Abstract
This study explores the mechanisms of Astragaloside A (AS-A), a significant active ingredient in Astragalus, This traditional Chinese medicine is both a medication and a food, combating lung adenocarcinoma using network pharmacology, molecular docking, molecular dynamics, and experimental validation. A protein–protein interaction (PPI) network was developed, identifying 10 key targets, including STAT3 and AKT1. GO and KEGG enrichment analyses indicated that these targets primarily participated in biological processes and pathways, including oxidative stress and the PI3K-Akt signalling pathway. Molecular docking and dynamic simulation evaluated AS-A's binding mode and stability with key targets. In molecular docking, 14 key targets of the HIF-1 signalling pathway had different binding energies with AS-A, such as the binding energy of PIK3R1 being -9.3. Kinetic simulations indicated the stability of the protein–ligand complex, as evidenced by RMSD values ranging from 0.2 to 0.4 nm. RMSF analysis showed that the protein residue flexibility characteristics were stable, the Rg values were stable, the number of hydrogen bonds was 10–20, and the solvent-accessible surface area was stable. Cell experiments showed that AS-A could regulate the expression of key signalling molecules such as STAT3 and AKT in lung adenocarcinoma models. This study provides insights into the mechanism of AS-A in treating lung adenocarcinoma. It proposes a new direction for anticancer research in traditional Chinese medicines, especially medications and foods.
Similar content being viewed by others
Introduction
Lung adenocarcinoma, a form of non-small cell lung cancer, poses significant treatment challenges due to its aggressive nature and the development of drug resistance1. Recent investigations have emphasized the potential of compounds derived from traditional Chinese medicine (TCM), mainly the active ingredients in medicinal and edible plants like Astragaloside A from Astragalus, in cancer therapy. Astragalus, a plant with both medicinal and edible value, yields Astragaloside A, a saponin extract recognized for its diverse pharmacological effects, including anti-inflammatory, immunomodulatory, and anticancer properties2.
To elucidate the regulatory mechanisms of Astragaloside A, an essential active ingredient in medicinal and edible plants, on lung adenocarcinoma, a network pharmacological approach can be employed3. This method integrates bioinformatics tools to identify the active compounds, their targets, and the associated signalling pathways4. By constructing a compound-target-disease network, researchers can visualize the interactions between Astragaloside A and its potential targets in lung adenocarcinoma, thereby gaining deeper insights into the mechanisms of medicinal and edible plants in cancer treatment.
Utilize the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and Swiss Target Prediction to identify the targets of Astragaloside A. Lung adenocarcinoma-related targets can be obtained from resources like GeneCards and OMIM5. Identifying the intersection of these targets will highlight the key proteins that Astragaloside A may affect in lung adenocarcinoma. Protein–protein interaction (PPI) network analysis can pinpoint crucial hub genes involved in disease progression. Enrichment analysis, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), offers insights into the biological processes and pathways influenced by Astragaloside A6. Astragaloside A can impact cancer progression by modulating PI3K/Akt and MAPK signalling pathways.
Given Astragaloside A’s potential antitumor activity and network pharmacology’s power in understanding drug-disease interactions, exploring the regulatory mechanism of Astragaloside A on lung adenocarcinoma based on network pharmacology holds great promise7,8. This study employs network pharmacology to focus on the regulatory effects of Astragaloside A, derived from the medicinal and edible plant Astragalus, on lung adenocarcinoma, potentially deepening the understanding of its pathogenesis and assisting in the development of novel therapeutic strategies9.
Material and methods
Screening of targets for Astragaloside A
Astragaloside A (AS-A) was chosen as the target compound, and its 2D structure was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The SMILES structure of AS-A was then input into various target prediction databases, including Swiss Target Prediction (http://www.swisstargetprediction.ch/), SEA Search Server (https://sea.bkslab.org/), and SuperPred (https://prediction.charite.de/). The 2D structure of AS-A was imported into the PharmMapper database (https://www.lilab-ecust.cn/pharmmapper/) to consolidate the predicted targets.After removing duplicates, the final list of potential targets for AS-A was obtained.
Screening of lung adenocarcinoma targets
Lung adenocarcinoma targets were identified using the GeneCards, OMIM, and CTD databases with the keyword 'lung adenocarcinoma.' Targets with a Relevance Score of at least 50 were selected from GeneCards, and those with a Reference Count of 15 or more were filtered from CTD. The final set of lung adenocarcinoma-related targets was obtained by merging the results and removing duplicates.
Identification of common targets of AS-A and lung adenocarcinoma
The event tool (https://jvenn.toulouse.inrae.fr/app/index.html) was used to identify common targets between AS-A and lung adenocarcinoma, highlighting potential molecular interaction points that AS-A might regulate in lung adenocarcinoma.
Development of a protein–protein interaction network linking AS-A with lung adenocarcinoma targets
The protein–protein interaction (PPI) network for AS-A and lung adenocarcinoma targets was developed using the STRING database (https://string-db.org), specifying human species and a medium confidence interaction score threshold of 0.400. Other parameters were kept at their default settings, and the resulting PPI network was analyzed to identify interactions between targets. The CentiScaPe module in Cytoscape (v3.7.0) was utilized to calculate the Matthews correlation coefficient (MCC), where higher scores indicate improved predictive accuracy and a stronger correlation between actual and predicted values. The key targets were identified as the top 10, and the network visualization was performed using Cytoscape software.
Functional enrichment and pathway network construction of AS-A targets in lung adenocarcinoma
The Metascape database (https://metascape.org) was used to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on AS-A targets in lung adenocarcinoma, with the species specified as human. The top 10 significantly enriched biological processes, cellular components, molecular functions, and KEGG pathways were selected and visualized based on p-values less than 0.05. Additionally, Cytoscape software was used to visualize the interactions between AS-A targets and the relationship between targets and signalling pathways for further analysis.
Molecular docking of AS-A with key targets
The 2D structure of AS-A was sourced from the PubChem database and processed in Chem3D for energy minimization and hydrogenation. The 3D X-ray crystal structure of key targets was obtained from the Protein Data Bank (PDB; https://www.rcsb.org/). The pure protein structure was extracted using PyMOL (Version 2.5.2) and imported into AutoDock Tools software (Version 1.5.6), and the structure was neutralized, hydrogenated, and non-standard amino acids were removed. The entire protein structure was set as the binding pocket for molecular docking. The resulting protein–ligand complex was visualized in 2D and 3D using LigPlus (version 2.2) and PyMOL software.
Molecular dynamics simulation
Perform molecular dynamics simulations on the Astragaloside A-protein complex with the highest docking score. The complexes 100 ns molecular dynamics simulations (MD) were performed using Gromacs 2022 software. Charmm 3610 was selected as the protein force field, Gaff2 was chosen as the ligand force field, and the TIP3P water model was selected to add solvents to the protein–ligand system and to create a water box with a periodic boundary of 1.2 nm11. The particle grid Ewald (PME) and Verlet algorithms deal with electrostatic interactions. Then 100,000 steps of isothermal isovolumetric ensemble equilibrium and isothermal isobaric ensemble equilibrium were simulated with a coupling constant of 0.1 ps and a duration of 100 ps. Both van der Waals and Coulomb interactions are calculated using 1.0 nm cutoff values. Finally, the system was simulated using Gromacs 2022 at constant temperature (310 K) and constant pressure (1 bar) for 100 ns.
MMPBSA binding free energy calculation
The free energy of the final state was computed by employing the Molecular Mechanics/Poisson—Boltzmann Surface Area (MMPBSA) method. Molecular dynamics trajectory data, obtained from previous simulations, were input into the gmx MMPBSA program, a component of the GROMACS software suite, to conduct these calculations12.
Cell culture and treatment
Human non-small lung cancer A549 cells were obtained from Shanghai Fuheng Biotechnology Co., Ltd. in China. A549 human lung cancer cells were maintained in 25 cm2 flasks using MEM with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin, incubated at 37 °C in an atmosphere of 95% air and 5% CO2.
RNA extraction and quantitative real-time PCR (qRT-PCR)
According to the manufacturer’s protocol, total RNA was isolated from cells using 1 mL of TRIzol reagent (Invitrogen). The solution was mixed vigorously for 15 s after adding 0.2 mL of chloroform, incubated at room temperature for 5 min, and then centrifuged at 12,000 rpm for 10 min at 4 °C. The aqueous phase was mixed with isopropanol (0.5 mL) to precipitate RNA. The RNA pellet was washed twice with 75% ethanol, air-dried, dissolved in 20–50 μL of DEPC-treated water, and stored at -80 °C.cDNA was synthesized by reverse transcribing 1 μg of RNA in a 20 μL reaction using All-in-One First-Strand Synthesis MasterMix (Applied Biosystems), according to the manufacturer’s instructions. The reaction conditions were 37 °C for 2 min, 55 °C for 15 min, and 85 °C for 5 min. The cDNA was stored at -20 °C.SYBR Green qPCR Premix (Takara) was utilized for qRT-PCR on an ABI 7500 system. The 20 μL reaction mixture comprised 10 μL of SYBR Green Premix, 0.4 μL each of forward and reverse primers (10 μM), 3 μL of cDNA, and 6.2 μL of RNase-free water. The cycling protocol involved 95 °C for 30 s, then 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The 2-ΔΔCt method was used to normalize gene expression to β-actin. The primers were synthesized by Hefei Yuanen Biotechnology Co., Ltd. (Hefei, China) and are listed in Table 1.
Statistical analysis
Experiments were conducted in triplicate, with results expressed as mean ± standard deviation (SD). GraphPad Prism software (version 8.1.1) was used for statistical analysis. The significance of differences between groups was assessed using one-way ANOVA followed by Tukey’s multiple comparison tests. A two-tailed unpaired Student’s t-test compared the two groups. P values less than 0.05 were considered statistically significant.
Results
Target identification of AS-A and lung adenocarcinoma
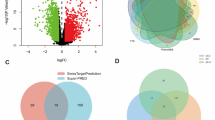
A total of 325 potential targets of AS-A were identified through various databases, including 106 targets from SwissTarget Prediction, 1 from SEA, and 61 from SuperPred (after merging and removing duplicates). For lung adenocarcinoma, 1147 potential targets were identified, including 461 targets from GeneCards, 475 from OMIM, and 364 from CTD (also after removing duplicates). Figure 1 illustrates 64 shared targets between AS-A and lung adenocarcinoma.
PPI network analysis of AS-A targets in lung adenocarcinoma
The STRING database was utilized to construct the PPI network of AS-A targets in lung adenocarcinoma, applying a minimum interaction confidence score of 0.400. After merging the data, 64 nodes and 741 edges were identified in the network. The network exhibited an average node degree of 23.2, an average local clustering coefficient of 0.678, and a PPI enrichment p-value of < 1.0e-16, demonstrating significant enrichment. The MCC of each target was calculated using the CentiScaPe module in Cytoscape. According to the MCC score ranking, the top 10 key targets were STAT3, AKT1, HSP90AA1, MTOR, NFKB1, ESR1, MDM2, HIF1A, EP300, and AR. Table 2 shows the MCC scores of these top-ranked proteins, and the PPI network visualization and the interaction map of the top 10 proteins are shown in Fig. 2. These results highlight the key roles of these proteins in the AS-A-lung adenocarcinoma interaction network.
GO and KEGG enrichment analysis of lung adenocarcinoma targets of AS-A
GO and KEGG pathway enrichment analyses were conducted on 64 lung adenocarcinoma targets of AS-A utilizing the microbial information database. The results revealed significant enrichment in BPs related to oxidative stress, protein kinase B signalling, and chemical stress responses. CCs analysis highlighted key locations such as the ruffle, vesicle lumen, and protein kinase complexes. MFs were enriched in protein kinase activity, hormone receptor binding, and transcription factor binding. The KEGG pathway analysis identified significant pathways such as PI3K-Akt signalling, Proteoglycans in cancer, HIF-1 signalling, and PD-L1/PD-1 checkpoint. The detailed enrichment results of GO and KEGG are shown in Fig. 3.
Network Analysis of AS-A in lung Adenocarcinoma: A compound-target pathway network was constructed to elucidate the interactions and mechanisms of AS-A in lung adenocarcinoma
A compound-target-pathway network was constructed to illustrate the interaction between AS-A, its 64 lung adenocarcinoma targets, and the top 10 most significantly enriched KEGG signalling pathways. Figure 4 illustrates the network visualized with Cytoscape software. In the network, AS-A is represented by a green diamond, while the targets not associated with the top 10 pathways are shown as blue circles. The pink circles represent targets linked to the top 10 KEGG pathways, and the red triangles represent these KEGG pathways. This network underscores the primary pathways AS-A potentially influences lung adenocarcinoma treatment, notably involving the PI3K-Akt signalling, HIF-1 signalling, PD-L1/PD-1 checkpoint pathway, and Proteoglycans in cancer. These findings provide insights into the potential molecular mechanisms of the effects of AS-A in regulating lung adenocarcinoma.
The green node in the center represents Astragaloside A. Pink nodes represent targets related to Astragaloside A in lung adenocarcinoma. Blue nodes represent various signalling pathways associated with these targets, such as the FoxO signalling pathway, HIF-1 signalling pathway, PI3K-Akt signalling pathway. Lines connecting the nodes indicate the interactions between astragaloside A, its targets, and the related pathways.
Molecular docking of key targets in the HIF-1 signaling pathway
The HIF-1 signalling pathway was selected for further molecular docking analysis, as shown in Fig. 5. The 14 key targets in the HIF-1 signalling pathway (PIK3R1,AKT1, EGFR, CREBBP, MTOR, TLR4, PIK3CA, AKT2, STAT3, SERPINE1, VEGFA, HIF1A, NFKB1, EP300) were submitted to the PDB database. The three-dimensional structure of the human protein bound to the ligand with the lowest resolution was chosen for each target. All protein structure Remove the chains that do not belong to the protein, remove water, Add hydrogen, neutralize charges, and eliminate non-standard amino acids; Since there is no co-crystal ligand within PIK3R1, MTOR, VEGFA, HIF1A, NFKB1, the entire protein structure was used as the docking pocket for blind docking with Astragaloside A. a blind docking was performed with Astragaloside A and AKT1, EGFR, CREBBP, TLR4, PIK3CA, AKT2, STAT3, SERPINE1, EP300 have co-crystal ligand, so the box coordinates and size of the small ligand binding pocket were obtained using the GetBox plugin in PyMOLand docking of the specific ligand’s pocket. The results of the molecular docking analysis are summarized in Table 3, and the interaction diagram is shown in Fig. 6 (A-M). These docking results provide an in-depth understanding of the binding affinity and interaction pattern between key proteins in the HIF-1 signalling pathway and potential small molecule ligands, laying the foundation for understanding the therapeutic potential of targeting the HIF-1 signalling pathway in lung adenocarcinoma.
Molecular docking interaction diagram between Astragaloside A and key targets in the HIF-1 signalling pathway. Diagram of the docking interaction between: (A) Astragaloside A and PIK3R1, (B)Astragaloside A and AKT1, (C) Astragaloside A and EGFR, (D) Astragaloside A and CREBBP, (E) Astragaloside A and MTOR, (F) Astragaloside A and TLR4, (G) Astragaloside A and PIK3CA, (H) Astragaloside A and AKT2, (I) Astragaloside A and STAT3, (J) Astragaloside A and SERPINE1, (K) Astragaloside A and VEGFA, (L) Astragaloside A and HIF1A, (M) Astragaloside A and NFKB1. In the depicted figure, the gray regions illustrate the protein structure. The pink moieties correspond to Astragaloside A. Meanwhile, the green-colored components represent the amino acids thatwith Astragaloside A and engage in hydrogen-bonding interactions, highlighting the specific molecular interactions at the atomic level. respectively. Binding modes of the drugs were rendered with the help of the academic version of PyMOL 2.x (https://pymol.org/educational v2.x) and LigPlot + v.2.2 (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/) packages.
Molecular dynamics simulation results
Perform molecular dynamics (MD) simulations on the complex of Astragaloside A and PIK3R1. Molecular dynamics simulations evaluated the temporal stability and interactions of the protein–ligand complex. The RMSD plot (Fig. 7A) demonstrates the stability of the small molecule, protein, and complex, with RMSD values ranging from 0.2 to 0.4 nm, suggesting a relatively stable conformation during the simulation. Figure 7B's root mean square fluctuation (RMSF) analysis illustrates the flexibility of individual protein residues. Most residues show slight fluctuation, except for a few residues near the C-terminus that exhibit higher flexibility. The radius of gyration (Rg) plot (Fig. 7C) shows the compactness of the protein–ligand complex over time, with relatively stable Rg values, indicating that no significant structural expansion or contraction occurred during the simulation. Hydrogen bond analysis (Fig. 7D) reveals that the ligand–protein hydrogen bonds fluctuate before stabilizing between 10 and 20, suggesting consistent interactions during the simulation. The solvent-accessible surface area (SASA) analysis (Fig. 7E) indicates stability in the total accessible surface area, with consistent values for both hydrophobic and hydrophilic regions that suggests that the protein–ligand complex consistently interacts stably with the solvent environment. The molecular dynamics simulations demonstrate that the protein–ligand complex remains stable with significant interactions.
Molecular dynamics simulation results for the Astragaloside A and PIK3R1 complex. A RMSD of Astragaloside A and PIK3R1 for 100 ns. B RMSF value of Astragaloside A and PIK3R1 for 100 ns. C Rg of Astragaloside A and PIK3R1 for 100 ns. D Hydrogen bond of Astragaloside A and PIK3R1 for 100 ns. E SASA value for all the systems of Astragaloside A and PIK3R1 for 100 ns.
MMPBSA binding free energy calculation
The binding free energy of the small molecule to the target protein was computed by MM/PBSA method using the binding conformation of the complex. The free energy of binding of the target protein to the small molecule was -34.381 kJ/mol. Negative values indicate that the molecule has a binding affinity for the target protein, and lower values indicate stronger binding. Therefore, both the small molecule and the target protein have high affinity. Subsequently, this study further computationally analyzed the amino acids that contribute significantly to the binding of the small molecule in the complex system. The results revealed that residues PRO418, TYR416, PHE384, and LYS419 contributed significantly to the binding interaction within the complex system (Fig. 8). This suggests that these amino acid residues may play an important role in the catalytic process.
Astragaloside A regulates key signalling pathways in the lung adenocarcinoma model
The expression levels of several key signalling molecules were measured in the regular lung adenocarcinoma model, and AS-A-treated groups to evaluate the therapeutic effects of AS-A. The bar graphs(Fig. 9) demonstrate significant upregulation of STAT3, AKT, and MTOR expression levels in the model group relative to the standard group, suggesting activation of these pathways under pathological conditions. AS-A treatment significantly reduced the expression of these molecules, demonstrating its effectiveness in mitigating the overactivation of the signalling pathways. Similarly, the levels of MDM2 and HIF1A were increased in the model group, consistent with increased tumour progression and hypoxia-related responses. AS-A treatment significantly reduced the expression of these markers, suggesting its potential to inhibit pathological processes linked to these molecules. Furthermore, the expression of AR, HSP90AA1, and NFKB1 was significantly increased in the model group, reflecting the abnormal activation of inflammation- and androgen-related signals. AS-A treatment reduced the expression of these molecules, indicating a partial therapeutic effect. Interestingly, the expression of ESR1 and EP300 was decreased in the model group, indicating disrupted estrogen receptor signalling and transcriptional regulation.AS-A treatment effectively restored the expression of these markers to levels closer to those of the regular group, suggesting that disease-associated alterations were reversed.
Effects of Astragaloside A on the expression of key signalling molecules in lung adenocarcinoma. It represents the relative expression levels of STAT3, AKT, MTOR, MDM2, HIF1A, AR, HSP90AA1, NFKB1, ESR1, and EP300 in three groups: standard (black bars), model (white bars), and Astragaloside A treatment (gray bars). Data are expressed as mean ± SD. Compared with the model group, Astragaloside A treatment significantly reduced the expression of STAT3, AKT, MTOR, MDM2, HIF1A, AR, HSP90AA1, and NFKB1, while restoring the levels of ESR1 and EP300 to levels closer to those of the standard group. These findings suggest that Astragaloside A regulates dysregulated signalling pathways associated with the lung adenocarcinoma model.
Discussion
Astragaloside does have a therapeutic effect on lung adenocarcinoma. Astragalus, a plant with a long history in traditional Chinese medicine, is renowned for its remarkable medicinal value and particularly unique feature due to its wide application in daily diet. As one of the main active ingredients of Astragalus, Astragaloside has shown anti-tumour activity, which not only provides new ideas for the treatment of lung adenocarcinoma but also further validates the value of the concept of "homology of medicine and food" in modern medicine. As a medicine-food homologous drug, Astragalus is characterized by high safety, easy availability and convenient use, making it a highly potential candidate for natural anti-cancer drugs. Research indicates that Astragaloside IV (AS-IV) inhibits tumour cell migration and the epithelial-mesenchymal transition (EMT), exerting anti-tumour effects. AS-IV effectively suppresses M2 macrophage polarization, diminishing tumour cell migration and invasion by inhibiting the TGF-β/Smad2/3 signaling pathway16,17,18. In addition, Astragalus polysaccharide (APS) has also shown antitumour activity against lung adenocarcinoma cells, reversing resistance to gefitinib and enhancing apoptosis by inhibiting epithelial-mesenchymal transformation19. Another study discovered that Astragaloside IV could hinder the advancement of hepatocellular carcinoma and maintain fibrosis development by modulating the pSmad3C/3L and Nrf2/HO-1 signalling pathways. The results indicate that Astragaloside and its derivatives may be applicable in treating lung adenocarcinoma.
This study successfully unveiled the interaction between AS-A and its 64 lung adenocarcinoma targets and the top 10 most significantly enriched KEGG signaling pathways by constructing a compound target pathway network20. This finding provides essential insights into the mechanism of AS-A in lung adenocarcinoma, carrying substantial theoretical and clinical significance21,22,23. In the network, the AS-A is represented by a green diamond, targets unrelated to the first 10 pathways are denoted by a blue circle, targets connected to the first 10 KEGG pathways are symbolized by a pink circle, and a red triangle represents the KEGG pathways themselves are distinguishable, facilitating further analysis24. These key pathways provide a clear direction for subsequent studies to further explore the regulatory mechanisms of AS-A on lung adenocarcinoma25. The PI3K-Akt pathway regulates cell growth, proliferation, survival, and metabolism26. AS-A may affect lung adenocarcinoma cell behaviour by modulating the PI3K-Akt signalling pathway, indicating a potential therapeutic target for this cancer type27,28,29. The HIF-1 pathway is crucial for tumour adaptation to hypoxia. The effect of AS-A on this pathway may contribute to improving the hypoxic microenvironment of lung adenocarcinoma tissue, thereby inhibiting tumour growth and progression30.
This study further focused on the HIF-1 signalling pathway to explore its potential role in treating lung adenocarcinoma31. The HIF-1 signalling pathway is essential for cellular adaptation to low oxygen levels and tumour development32. Therefore, we selected 14 key targets in the HIF-1 signalling pathway (MTOR, AKT1, PIK3R1, AKT2, TLR4, EGFR, VEGFA, HIF1A, CREBBP, PIK3CA, STAT3, SERPINE1, NFKB1, and EP300) for molecular docking analysis33,34,35,36,37,38,39,40,41,42,43,44,45,46,47. We optimized the structures of the proteins and ligands and performed molecular docking analysis48. These targets are crucial in the onset and progression of various cancers. HIF-1α, as a hypoxia-inducing factor, regulates the metabolism and growth of tumour cells49,50,51,52. Activation of the STAT3 and AKT pathways is closely related to tumour proliferation and survival. HSP90AA1 is an important chaperone regulating various various signalling pathways, including NF-κB and HIF-1α stability. The MTOR pathway is crucial for cell growth and metabolism, particularly in cancer cells. Astragaloside may significantly contribute to anti-tumour therapy by regulating these targets.
RMSD values in the range of 0.2–0.4 nm indicate high stability of protein–ligand complexes over simulated timescales, which has significant implications for drug design53. As can be seen in Fig. 7D, the number of hydrogen bonds fluctuates significantly over time. The formation and breaking of hydrogen bonds can affect the interactions between molecules, The interactions between solvent molecules and solute molecules also affect the system’s stability. The change in the solvent-accessible surface area (Figure E) indicates that the interactions between the solvent and solute are constantly adjusting, which may indirectly lead to fluctuations in the 25 ns RMSD.RMSF analysis revealed the flexibility of residues in proteins. these flexible regions can be optimized in drug design to improve ligands’ binding affinity and specificity to proteins54. The Rg diagram shows that the compactness of the complex is relatively stable, with no apparent structural expansion or contraction. The stable structure helps to reduce potential energy loss and conformational changes, thereby improving the stability of the complex. In drug design, it is possible to consider designing ligands with specific structures to maintain the compact structure of the complex and enhance its biological activity55. Hydrogen bonds are crucial in protein–ligand interactions and are a key reference for designing high-affinity ligands56. The drug’s efficacy can be improved by optimizing the structure of the ligand and increasing the number and stability of hydrogen bonds with the protein. SASA analysis indicated that the protein–ligand complex consistently interacted stably with the solvent environment57. The solvent environment has an essential influence on the stability and bioactivity of the complex. Stable solvent interaction helps maintain the structure and function of the complex58.
The MM/PBSA—based binding free energy calculation for the Astragaloside A—PIK3R1 complex reveals a high binding affinity with a magnitude of—34.381 kJ/mol59, indicating a stable association as negative magnitudes indicate binding propensity and lower magnitudes mean stronger binding60. Analysis of amino—acid contributions shows that PRO418, TYR416, PHE384, and LYS419 have high contribution magnitudes, likely playing a key role in the catalytic process61. These residues may be crucial for stabilizing the complex through interactions like hydrogen—bonding or electrostatic forces, providing potential hotspots for studying the interaction mechanism, which is important for drug design and new therapeutic strategies62.
This study assessed AS-A's therapeutic impact by measuring key signalling molecule expression levels across the standard, lung adenocarcinoma model, and AS-A treatment groups. It provides new insights and potential therapeutic strategies for treating lung adenocarcinoma63. Elevated levels of MDM2 and HIF1A in the model group are consistent with accelerated tumour progression and hypoxia-related responses, which align with the pathological features of lung adenocarcinoma64. However, the expression of these markers is significantly reduced by AS-A treatment, suggesting that AS-A can effectively alleviate the overactivation of the signalling pathways associated with these molecules and may have an inhibitory effect on the development of lung adenocarcinoma58,63. This finding provides critical clues for further research on the mechanism of AS-A therapy for lung adenocarcinoma65. These findings not only provide a theoretical basis for applying AS-IV treating lung adenocarcinoma, but also further emphasize the potential of medicine-food homologous drugs in cancer treatment. It is worth noting that as a medicine-food homologous drug, the curative effect of Astragalus is not achieved overnight. Instead, long-term and appropriate intake is required to achieve the best results which coincides with the concept of "preventive treatment of diseases" in traditional Chinese medicine, preventing diseases and strengthening the body through daily dietary conditioning.
Conclusion
This study, leveraging network pharmacology, delves deeply into the regulatory mechanism of astragaloside A on lung adenocarcinoma. This research not only highlights the medicinal value of Astragalus but also further confirms its unique advantages as a medicine-food homology substance. This discovery offers us novel therapeutic insights and opens up new directions for cancer treatment research based on the traditional Chinese medicine theory of medicine-food homology. This dual functionality broadens its application prospects in cancer prevention and treatment, providing robust support for developing more medicine-food homology products with medicinal value.
Data availability
Data is provided within the manuscript.
References
Auyeung, K. K. et al. Astragalus saponins modulate cell invasiveness and angiogenesis in human gastric adenocarcinoma cells. J. Ethnopharmacol. 141, 635–641. https://doi.org/10.1016/j.jep.2011.08.010 (2012).
Li, L. et al. Astragaloside IV enhances the sensibility of lung adenocarcinoma cells to bevacizumab by inhibiting autophagy. Drug Dev. Res. 83, 461–469. https://doi.org/10.1002/ddr.21878 (2022).
Zhang, X. et al. Systems pharmacology unravels the synergic target space and therapeutic potential of Rhodiola rosea L. for non-small cell lung cancer. Phytomedicine 79, 153326. https://doi.org/10.1016/j.phymed.2020.153326 (2020).
Zheng, S. et al. Anti-inflammatory mechanism research of tanshinone II A by module-based network analysis. Biomed. Mater. Eng. 24, 3815–3824. https://doi.org/10.3233/BME-141211 (2014).
Song, W. et al. Uncovering the mechanism of maxing ganshi decoction on asthma from a systematic perspective: A network pharmacology study. Sci. Rep. 8, 17362. https://doi.org/10.1038/s41598-018-35791-9 (2018).
Li, A. P. et al. Uncovering the mechanism of astragali radix against nephrotic syndrome by intergrating lipidomics and network pharmacology. Phytomedicine 77, 153274. https://doi.org/10.1016/j.phymed.2020.153274 (2020).
Hao, J. et al. Metabolomics combined with network pharmacology reveals the protective effect of astragaloside IV on alcoholic liver disease. Phytomedicine 135, 156032. https://doi.org/10.1016/j.phymed.2024.156032 (2024).
Wang, H. et al. Revealing the active ingredients and mechanisms of xiatianwu against hepatocellular carcinoma: a study based on network pharmacology and bioinformatics. Naunyn Schmiedebergs Arch. Pharmacol. https://doi.org/10.1007/s00210-024-03278-2 (2024).
Chen, D. et al. Network pharmacology and experimental validation to reveal the pharmacological mechanisms of astragaloside IV in treating intervertebral disc degeneration. Eur. J. Pharmacol. 982, 176951. https://doi.org/10.1016/j.ejphar.2024.176951 (2024).
Jo, S. et al. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865. https://doi.org/10.1002/jcc.20945 (2008).
Mark, P. et al. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 105, 9954–9960. https://doi.org/10.1063/1.4872239 (2001).
Kumari, R. et al. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 54, 1951–1962. https://doi.org/10.1021/ci500020m (2014).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Gao, P. et al. Exploring the Mechanism of hepatotoxicity induced by dictamnus dasycarpus based on network pharmacology molecules molecular docking and experimental. Pharmacology https://doi.org/10.3390/molecules28135045 (2023).
Zhang, Y. C. et al. Network pharmacology and molecular docking analysis on molecular targets and mechanisms of fei jin sheng formula in the treatment of lung cancer. Curr. Pharm. Des. 29, 1121–1134. https://doi.org/10.2174/1381612829666230503164755 (2023).
Wu, J. et al. Network pharmacology-based study on the mechanism of action of Trollius chinensis capsule in the treatment of upper respiratory tract infection. Medicine (Baltimore) 103, e35529. https://doi.org/10.1097/MD.0000000000035529 (2024).
Wang, Z. et al. To study the mechanism of scutellariae radix and astragaloside in the treatment of lung cancer based on network pharmacology. Med. (Baltimore) 101, e29729. https://doi.org/10.1097/MD.0000000000029729 (2022).
Kim, Y. W. et al. Systematic exploration of therapeutic effects and key mechanisms of panax ginseng using network-based approaches. J. Ginseng Res. 48, 373–383. https://doi.org/10.1016/j.jgr.2024.01.005 (2024).
Li, R. et al. Study on the mechanism of anti-acute lung injury of Shuanghuanglian oral liquid based on identification of transitional components in blood and network pharmacology. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 1212, 123498. https://doi.org/10.1016/j.jchromb.2022.123498 (2022).
Fang, T. et al. Caizhixuan hair tonic regulates both apoptosis and the PI3K/Akt pathway to treat androgenetic alopecia. PLoS One 18, e0282427. https://doi.org/10.1371/journal.pone.0282427 (2023).
Huang, L. P. et al. Platycodin D relieves rheumatoid arthritis by promoting apoptosis of mitochondria to inhibit activation of hedgehog pathway. Autoimmunity 56, 2205053. https://doi.org/10.1080/08916934.2023.2205053 (2023).
Liu, J. et al. Mechanistic insights into xanthomicrol as the active anti-HCC ingredient of phytolacca acinosa roxb a network pharmacology analysis and transcriptomics integrated experimental verification. J. Ethnopharmacol. 333, 118467. https://doi.org/10.1016/j.jep.2024.118467 (2024).
Zhang, S. Q. et al. Identification of the active compounds and significant pathways of artemisia annua in the treatment of non-small cell lung carcinoma based on network pharmacology. Med. Sci. Monit. 26, e923624. https://doi.org/10.12659/MSM.923624 (2020).
Lee, I. Y. et al. Astragalus polysaccharides and metformin may have synergistic effects on the apoptosis and ferroptosis of lung adenocarcinoma A549 cells. Curr. Issues Mol. Biol. 46, 7782–7794. https://doi.org/10.3390/cimb46080461 (2024).
Lin, W. et al. Network pharmacology study of the hepatoprotective effects of quercetin-containing traditional chinese medicine, anoectochilus roxburghii, and validation of quercetin as an anti-liver injury agent in a mouse model of liver injury. Med. Sci. Monit. 26, e923533. https://doi.org/10.12659/MSM.923533 (2020).
Yao, J. et al. L-ascorbate alleviates chronic obstructive pulmonary disease through the EGF/PI3K/AKT signaling axis. Curr. Med. Chem. https://doi.org/10.2174/0109298673302394240823114448 (2024).
Zhang, K. et al. Exploring the mechanism of zilongjin in treating lung adenocarcinoma based on network pharmacology combined with experimental verification. Crit. Rev. Immunol. 44, 27–40. https://doi.org/10.1615/CritRevImmunol.2024051316 (2024).
Jia, L. et al. Astragaloside IV inhibits the progression of non-small cell lung cancer through the Akt/GSK-3beta/beta-catenin pathway. Oncol. Res. 27, 503–508. https://doi.org/10.3727/096504018X15344989701565 (2019).
Yuan, S. et al. Integrating network pharmacology and experimental validation to explore the pharmacological mechanism of astragaloside IV in treating bleomycin-induced pulmonary fibrosis. Drug Des. Devel. Ther. 17, 1289–1302. https://doi.org/10.2147/DDDT.S404710 (2023).
Liu, X. H. et al. Component analysis and structure identification of active substances for anti-gastric ulcer effects in radix astragali by liquid chromatography and tandem mass spectrometry. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 960, 43–51. https://doi.org/10.1016/j.jchromb.2014.04.020 (2014).
Wang, Y. C. et al. Mechanism of key ingredient of astragalus membranaceus on lung adenocarcinoma via PI3K/AKT signaling clarified by utilizing network pharmacology approach and experimental validation. Chin. J. Integr. Med. 29, 244–252. https://doi.org/10.1007/s11655-022-3681-x (2023).
Feng, Y. Y. et al. Network pharmacology based elucidation of molecular mechanisms of laoke formula for treatment of advanced non-small cell lung cancer. Chin. J. Integr. Med. 30, 984–992. https://doi.org/10.1007/s11655-024-3717-5 (2024).
Priyamvada, P. et al. Unravelling the molecular mechanistic pathway underlying the anticancer effects of kaempferol in colorectal cancer: A reverse pharmacology network approach. Mol. Divers https://doi.org/10.1007/s11030-024-10890-0 (2024).
Chen, Y. et al. Network pharmacology-based investigation of protective mechanism of aster tataricus on lipopolysaccharide-induced acute lung injury. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20030543 (2019).
Li, A. et al. Exploration of the main effective constituent and the mechanism in astragali radix in the treatment for doxorubicin-induced nephropathy by integrating metabolomics and molecular docking. J. Ethnopharmacol. 305, 116074. https://doi.org/10.1016/j.jep.2022.116074 (2023).
Bing, P. et al. Study on the mechanism of astragalus polysaccharide in treating pulmonary fibrosis based on “Drug-Target-Pathway” network. Front. Pharmacol. 13, 865065. https://doi.org/10.3389/fphar.2022.865065 (2022).
Cao, B. et al. Mechanisms exploration of xiaojin pills on lung cancer based on metabolomics and network pharmacology. J. Pharm. Pharmacol. 73, 1071–1079. https://doi.org/10.1093/jpp/rgab050 (2021).
Fan, Q. et al. Optimizing component formula suppresses lung cancer by blocking DTL-mediated PDCD4 ubiquitination to regulate the MAPK/JNK pathway. J. Ethnopharmacol. 299, 115546. https://doi.org/10.1016/j.jep.2022.115546 (2022).
Zhang, C. et al. Astragaloside IV inhibits hepatocellular carcinoma by continually suppressing the development of fibrosis and regulating pSmad3C/3L and Nrf2/HO-1 pathways. J. Ethnopharmacol. 279, 114350. https://doi.org/10.1016/j.jep.2021.114350 (2021).
Yang, L. et al. Comprehensive investigation of mechanism and effective ingredients of fangji huangqi tang by serum pharmacochemistry and network pharmacology. Biomed. Chromatogr. 34, e4785. https://doi.org/10.1002/bmc.4785 (2020).
Xiao, Z. et al. Clinical efficacy and safety of aidi injection combination with vinorelbine and cisplatin for advanced non-small-cell lung carcinoma: A systematic review and meta-analysis of 54 randomized controlled trials. Pharmacol. Res. 153, 104637. https://doi.org/10.1016/j.phrs.2020.104637 (2020).
Liu, X. H. et al. Tissue distribution of six major bio-active components after oral administration of zhenqi fuzheng capsules to rats using ultra-pressure liquid chromatography-tandem mass spectrometry. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 986–987, 44–53. https://doi.org/10.1016/j.jchromb.2015.01.033 (2015).
Cheng, C. et al. Astragaloside IV targets PRDX6, inhibits the activation of RAC subunit in NADPH oxidase 2 for oxidative damage. Phytomedicine 114, 154795. https://doi.org/10.1016/j.phymed.2023.154795 (2023).
Xiao, Z. et al. Clinical efficacy and safety of Aidi injection plus paclitaxel-based chemotherapy for advanced non-small cell lung cancer: A meta-analysis of 31 randomized controlled trials following the PRISMA guidelines. J. Ethnopharmacol 228, 110–122. https://doi.org/10.1016/j.jep.2018.09.024 (2019).
Liu, J. et al. AS-IV enhances the antitumor effects of propofol in NSCLC cells by inhibiting autophagy. Open Med. (Wars) 18, 20230799. https://doi.org/10.1515/med-2023-0799 (2023).
He, X. R. et al. Chinese medicine bu-fei decoction attenuates epithelial-mesenchymal transition of non-small cell lung cancer via inhibition of transforming growth factor beta1 signaling pathway in vitro and in vivo. J. Ethnopharmacol. 204, 45–57. https://doi.org/10.1016/j.jep.2017.04.008 (2017).
Jin, L. et al. Stephania tetrandra and ginseng-containing chinese herbal formulation NSENL reverses cisplatin resistance in lung cancer xenografts. Am. J. Chin. Med. 45, 385–401. https://doi.org/10.1142/S0192415X17500240 (2017).
Wang, J. et al. Aidi injection plus platinum-based chemotherapy for stage IIIB/IV non-small cell lung cancer: A meta-analysis of 42 RCTs following the PRISMA guidelines. J. Ethnopharmacol. 221, 137–150. https://doi.org/10.1016/j.jep.2018.04.013 (2018).
Gu, X. et al. Astragaloside IV and saponins of rhizoma polygonati cure cyclophosphamide-induced myelosuppression in lung adenocarcinoma via down-regulating miR-142-3p. Front. Oncol. 11, 630921. https://doi.org/10.3389/fonc.2021.630921 (2021).
Wang, C. Q. et al. The optimal adjuvant strategy of aidi injection with gemcitabine and cisplatin in advanced non-small cell lung cancer: A meta-analysis of 70 randomized controlled trials. Front. Pharmacol. 12, 582447. https://doi.org/10.3389/fphar.2021.582447 (2021).
Meng, Y. et al. GBP1 facilitates indoleamine 2,3-dioxygenase extracellular secretion to promote the malignant progression of lung cancer. Front. Immunol. 11, 622467. https://doi.org/10.3389/fimmu.2020.622467 (2020).
Yang, M. et al. Chinese patent medicine Aidi injection for cancer care: An overview of systematic reviews and meta-analyses. J. Ethnopharmacol. 282, 114656. https://doi.org/10.1016/j.jep.2021.114656 (2022).
Wu, T. et al. Astragaloside IV augments anti-PD-1 therapy to suppress tumor growth in lung cancer by remodeling the tumor microenvironment. Eur. J. Histochem. https://doi.org/10.4081/ejh.2024.4098 (2024).
Yang, B. et al. An integrated strategy for effective-component discovery of astragali radix in the treatment of lung cancer. Front. Pharmacol. 11, 580978. https://doi.org/10.3389/fphar.2020.580978 (2020).
Xia, D. et al. Astragaloside IV, as a potential anticancer agent. Front. Pharmacol. 14, 1065505. https://doi.org/10.3389/fphar.2023.1065505 (2023).
Kollman, P. A. et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem. Res. 33, 889–897. https://doi.org/10.1021/ar000033j (2000).
Wang, E. et al. Development and evaluation of MM/GBSA based on a variable dielectric GB model for predicting protein-ligand binding affinities. J. Chem. Inf. Model. 60, 5353–5365. https://doi.org/10.1021/acs.jcim.0c00024 (2020).
Dubey, K. D. et al. Recent advances in protein-ligand interactions:molecular dynamics Simulations and binding free energy. Curr. Comput. Aided Drug Des. 9, 518–531. https://doi.org/10.2174/15734099113096660036 (2013).
Wan, S. et al. Rapid, accurate, precise and reproducible ligand-protein binding free energy prediction. Interface Focus 10, 20200007. https://doi.org/10.1098/rsfs.2020.0007 (2020).
Su, C. C. et al. Astragaloside IV increases MMP-2 mRNA and protein expression in human lung cancer A549 cells. Mol. Med. Rep. 2, 107–113. https://doi.org/10.3892/mmr_00000070 (2009).
Xiao, W. et al. Network module analysis and molecular docking-based study on the mechanism of astragali radix against non-small cell lung cancer. BMC Complement Med. Ther. 23, 345. https://doi.org/10.1186/s12906-023-04148-9 (2023).
Zhang, L. L. et al. MCM5 Aggravates the HDAC1-mediated malignant progression of lung cancer. Front. Cell. Dev. Biol. 9, 669132. https://doi.org/10.3389/fcell.2021.669132 (2021).
Tan, B. et al. Astragaloside attenuates the progression of prostate cancer cells through endoplasmic reticulum stress pathways. Oncol. Lett. 16, 3901–3906. https://doi.org/10.3892/ol.2018.9071 (2018).
Funding
This study was supported by the Key Support Project of Regional Innovation and Development Joint Fund of National Natural Science Foundation of China (U20A20398), the National Natural Science Foundation of China (82104454, 82374399), the Clinical Medical Research Transformation Project of Anhui Province (202304295107020111) and the Natural Science Research Key Project of Anhui Provincial Department of Education (KJ2021A0542). The study funders were not involved in the design of this study, data collection, analysis, interpretation, or in writing the report.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.D and Q.X; methodology, W.G; validation, J.D, Q.X and W.G; formal analysis, G.C; data curation, L.Z and T.H; writing—original draft preparation, J.D.; writing—review and editing, D.W, J.T, C.Y, Y.G and Z.L; All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ding, J., Xue, Q., Guo, W. et al. Mechanism of astragaloside A against lung adenocarcinoma based on network pharmacology combined with molecular dynamics simulation technique. Sci Rep 15, 12033 (2025). https://doi.org/10.1038/s41598-025-94793-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-94793-6