mendoza, P. R. et al. Histopathologic grading of anaplasia in retinoblastoma. Am. J. Ophthalmol. 159, 764–776. https://doi.org/10.1016/j.ajo.2014.12.014 (2015).
Hussain, Z. Impact of laterality on cumulative survival in patients diagnosed with retinoblastoma: a retrospective cohort analysis of 1925 cases in the surveillance, epidemiology, and end results (SEER) program. Clin. Ophthalmol. 15, 991–1001. https://doi.org/10.2147/opth.S298209 (2021).
Soliman, S. E., Racher, H., Zhang, C., MacDonald, H. & Gallie, B. L. Genetics and molecular diagnostics in retinoblastoma—an update. Asia-Pac. J. Ophthalmol. 6, 197–207. https://doi.org/10.22608/apo.201711 (2017).
Liu, Y. et al. p16(INK4a) expression in retinoblastoma: a marker of differentiation grade. Diagn. Pathol. 9, 180. https://doi.org/10.1186/s13000-014-0180-1 (2014).
Dimaras, H. et al. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 17, 1363–1372. https://doi.org/10.1093/hmg/ddn024 (2008).
Alvarez-Suarez, D. E. et al. Discovery of a transcriptomic core of genes shared in 8 primary retinoblastoma with a novel detection score analysis. J. Cancer Res. Clin. Oncol. 146, 2029–2040. https://doi.org/10.1007/s00432-020-03266-y (2020).
Hudson, L. E. et al. Distinct gene expression profiles define anaplastic grade in retinoblastoma. Am. J. Pathol. 188, 2328–2338. https://doi.org/10.1016/j.ajpath.2018.06.013 (2018).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. https://doi.org/10.1186/gb-2004-5-10-r80 (2004).
Huber, W. et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat. Methods 12, 115–121. https://doi.org/10.1038/nmeth.3252 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinform. (Oxf., Engl.) 27, 1739–1740. https://doi.org/10.1093/bioinformatics/btr260 (2011).
Raddatz, B. B. et al. Microarray-based gene expression analysis for veterinary pathologists: a review. Vet. Pathol. 54, 734–755. https://doi.org/10.1177/0300985817709887 (2017).
Liu, J. et al. A high-risk retinoblastoma subtype with stemness features, dedifferentiated cone states and neuronal/ganglion cell gene expression. Nat. Commun. 12, 5578. https://doi.org/10.1038/s41467-021-25792-0 (2021).
Dimaras, H. et al. Retinoblastoma. Nat. Rev. Dis. Prim. 1, 15021. https://doi.org/10.1038/nrdp.2015.21 (2015).
DiCiommo, D., Gallie, B. L. & Bremner, R. Retinoblastoma: the disease, gene and protein provide critical leads to understand cancer. Semin. Cancer Biol. 10, 255–269. https://doi.org/10.1006/scbi.2000.0326 (2000).
Salviat, F. et al. Association between genotype and phenotype in consecutive unrelated individuals with retinoblastoma. JAMA Ophthalmol. 138, 843–850. https://doi.org/10.1001/jamaophthalmol.2020.2100 (2020).
Brichard, B. et al. Unilateral retinoblastoma, lack of familial history and older age does not exclude germline RB1 gene mutation. Eur. J. Cancer (Oxf., Engl. 1990) 42, 65–72. https://doi.org/10.1016/j.ejca.2005.07.027 (2006).
Funding
This research was supported by the Natural Science Foundation of Hubei Province (Grant No.2022CFB182).
Author information
Authors and Affiliations
Contributions
W.L. reorganized the database and conducted an analysis of gene expression. KK.X. was a major contributor in writing the manuscript. F.C. conceived the idea for the experiment and reviewed the results. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, W., Xu, K. & Chen, F. Gene expression study in retinoblastoma patients with different anaplasias and lateralities. Sci Rep 15, 9925 (2025). https://doi.org/10.1038/s41598-025-94225-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-94225-5