Abstract
Cadmium (Cd) is a toxic heavy metal whose contamination in soil threatens food safety, agricultural production, and human health. To date, phytoremediation is a low-cost and environmentally friendly method for eliminating Cd contamination. In this study, we report a gene from ramie (Boehmeria nivea) that encodes a dehydration responsive element binding (DREB) factor associated with plant tolerance to Cd, namely BnDREB1. The open reading frame of BnDREB1 comprises 873 bp encoding 290 amino acids and includes a characteristic AP2 domain. Its cloned promoter sequence contains various hormone and stress responsive elements. Quantitative RT-PCR analysis showed that BnDREB1 is expressed in different organs of ramie. Treatments with polyethylene glycol (PEG), abscisic acid (ABA), and Cd upregulated the expression of BnDREB1. Confocal microscopic analysis revealed that BnDREB1 is mainly localized in the nucleus. Overexpression of BnDREB1 in Arabidopsis thaliana increased the tolerance of transgenic plants to Cd, thereby protecting plant growth from its toxicity. Biochemical analysis revealed that overexpression of BnDREB1 reduced the levels of Cd induced malonaldehyde and hydrogen peroxide, inhibited the reduction of Cd caused soluble protein contents, increased the Cd accumulation, and enhanced Cd translocation in transgenic plants. Taken together, these findings suggest that BnDREB1 is an appropriate candidate gene for phytoremediation of Cd-contaminated soil .
Similar content being viewed by others
Introduction
Cadmium (Cd) is a toxic heavy metal that contaminates the soil ecosystem and affects plant growth1. Cd pollution of agricultural soil is a major environmental problem2,3,4. It has been reported that 5.6–38 thousand metric tons of Cd contaminate soil annually worldwide5. Traditionally, the remediation of Cd contamination in soil has been costly and environmentally unfriendly. Phytoextraction, a phytoremediation method, uses the plant root system to remove contaminants from the soil4,6.This technique primarily relies on hyperaccumulating plants to extract heavy metals from the soil1. However, the use of natural hyperaccumulating plants is limited due to their low biomass and slow growth, which restricts their effectiveness in metal extraction7. To overcome this limitation, genetic engineering techniques have been explored to enhance phytoextraction abilities of plants. A study has reported that new genetic engineering techniques can improve the capability to extract Cd, resulting in a more efficient and time-saving extraction process8. More importantly, multiple past studies have reported that transcription factors, such as DREB (Dehydration-responsive element–binding), MYB (Myeloblastosis ), bZIP (basic leucine zipper), etc., are appropriate candidates to effectively enhance plant resistance to heavy and other abiotic stresses9.
DREBs belong to the ERF family of transcription factors. DREB factors constitute a subfamily of the AP2/ethylene-responsive factor (ERF) and contain single AP2 DNA-binding domains, which recognize the six nucleotides (A/G)CCGAC of the dehydration-responsive element/C-repeat (DRE/CRT)10. Numerous DREB/CBF members have been reported to enhance the stress tolerance of transgenic plants. ScDREB8, an A-5 type of DREB gene cloned from Syntrichia caninervis, has been shown to upregulate the expression of stress-related upstream genes when overexpressed, thereby enhancing the plant’s ability to eliminate reactive oxygen species (ROS) and increasing the salt tolerance of transgenic Arabidopsis11. Similarly, the overexpression of StDREB has been reported to improve the adaptability of potato plants to Cd pollution, promote plant growth, increase the production of proline and various antioxidants, and reduce the oxidative stress-related damage12. These past studies have demonstrated that DREB factors are crucial regulators of plants’ responses to abiotic stress.
Ramie is a bast fiber producing perennial plant that belongs to the Urticaceae family. It grows extensively in China, India, Southeast Asia, and the Pacific Rim13. Ramie is not only a crop for fiber production with high agricultural economy but also a plant for phytoremediation of soil contaminated by multiple metals14,15. Furthermore, ramie can be easily cultivated because of its stress resistance and high environmental adaptability. More importantly, ramie is resistant to the toxicity of Cd16,17. To date, several genes have been cloned from ramie and shown to encode PCS (phytochelatins), MYB (Myeloblastosis), WRKY, XTH (xyloglucan endotransglucosylase/hydrolase) and others. Functional analyses have demonstrated that these genes can improve the adaption of ramie to Cd stress18,19,20,21. These findings suggest that ramie uses different genes to adapt to heavy metal stresses and indicate that the continuous identification of genes of interest can enhance our understanding of ramie’s resistance to Cd and other heavy metal stresses.
In this study, we screened other TF candidates from the ramie genome and identified a DREB transcription factor, which we named BnDREB1. We characterized its function by examining its expression pattern, promoter sequence, subcellular localization, and conducting transgenic and Cd tests. The results indicate that BnDREB1 regulates plant adaptation to Cd stress and suggest that it is an appropriate candidate for phytoremediation of Cd-contaminated soil.
Materials and methods
Plant materials and stress treatments
Xiang Zhu No. 3 is an elite ramie cultivar widely grown in south China. It has been cultivated for seeds and cutting materials at the Yunyuan Research Station at Hunan Agricultural University, Changsha, China.
The conventional propagation of ramie primarily depends on rhizomes; however, this approach is labor-intensive and time-consuming. To accelerate the acquisition of planting materials, ramie is predominantly propagated through asexual means, with seedlings obtained via the selection of ramie shoots or branch tip cuttings.For this study, we planted 20-day-old ramie seedlings in pots containing perlite and fertilized them once with a half-strength Hoagland solution. The seedlings were treated with Cd, polyethylene glycol (PEG)-6000, and abscisic acid (ABA), respectively. Cd, a heavy metal, was added to the half-strength Hoagland solution to achieve a final concentration of 200 µM, and PEG-6000 was added to the half-strength Hoagland solution to achieve a final concentration of 20%. After treating the ramie seedlings with Cd and PEG for 6, 9, and 12 h, we collected the roots, stems, leaves, and stem tips, respectively, and quickly transferred them into liquid nitrogen for freezing and preservation. Samples were collected before the application of Cd and PEG as 0 h treatment. For the ABA treatment, this plant hormone was dissolved in half-strength Hoagland solution to obtain a concentration of 100 µM, which was then sprayed onto ramie seedlings. Leaves were collected after 6, 9, and 12 h of ABA treatment, as described above. Leaves were collected before spraying as 0 h treatment. Three biological replicates were collected for each treatment.
Identification of BnDREB1 gene
Sequence alignment was performed using the Basic Local Alignment Search Tool (BLAST) from National Center for Biotechnology Information (NCBI). Based on the ramie transcriptome analysis, 83 AP2/ERF family genes were identified and analyzed their bioinformatics and expression patterns22; Based on previous experimental results, the Unigene22140 gene were selected for assessment of involvement in growth and stress tolerance of ramie.
Synthesis of first-strand cDNA
The TRIzol kit was used to extract total RNA from different samples according to the kit’s instructions. All RNA samples were treated with DNase following the DNase protocol. The quality of all RNA samples was examined on a 1% agarose gel, and their concentrations were measured using spectrophotometry. The resulting DNA-free RNA samples were reverse-transcribed into cDNA using the Trans Script First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) according to the manufacturer’s protocol.
Isolation of full-length BnDREB1 cDNA
The first-strand cDNA was used as a template for polymerase chain reaction (PCR). Forward and reverse primers were synthesized for the amplification of the entire open reading frame (ORF) of Unigene22140. Primers for the full-length cDNA were manually designed using Primer (version 5.0). The PCR protocol was as follows: initial denaturation at 95 °C for 5 min; 33 cycles of denaturation at 95 °C for 30 s, annealing at 57 °C for 30 s, extension at 72 °C for 90 s; and final extension at 72 °C for 10 min. The PCR product was examined on a 1.5% agarose gel and subsequently purified using the EasyPure Quick Gel Extraction Kit (TransGen Biotech). The purified PCR product was inserted into a cloning vector (pEASY-Blunt Zero Cloning Vector) to develop a fusion construct (pEASY-Blunt Zero-BnDREB1). The plasmid containing the insert was then introduced into competent cells (Trans1-T1 Phage Resistant Chemically Competent Cell; TransGen Biotech). Positive colonies were selected and cultured to isolate the recombinant plasmid, which was used for sequencing (Shanghai Shenggong, Shanghai, China). A full-length BnDREB1 sequence was obtained and then submitted to GenBank (accession number: OQ821767).
Isolation of the BnDREB1 promoter
Genomic DNA was extracted from the leaves of Xiang Zhu No.3. After evaluating the DNA quality via 0.8% agarose gel electrophoresis, a high-quality DNA sample was used as the template to clone a 1000 bp sequence immediately upstream of the start codon using a pair of primers. The PCR protocol was as follows: initial denaturation at 95 °C for 5 min; 32 cycles of denaturation at 95 °C for 30 s, annealing at 57 °C for 30 s, and extension at 72 °C for 90 s; and final extension at 72 °C for 10 min. After examining the PCR product via 1.2% agarose gel electrophoresis, it was purified using the EasyPure Quick Gel Extraction Kit according to the manufacturer’s protocol. The DNA samples were sequenced at Shanghai Shenggong Inc (Shanghai, China). Potential cis-acting elements were identified with the PlantCARE database.
BnDREB sequence and phylogenetic analyses
Nucleotide sequence alignment was performed using the NCBI database. BlastP was used for protein sequence alignment and domain prediction. The basic physicochemical properties of BnDREB was identified using ProtParam (ExPASy website). Sequence analysis was conducted using MEGA (version 5.05), and multiple sequence alignment was performed using ClustalW. Subsequently, a phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replications. The phylogenetic analysis included BnDREB1 and 10 DREB factors from other plants (identified from the NCBI GenBank database). The tree was constructed using Kimura’s two parameter model. WoLF PSORT was used to predict the subcellular localization of BnDREB.
Quantitative real-time PCR assay
The first strand cDNA described above was used as template for quantitative real-time PCR (qRT-PCR). A pair of gene-specific primers, DREB-QF and DREB-QR, were designed for qRT-PCR. Actin was used as an internal control for normalizing expression levels. All primers are listed in Supplementary Table S1. The PCR protocol was as follows: 50 °C for 2 min, 95 °C for 10 min, 95 °C for 30s, 60 °C for 60s and by 40 cycles. Relative gene expression levels were evaluated using the 2−△△Ct method23. Each sample inclueded three independent technical replicates and three biological replicates.
Subcellular localization of BnDREB1
A pair of primers, DREB-SF and DREB-SR (Supplementary Table S1), were designed to amplify the ORF of BnDREB1. The PCR product was digested with a restriction endonuclease and cloned to fuse it with a green fluorescence protein (GFP) reporter gene driven by a 35 S promoter in the pAN580 expression vector. This cloning generated the vector 35 S-GFP-BnDREF1. The 35 S::GFP vector was used as a control, and the 35 S::SV40-mCherry plasmid was used as a nuclear marker control. Three different constructs were used to transfect Arabidopsis protoplasts according to a protocol described by Yoo et al.24. Briefly, mesophyll protoplasts were extracted from the rosette leaves of 3- to 4-week-old wild type (WT) Arabidopsis thaliana. The protoplasts were transfected with three vectors, 35 S::SV40-mCherry, 35 S::BnDREB1-GFP, and 35 S-GFP-BnDREF1, respectively. A Nikon A1 HD25 laser confocal microscope was used for this experiment. The image analysis software is ImageJ and the magnification is 100x.
Overexpression of BnDREB1 in Arabidopsis
The BnDREB1 ORF was amplified via PCR using primers DREB-F/R (BamHI/SacI sites; Table S1) with a thermal profile of 33 cycles (95 °C/30 s, 57 °C/30 s, 72 °C/60 s). The PCR product and pBI121 vector were digested with BamHI/SacI, ligated with T4 ligase, and transformed into DH5α E. coli for kanamycin-resistant colony selection. Verified recombinant plasmid pBI121-BnDREB (Qiagen kit isolation, sequenced by Shanghai Shenggong) was transformed into Agrobacterium GV3101. For Arabidopsis transformation, GV3101 cultures (LB + 30 mg/L kanamycin) were expanded, centrifuged (5500 g, 20 min), and resuspended in 1/2 MS medium with 0.03% Silwet L-77 (OD600 = 0.8). Inflorescences of Arabidopsis Col were dipped in the suspension for 5 min, incubated in darkness (12 h), then grown to maturity. Seeds were sterilized and screened on kanamycin-MS agar. Kanamycin-resistant T1 plants underwent genotyping via PCR (protocol: 95℃/5 min; 33 cycles of 95℃/30 s, 57℃/30 s, 72℃/30 s; 72℃/7 min) and qRT-PCR. Primers are listed in Table S1.
Cd treatment of transgenic Arabidopsis plants
-
(1)
Seeds of T3 transgenic and WT Arabidopsis were sterilized with 75% alcohol solution containing 6% sodium hypochlorite for 6 min and sown on MS medium with 0, 100, and 150 µM Cd. After stratification at 4℃ for 4 days, they were moved to a growth chamber (16/8 hr light/dark, 25℃) to observe germination, growth, root length, and fresh weight. This experiment was repeated three times.
-
(2)
For pot treatment, seedlings of T3 transgenic and WT Arabidopsis plants were grown in Cd-free vermiculite, irrigated with half-strength Hoagland solution for 3 weeks, then treated with 50 µM Cd for 14 days. Root and shoot samples were collected, dried, digested with HNO3-HClO4 and analyzed for Cd content using flame atomic absorption spectroscopy to calculate the translocation factor (TF). Three biological replicates were analyzed.
-
(3)
To evaluate the plant’s tolerance to Cd treatment, T3 transgenic and WT Arabidopsis plants were grown in Cd-free soil for 3 weeks and sprayed with 0, 50, 100, and 150 µM Cd for 1 week. H2O2, MDA, and soluble protein content were measured using the method described previously25,26. Each treatment had three biological replicates and three technical replicates.
Statistical analysis
Statistical analyses were performed using SPSS (version 22.0) and Excel (version 2013; Microsoft Corporation). Data are presented as the mean values of three biological replicates evaluated with standard deviation. The significance of group comparisons was assessed using one-way analysis of variance (ANOVA) and Student’s t test. The significance level was set at 0.05 GraphPad Prism (version 9) was used for data visualization.
Results
Molecular characterization of BnDREB1
Sequence mining obtained 1686 bp of nucleotides for BnDREB1. Further sequence analysis revealed an 873 bp ORF, which was deduced to encode 290 amino acids. The molecular weight of BnDREB1 was predicted to be 32.09 kDa, and the theoretical isoelectric point of the protein was estimated to be 6.41 (Supplementary Fig. 1). Analyses performed using SignalP (version 5.0) and TMHMM indicated that the 290 amino acids of BnDREB1 did not contain a signal peptide or transmembrane domain. Analysis with WoLF PSORT predicted the nucleus localization of BnDREB1. Functional domain analysis revealed an AP2 domain located between amino acids 142 and 201 (Fig. 1a).
Nine homologous sequences of BnDREB1 were obtained from GenBank and analyzed using MEGA 7.0 software (Fig. 1b). Sequence alignment was completed for BnDREB1, MnDREB (Morus notabilis, XP_010102588.1), HaDREB (Hibiscus syriacus, XP_038997909.1), BpDREB (Broussonetia papyrifera, ABB89755.1), and BnaDREB (Brassica napus, XP_013702273.1). The results showed high similarity in amino acid sequences and functional domains (Fig. 1a). A phylogenetic tree was constructed with 10 homologous sequences using a neighbor-joining method. The resulting tree clustered BnDREB1 with MnDREB (Morus notabilis) and BpDREB (Broussonetia papyrifera). This tree also indicated the potential evolutionary relevance between BnDREB1 and DREB factors (Tamura et al. 2007).
Sequence alignment of BnDREB1 and three DREB factor homologs and phylogenetic analysis. (a) Amino acid sequence alignment of BnDREB1 and three homologs from three plants. The conserved residues are highlighted in black color. Sequences highlighted with an underline are the DREB DNA-binding domain (AP2) conserved in the four homologs. (b) An unrooted phylogenetic tree constructed with 10 homologs. The scale bar of 0.05 represents a single amino acid substitution per site. The GenBank accession numbers of these proteins are HsDREB (XP_038997909.1; Hibiscus syriacus), MnDREB (XP_010102588.1; Morus notabilis), TjPCS1 (BAB93119.1; Thlaspi japonicum), NcPCS1 (BAB93120.1; Noccaea caerulescens), PbPCS1(AEY68568.1; Pyrus betulifolia), GrDREB (XP_012469770.1; Gossypium raimondii), GhDREB (XP_016740805.1; Gossypium hirsutum), GaDREB (XP_017615755.1; Gossypium arboreum, McDREB (XP_022133397.1; Momordica charantia), BpDREB (ABB89755.1; Broussonetia papyrifera), BnaDREB (XP_013702273.1; Brassica napus), and CiDREB (XP_042985052.1; Carya illinoinensis).
Characteristics of the BnDREB1 promoter
A 2000 bp fragment containing the BnDREB1 promoter was amplified from the genomic DNA of ramie via PCR (Supplementary Tables 1 and Supplementary Fig. 2). Analysis of the promoter sequence revealed the presence of TATA-box and CAAT-box cis-elements, as well as stress-responsive elements such as aerobic induction elements, drought response elements, MeJA (TGACG motif), ABA, auxin (TGA-element), and gibberellin response elements. These features suggest that the expression of BnDREB1 in plants may be regulated by different growth conditions (Supplementary Table 2).
Nucleus localization of BnDREB1
To determine the subcellular localization of BnDREB1, a construct fusing BnDREB1 to the N-terminus of GFP was developed for transient expression and transfected into Arabidopsis protoplasts. Additionally, GFP alone and a positive vector were transfected as controls. Confocal microscopy revealed that the BnDREB1-GFP fusion localized to the nucleus, similar to the positive vector, while GFP alone was localized to the cytoplasm (Fig. 2). These results indicate that BnDREB1 is mainly localized in the nucleus (Fig. 2).
Subcellular localization of BnDREB1.Three plasmids, 35 S::SV40-mCherry, recombinant35S::BnDREB1-GFP, and vector 35 S::GFP were transfected into Arabidopsis protoplasts, respectively. The fluorescence signals emitted by mCherry and GFP were detected with a laser-powered scanning microscope. (a) 35 S::BnDREB1-GFP, (b) 35 S::SV40-mCherry, (c) chloroplast autofluorescence, (d) bright field, (e) overlapping images (a–d), (f) 35 S::GFP, (g) 35 S::SV40-mCherry, (h) chloroplast autofluorescence, (i) bright field, (j) overlapping images (f–i).
Effects of cd, PEG, and ABA on the expression patterns of BnDREB1 in different Ramie organs
The expression patterns of BnDREB1 in various organs of 22-day-old ramie plants grown under different conditions were evaluated using qRT-PCR. BnDREB1 was expressed in roots, shoots, and leaves under normal growth conditions, with the highest expression observed in roots (Fig. 3a). Treatments with ABA, Cd, and PEG altered these expression patterns (Fig. 3b-h). ABA treatment resulted in peak induction of BnDREB1 expression after 6 h, which then decreased but remained higher than in controls (Fig. 3b). Cd treatment stimulated BnDREB1 expression in a time-dependent manner in roots and stems (Fig. 3c and d), with elevated expression in leaves observed only after 12 h (Fig. 3e). PEG treatment induced BnDREB1 expression in stems and leaves, peaking at 6 h and decreasing thereafter (Fig. 3g and h).
Effects of Cd, polyethylene glycol (PEG), and abscisic acid (ABA) treatments on expression patterns of BnDREB1. (a) the expression levels of BnDREB1 in the roots, shoots, and leaves of ramie seedlings in a regular growth condition. (b) 100 µM ABA enhanced the expression levels of BnDREB1 in the leaves of ramie seedlings in 12 h of treatment. 200 µM Cd enhanced the expression levels of BnDREB1 in the roots (c), shoots (d), and leaves (e) of ramie seedlings in 12 h of treatment. (f–h) the treatment of 20% PEG-6000 differentially altered the expression levels of BnDREB1 in the roots (f), shoots (g), and leaves (h) of ramie seedlings. Data represent the average value of three biological replicates and standard errors (indicated as vertical bars) are used to indicate the variation. “*” denotes the significant differences between treated and untreated samples.
Overexpression of BnDREB1 enhances the tolerance of Transgenic Arabidopsis plants to cd toxicity and the accumulation of cd in tissues
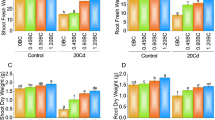
Four transgenic Arabidopsis lines (L1-L4) overexpressing BnDREB1 were obtained (Supplementary Fig. 4). Among these, L1 plants exhibited the highest transgene expression, followed by L4. Seeds of T3 transgenic plants and wild-type controls were inoculated on MS media containing 0 (control), 100, and 150 µM Cd. In the absence of Cd, no significant differences were observed among WT, L1, and L4 plants (Fig. 4a). However, Cd treatments inhibited the growth of both transgenic and wild-type seedlings, with BnDREB1 overexpression significantly enhancing Cd tolerance in L1 and L4 plants. In response to 100 µM Cd stress, root lengths of L1 and L4 plants were 1.2 and 1.4 times longer than those of WT plants, respectively (Fig. 4b), the fresh weights of L1 and L4 plants were 1.4 and 1.1 times higher than those of WT plants, respectively (Fig. 4c); In response to 150 µM Cd stress, root lengths of L1 and L4 plants were 1.9 and 1.7 times longer than those of WT plants, respectively (Fig. 4b). The fresh weights of L1 and L4 plants were 3.2 and 2.7 times higher than those of WT plants, respectively (Fig. 4c).
To investigate the effects of BnDREB1 overexpression on Cd accumulation, Cd content in roots and shoots was measured. The Cd content in the shoots of L1 and L4 plants was approximately 1.5 times higher than that in WT plants (Fig. 4d). Additionally, BnDREB1 overexpression increased Cd translocation from roots to shoots, as indicated by higher translocation factor (TF) values of 0.81 and 0.72 for L1 and L4 plants, respectively, compared to 0.56 for WT plants (Fig. 4e).
Enhancement of Arabidopsis thaliana to Cd tolerance by overexpression of BnDREB1. BnDREB1 transgenic and WT Arabidopsis seedlings were cultivated for 2 weeks on agar-solidified half-strength MS medium supplemented with Cd (0, 100, and 150 µM). (A) Images of phenotypes show that transgenic seedlings are more tolerant that WT seedlings grown on the media supplemented with 100 and 150 µM Cd, respectively. (b and c) Both 100 and 150 µM concentrations of Cd decreased more fresh weights of WT roots and leaves that those of transgenic roots (B) and leaves (C). (D) Cd contents were higher in transgenic shoots than in WT shoots, while were similar in roots of both WT and transgenic seedlings. Fresh weights were measured after transgenic and WT Arabidopsis plants were treated with 50 µM Cd once time. (E) The contents of the Cd translocation factor were higher in transgenic seedlings than in WT seedlings. WT, wild-type.
BnDREB1 overexpression decreases the levels of H2O2 and MDA induced by cd
Heavy metal exposure leads to an increase in ROS levels in plants, which causes cellular damage through oxidative stress27. H2O2 is an ROS, and MDA is the primary byproduct of ROS-dependent lipid peroxidation. These two molecules serve as biomarkers for measuring oxidative stress intensity in plants. Measurement showed no significant difference in the levels of these two molecules between transgenic and WT plants in the absence of Cd (Fig. 5a and b). However, Cd treatments (100 and 150 µM) resulted in an increase in the levels of both molecules, H2O2 and MDA, in plants. In comparison, the levels of these two molecules were significantly lower in transgenic plants than in WT plants. These data indicate that the overexpression of BnDREB1 enhances the oxidative tolerance of plants to Cd stress.
BnDREB1 overexpression improves the contents of total soluble proteins in Transgenic Arabidopsis plants treated by cd
The contents of total soluble proteins are associated with the capacity of osmoregulation in plants28,29. We measured the contents of soluble proteins in both transgenic and WT Arabidopsis plants before and after the Cd treatments. The results showed that the Cd treatments led to the reduction in total soluble protein contents in both genotypes of plants. In comparison, the contents of soluble proteins were higher in the two transgenic lines than in the WT plants under all tested conditions (Fig. 5c). These data indicate that overexpression of BnDREB1 mitigates the reduction in soluble protein contents in the transgenic plants caused by the Cd stress.
Effects of the overexpression of BnDREB1 on H2O2, malonaldehyde, and soluble protein contents in different Cd conditions. BnDREB1 transgenic and wild-type plants were treated with 0, 50, 100, and 150 µM Cd for 7 days, respectively. a, the overexpression of BnDREB1 decreased the contents of H2O2 that were increased by the treatment of Cd. (b) The overexpression of BnDREB1 decreased the contents of malonaldehyde that were increased by the treatment of Cd. (c) The overexpression of BnDREB1 slowed down the reduction trend of soluble protein contents, which were caused by the treatment of Cd.
Prediction analysis of proteins interacting with BnDREB1
Plant stress response has an important impact on plant stress tolerance. Upon stress plants activate the expression of a series of stress-related genes and synthesise proteins that reduce damage caused by oxidative stress, thereby enhancing plant tolerance30.To identify the stress response–related proteins that are impacted by BnDREB1, we used STRING (https://cn.string-db.org/) to analyze proteins that interacts with BnDREB1 and its homolog (AtERF53) in A. thaliana (Fig. 6). The resulting data revealed interactions between AtERF53 and several metal ion–binding proteins, such as RGLG1, RGLG2, EMB1467, SDH2-1, and SDH3-1. Additionally, this analysis revealed that AtERF53 interacts with a few stress response–related proteins. These data indicate that BnDREB1 regulates the response to Cd stress via other stress-related proteins.
Discussion
DREB transcription factors play a crucial role in enhancing plant tolerance to various abiotic stresses, including salinity, dehydration, cold, and oxidative stress31,32. For instance, overexpression of OsDREB1A in transgenic Arabidopsis has been shown to improve resilience to severe drought, high salinity, and low temperatures33,34. However, the involvement of DREB factors in heavy metal stress responses, particularly Cd stress, is less understood. One report indicated that the treatments of zinc and lead considerably increased the expression level of AmCBF2 in leaves of Avicennia marina, while the Cd treatment only slightly upregulated AmCBF2 expression. Gene expression analysis have indicated that the transcription of AmCBF2 is induced by heavy metals, with a stronger response to Pb2+ or Zn2+ than to Cd2+35. BjDREB1B is a member of DREB and its expression is responsive to zinc, Cd, and nickel exposure36. In addition, StDREB is another member of DREB that has been reported to respond Cd stresses12. In our study, we characterized that BnDREB1 was responsive to Cd stresses. Its overexpression increased the tolerance of the transgenic Arabidopsis plants to Cd. The overexpression of BnDREB1 not only increases Cd accumulation and translocation but also protects plants from Cd-induced oxidative damage by reducing lipid peroxidation and maintaining higher levels of soluble proteins (Fig. 7). These findings suggest that BnDREB1 could be a promising candidate for phytoremediation of Cd-contaminated soils.
The protein and promoter sequences of BnDREB1 support its function in enhancing plant tolerance to Cd stress. The expression pattern of BnDREB1 in different organs indicates that its expression level is highest in roots and is significantly upregulated under Cd, PEG, and ABA treatments (Fig. 3). Subcellular localization analysis revealed that BnDREB1 is primarily localized in the nucleus (Fig. 2), which is consistent with its function as a transcription factor. Additionally, the conserved AP2 domain in DREB transcription factors is known to bind specifically to DRE/CRT cis-acting elements on the promoters of stress-related genes37,38. Our sequence analysis confirmed the presence of this domain in BnDREB1 (Fig. 1). Additionally, the promoter analysis revealed several stress-responsive elements, which were validated by the upregulation of BnDREB1 under Cd, PEG, and ABA treatments. These elements likely contribute to the enhanced expression of BnDREB1 under stress conditions. Although the exact mechanisms of how BnDREB1 interacts with other genes in response to Cd stress remain to be elucidated, our analysis suggests that it plays a significant role. Our analysis of its homolog AtERF53 in Arabidopsis suggests that it interacts with stress-responsive proteins involved in metal ion binding (Fig. 6). This interaction network indicates that BnDREB1 may regulate Cd stress responses through interactions with other regulatory proteins. Future studies should focus on elucidating these interactions and their roles in Cd tolerance.
Heavy metal stress causes an increase of ROS and a reduction in soluble proteins in plants36,39. Two molecules, MDA and H2O2, serve as biomarkers of the increase in ROS and reduction of total protein. For example, the overexpression of AmDREB3 has been shown to antagonize the increase of two molecules caused by salinity in plants40,41. Our data supported this finding, showing that the overexpression of BnDREB1 counteracts the increase in MDA and the reduction of total soluble proteins caused by Cd stresses. Additionally, the overexpression of BnDREB1 changes the accumulation patterns and transport of Cd. These data suggest that BnDREB1 is an appropriate candidate gene for phytoremediation of Cd-contaminated soil.
This study, through the identification and functional analysis of the BnDREB1 gene, has revealed its important role in plant tolerance to cadmium (Cd) stress. The overexpression of BnDREB1 significantly enhances plant tolerance to Cd by increasing Cd accumulation and translocation, reducing oxidative damage, and maintaining higher levels of soluble proteins. These results indicate that BnDREB1 is a key factor in Cd stress response and may regulate plant Cd tolerance by interacting with other regulatory proteins. Future studies will further elucidate its molecular mechanisms in Cd stress response and explore its potential application in the phytoremediation of Cd-contaminated soils.
Data availability
The datasets generated and/or analysed during the current study are available in the NCBI repository, [ https://preview.ncbi.nlm.nih.gov/nuccore/OQ821767 ]”.
Abbreviations
- Cd:
-
Cadmium
- ERF:
-
AP2/ethylene-responsive factor
- DREB:
-
Dehydration responsive element binding
- MYB:
-
Myeloblastosis
- bZIP:
-
Basic leucine zipper
- ORF:
-
Open reading
- cDNA:
-
Complementary DNA
- NCBI:
-
National Center for Biotechnology Information
- qRT-PCR:
-
Real-time quantitative polymerase chain reaction
- GFP:
-
Green fluorescent protein
- ABA:
-
Abscisic acid
- H2O2 :
-
Hydrogen peroxide
- MDA:
-
Malondialdehyde
- ROS:
-
Reactive oxygen species
- WT:
-
Wild-type
- TF:
-
Translocation factor
References
Li, R. et al. Heterologous expression of the tobacco Metallothionein gene NtMT2F confers enhanced tolerance to cd stress in Escherichia coli and Arabidopsis thaliana. Plant. Physiol. Biochem. 195, 247–255. https://doi.org/10.1016/j.plaphy.2023.01.027 (2023).
Zhang, Z. et al. The overexpression of LOW PHOSPHATE ROOT 1 (LPR1) negatively regulates Arabidopsis growth in response to cadmium (Cd) stress. Plant. Physiol. Biochem. 196, 556–566. https://doi.org/10.1016/j.plaphy.2023.02.003 (2023).
Kubier, A., Wilkin, R. T. & Pichler, T. Cadmium in soils and groundwater: a review. Appl. Geochem. 108, 1–16. https://doi.org/10.1016/j.apgeochem.2019.104388 (2019).
Dixit, R. et al. Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustain.-Basel 7, 2189–2212. https://doi.org/10.3390/su7022189 (2015).
Jadia, C. D. & Fulekar, M. H. Phytoremediation of heavy metals: recent techniques. Afr. J. Biotechnol. 8, 921–928 (2009).
Sharma, J. K., Kumar, N., Singh, N. P. & Santal, A. R. Phytoremediation technologies and their mechanism for removal of heavy metal from contaminated soil: an approach for a sustainable environment. Front. Plant. Sci. 14, 1076876. https://doi.org/10.3389/fpls.2023.1076876 (2023).
Das, N., Bhattacharya, S. & Maiti, M. K. Enhanced cadmium accumulation and tolerance in Transgenic tobacco overexpressing rice metal tolerance protein gene OsMTP1 is promising for phytoremediation. Plant. Physiol. Bioch. 105, 297–309. https://doi.org/10.1016/j.plaphy.2016.04.049 (2016).
Sarkar, T., Thankappan, R., Mishra, G. P. & Nawade, B. D. Advances in the development and use of DREB for improved abiotic stress tolerance in Transgenic crop plants. Physiol. Mol. Biol. Plants. 25, 1323–1334. https://doi.org/10.1007/s12298-019-00711-2 (2019).
Agarwal, P. K., Agarwal, P., Reddy, M. K. & Sopory, S. K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant. Cell. Rep. 25, 1263–1274. https://doi.org/10.1007/s00299-006-0204-8 (2006).
Wu, Y. et al. Genome-wide analysis of the DREB family genes and functional identification of the involvement of BrDREB2B in abiotic stress in Wucai (Brassica campestris L.). BMC Genom. 23, 598. https://doi.org/10.1186/s12864-022-08812-1 (2022).
Liang, Y. et al. ScDREB8, a novel A-5 type of DREB gene in the desert moss syntrichia Caninervis, confers salt tolerance to Arabidopsis. Plant. Physiol. Biochem. 120, 242–251. https://doi.org/10.1016/j.plaphy.2017.09.014 (2017).
Charfeddine, M., Charfeddine, S., Bouaziz, D., Ben Messaoud, R. & Bouzid, R. G. The effect of cadmium on Transgenic potato (Solanum tuberosum) plants overexpressing the StDREB transcription factors. Plant. Cell. Tiss Org. 128, 521–541. https://doi.org/10.1007/s11240-016-1130-2 (2017).
An, X. et al. Transcriptome profiling and identification of transcription factors in Ramie (Boehmeria nivea L. Gaud) in response to PEG treatment, using illumina Paired-End sequencing technology. Int. J. Mol. Sci. 16, 3493–3511. https://doi.org/10.3390/ijms16023493 (2015).
Wang, X. et al. Subcellular distribution and chemical forms of cadmium in Boehmeria nivea (L.) Gaud. Environ. Exp. Bot. 62, 389–395. https://doi.org/10.1016/j.envexpbot.2007.10.014 (2008).
Yang, B. et al. Constitutional tolerance to heavy metals of a fiber crop, Ramie (Boehmeria nivea), and its potential usage. Environ. Pollut. 158, 551–558. https://doi.org/10.1016/j.envpol.2009.08.043 (2010).
Tang, H. et al. Effects of selenium and silicon on enhancing antioxidative capacity in Ramie (Boehmeria nivea (L.) Gaud.) under cadmium stress. Environ. Sci. Pollut Res. Int. 22, 9999–10008. https://doi.org/10.1007/s11356-015-4187-2 (2015).
Ma, Y. S. et al. Identification of the Xyloglucan endotransglycosylase/hydrolase (XTH) gene family members expressed in Boehmeria nivea in response to cadmium stress. Int. J. Mol. Sci. 23, 56. https://doi.org/10.3390/ijms232416104 (2022).
Zhu, S. J., Shi, W. J. & Jie, Y. C. Overexpression of BnPCS1, a novel phytochelatin synthase gene from ramie (Boehmeria nivea), enhanced Cd tolerance, accumulation, and translocation in Arabidopsis thaliana. Front Plant Sci 12, 639189 (2021).
Feng, X. K. et al. Genome-wide analysis of R2R3-MYB transcription factors in Boehmeria nivea (L.) gaudich revealed potential cadmium tolerance and anthocyanin biosynthesis genes. Front. Genet. 14,1080909 (2023).
Ma, Y. S. et al. Identification of the Xyloglucan Endotransglycosylase/Hydrolase (XTH) gene family members expressed in Boehmeria nivea in response to cadmium stress. Int. J. Mol. Sci. 23, 232416104 (2022).
Feng, X. K. et al. Analysis of WRKY resistance gene family in Boehmeria nivea (L.) Gaudich: crosstalk mechanisms of secondary cell wall thickening and cadmium stress. Front. Plant Sci. 13, 812988 (2022).
Zhang, X., Peng, W., Chen, H. & Xing, H. BnAP2-12 overexpression delays Ramie flowering: evidence from AP2/ERF gene expression. Front. Plant. Sci. 15, 1367837. https://doi.org/10.3389/fpls.2024.1367837 (2024).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. https://doi.org/10.1038/nprot.2007.199 (2007).
del Carmen Pinto, M., Tejeda, A., Duque, A. L. & Macías, P. Determination of Lipoxygenase activity in plant extracts using a modified ferrous oxidation-xylenol orange assay. J. Agr Food Chem. 55, 5956–5959. https://doi.org/10.1021/jf070537x (2007).
Wu, S. Q. et al. Exogenous melatonin improves physiological characteristics and promotes growth of strawberry seedlings under cadmium stress. Hortic. Plant. J. 7, 13–22. https://doi.org/10.1016/j.hpj.2020.06.002 (2021).
Yang, C. et al. Phosphorus influence Cd phytoextraction in Populus stems via modulating xylem development, cell wall Cd storage and antioxidant defense. Chemosphere 242, 125154. https://doi.org/10.1016/j.chemosphere.2019.125154 (2020).
Farhangi-Abriz, S. & Torabian, S. Antioxidant enzyme and osmotic adjustment changes in bean seedlings as affected by Biochar under salt stress. Ecotox Environ. Safe. 137, 64–70. https://doi.org/10.1016/j.ecoenv.2016.11.029 (2017).
Ozturk, M. et al. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 172, 1321–1335. https://doi.org/10.1111/ppl.13297 (2021).
Yan, J. et al. Overexpression of AtbZIP69 in transgenic wheat confers tolerance to nitrogen and drought stress. Planta 261, 25. https://doi.org/10.1007/s00425-024-04605-w (2025).
Ali, N. & Hadi, F. CBF/DREB transcription factor genes play role in cadmium tolerance and phytoaccumulation in ricinus communis under molybdenum treatments. Chemosphere 208, 425–432. https://doi.org/10.1016/j.chemosphere.2018.05.165 (2018).
Singh, K. & Chandra, A. DREBs-potential transcription factors involve in combating abiotic stress tolerance in plants. Biologia 76, 3043–3055. https://doi.org/10.1007/s11756-021-00840-8 (2021).
Ito, Y. et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in Transgenic rice. Plant. Cell. Physiol. 47, 141–153. https://doi.org/10.1093/pcp/pci230 (2006).
Chen, J. Q., Meng, X. P., Zhang, Y., Xia, M. & Wang, X. P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 30, 2191–2198. https://doi.org/10.1007/s10529-008-9811-5 (2008).
Peng, Y. L. et al. Characterization and expression analysis of three CBF/DREB1 transcriptional factor genes from Mangrove avicennia Marina. Aquat. Toxicol. 140, 68–76. https://doi.org/10.1016/j.aquatox.2013.05.014 (2013).
Cong, L., Chai, T. Y. & Zhang, Y. X. Characterization of the novel gene BjDREB1B encoding a DRE-binding transcription factor from Brassica juncea L. Biochem. Biophys. Res. Commun. 371, 702–706. https://doi.org/10.1016/j.bbrc.2008.04.126 (2008).
Lim, C. W., Lim, J. & Lee, S. C. The pepper AP2 domain-containing transcription factor CaDRAT1 plays a negative role in response to dehydration stress. Environ. Exp. Bot. 164, 170–180. https://doi.org/10.1016/j.envexpbot.2019.05.010 (2019).
Agarwal, P. K. & Jha, B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 54, 201–212. https://doi.org/10.1007/s10535-010-0038-7 (2010).
Gu, C. S. et al. Overexpression of iris. Lactea Var. Chinensis Metallothionein llMT2a enhances cadmium tolerance in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 105, 22–28. https://doi.org/10.1016/j.ecoenv.2014.04.002 (2014).
Zhang, Z., Li, W., Gao, X., Xu, M. & Guo, Y. DEAR4, a member of DREB/CBF family, positively regulates leaf senescence and response to multiple stressors in Arabidopsis thaliana. Front. Plant. Sci. 11, 367. https://doi.org/10.3389/fpls.2020.00367 (2020).
Ren, M. et al. Constitutive expression of an A-5 subgroup member in the DREB transcription factor subfamily from ammopiptanthus mongolicus enhanced abiotic stress tolerance and anthocyanin accumulation in Transgenic Arabidopsis. PLoS One. 14, e0224296. https://doi.org/10.1371/journal.pone.0224296 (2019).
Funding
This work was funded by the National Natural Science Foundation of China (Grant Number 32272137).
Author information
Authors and Affiliations
Contributions
Xiaoyang Zhang: Formal analysis, Data curation, Software, Writing – original draft, Writing – review & editing. Mingyu Shao: Data curation, Software, Writing – original draft. Wenxian Peng: Investigation, Formal analysis, Data curation, Software. Hongyue Qu: Investigation. Xinran Han: Investigation. Hucheng Xing: Funding acquisition, Writing – review & editing, Supervision. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, X., Shao, M., Peng, W. et al. BnDREB1 confers cadmium tolerance in ramie. Sci Rep 15, 11662 (2025). https://doi.org/10.1038/s41598-025-96051-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-96051-1