Abstract
Sarcopenia and body composition metrics are strongly associated with patient outcomes. In this study, we developed and validated a flexible, open-access pipeline integrating available deep learning-based segmentation models with pre- and postprocessing steps to extract body composition measures from routine computed tomography (CT) scans. In 337 surgical oncology patients, total skeletal muscle tissue (SMtotal), psoas muscle tissue (SMpsoas), visceral adipose tissue (VAT), and subcutaneous adipose tissue (SAT) were quantified both manually and using the pipeline. Automated and manual measurements showed strong correlations (SMpsoas: r = 0.776, VAT: r = 0.993, SAT: r = 0.984; all P < 0.001). Measurement discrepancies primarily resulted from segmentation errors, anatomical anomalies or image irregularities. SMpsoas measurements showed substantial variability depending on slice selection, whereas SMtotal, averaged across all L3 levels, provided greater measurement stability. Overall, SMtotal performed comparably to SMpsoas in predicting overall survival (OS). In summary, body composition measures derived from the pipeline strongly correlated with manual measurements and were prognostic for OS. The increased stability of SMtotal across vertebral levels suggests it may serve as a more reliable alternative to psoas-based assessments. Future studies should address the identified areas of improvement to enhance the accuracy of automated segmentation models.
Similar content being viewed by others
Introduction
Sarcopenia, characterized by the loss of skeletal muscle mass, has gained significant relevance in both oncology and oncological surgery1,2,3,4. Affecting approximately 35% of oncology patients5, it is associated with poorer overall survival (OS), reduced disease-free survival, and diminished response to cancer therapies2,6,7. In surgical contexts, it is consistently linked to higher risk of postoperative complications8,9,10. For instance, in pancreatic cancer, sarcopenia is associated with higher perioperative mortality and shorter OS11. Early recognition of sarcopenia is therefore critical, as it allows targeted prehabilitation strategies—such as structured exercise programs and nutritional optimization—that may improve both short- and long-term outcomes11,12. Identifying sarcopenic patients before surgery provides an opportunity for timely intervention, potentially mitigating the clinical impact of muscle loss and improving patient prognosis.
Sarcopenia or its surrogate parameters can be determined using various methods, such as dual-energy X-ray absorptiometry, bioelectrical impedance analysis, hand grip tests, clinical scores, or the analysis of imaging studies4. The evaluation of computed tomography (CT) scans is particularly well established due to its widespread availability and the standardized, reproducible nature of measurements4,13. However, manual measurements are labor-intensive, limiting their practicality in clinical routine.
Given the advances in deep learning-based semantic segmentation techniques, applying such methods to determine body composition parameters is promising. Several studies have demonstrated the feasibility of using deep learning models to segment muscle and adipose tissue areas, showing strong correlations with manual measurements and clinical outcomes14,15,16,17,18,19. However, widespread adoption of these models is hampered by limited accessibility15,20,21.
In response, we developed a flexible pipeline that integrates existing segmentation models with pre- and postprocessing steps to assess sarcopenia and body composition measures. This study aimed to validate this automated pipeline against manual measurements in a real-world cohort of surgical oncology patients, with particular emphasis on identifying sources of discrepancy and assessing measurement reliability across different skeletal muscle metrics.
Materials and methods
Study design
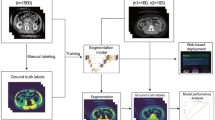
This study is a non-interventional, retrospective analysis of 382 patients who underwent surgery for hepatic or pancreatic disease between 2003 and 2016 (Fig. 1a). We included patients with underlying oncological disease and available CT scans of adequate quality and coverage performed up to two months prior to surgery (see Online Resource 1, Image Quality Criteria). Manual measurements were derived from earlier, unpublished analyses. Five patients were secondarily excluded from deep learning-based measurements due to missing imaging data (n = 2), metadata errors (n = 2) preventing accurate image orientation, or incomplete imaging data (n = 1) caused by series reconstruction issues (see Online Resource 2, Secondary Exclusions). These exclusions were unrelated to segmentation accuracy but based on data availability only.
The study received ethical approval from the institutional review board (IRB) of the medical faculty of the LMU Munich (23-0891) and was registered at the Clinical Study Center of the LMU University Hospital (102402). Data screening was performed by authors who were obligated to confidentiality. CT scans, OS and body height of the included patients were irreversibly anonymized, and all analyses were conducted thereafter. The IRB determined that informed consent was not necessary, as all data utilized in the study were initially collected for clinical purposes, the study design was entirely retrospective, and all analyses were performed only in the irreversibly anonymized dataset.
Imaging
CT scans were performed either at LMU University Hospital or at referring institutions, using a variety of CT scanners from four manufacturers. A weight-adapted dose of iodinated contrast agent was administered intravenously in all cases, and portal venous phase scans were preferred for analysis. Image data were reconstructed using a standard soft tissue kernel with a maximum slice thickness of 8 mm.
Definitions
In this study, we distinguished between compartments and tissue (Table 1). Compartments are defined by anatomical boundaries such as skin, fascia, bone, vessels, and organs. Tissue refers to the actual tissue composition within each compartment and was assigned based on Hounsfield Units (HU) using established thresholds3,4,7,17. We determined the cross-sectional area (CSA) of total skeletal muscle tissue (SMtotal), psoas muscle tissue (SMpsoas), visceral adipose tissue (VAT), and subcutaneous adipose tissue (SAT).
Previous studies have demonstrated that the CSA of total muscle at the third lumbar vertebra (L3) correlates well with overall muscle mass4,13. Some authors have used psoas muscle CSA as a simplified surrogate for total muscle mass22,23,24, although this approach remains under debate22,25,26. Therefore, the pipeline was constructed to determine deep learning-based measurements for both SMtotal and SMpsoas.
Manual measurement
Manual measurements were obtained from earlier, unpublished analyses. They were performed using the SliceOmatic software (version 5.0; Tomovision, Magog, Quebec, Canada) by three readers (2 medical students under supervision of CH) under the guidance of a board-certified radiologist (ABG). First, the first lumbar vertebra (L1) was identified as the most cranial lumbar vertebra without rib attachments. The readers then counted downward to locate L3. The axial slice at L3 where both transverse processes were clearly visible was selected for analysis. In this slice, standard HU thresholds were applied to measure the respective tissue CSA. Segmentation was performed using either manually marking the relevant areas (“Paint method”) or selecting a single pixel and automatically including all connected pixels (“Grow 2D method”).
Automated measurement
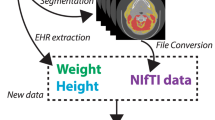
Automated measurements were performed by mirroring the manual measurement workflow (Fig. 1b). The developed pipeline integrates TotalSegmentator27, with specific pre- and postprocessing steps (Fig. 1b). The pipeline’s source code and detailed documentation are available at https://github.com/fohofmann/BodyComposition/releases/tag/v0.1.
CT scans of the included patients were exported with standardized image formatting of 512 × 512 voxels, and the DICOM files were converted to NIfTI format. First, segmentations of the spine (vertebrae T1 to L5, S1 and os sacrum) and compartments (muscle, visceral, and subcutaneous) were performed using TotalSegmentator. Segmentations of the body trunk, psoas muscle, and vertebral bodies (excluding the spinous and transverse processes) were created separately. Using the latter, labels of the spine were reduced to include only the vertebral bodies, excluding the vertebral arch and spinous processes. The center of each vertebral body was then determined by calculating the center of mass of the assigned voxels. Compartment labels were confined to the body trunk, excluding extremities, head and neck. Subsequently, the tissue composition of the specific compartments was determined using established thresholds (Table 1). For each CT slice and tissue type, the CSA was calculated.
Qualitative and statistical analysis
The relationship between manual and automated measurements of tissue CSA was assessed using Bland–Altman analysis and correlation metrics. Since manual segmentations were not systematically stored in a format enabling voxel-wise comparison, calculation of the Dice similarity coefficient was not feasible. In a subset of patients who underwent pancreatic surgery, measurement inconsistencies and outliers (defined as deviations beyond the 95% confidence interval) were qualitatively explored, categorized, and described. The colorectal cancer cohort was not included in the discrepancy analysis, as manual measurements lacked documentation regarding the specific slice selection. Exemplary visualizations were created using 3D slicer, integrating labels obtained through manual HU thresholding with those derived from the pipeline28. To assess measurement variability across vertebral levels, cases with segmentation errors in L2, L3 or L4 were excluded, and CSA values (per slice and per vertebra) at L2 and L4 were compared to their respective reference at L3. A linear mixed effects model was developed, including the vertebral level as a fixed effect and a random intercept for each patient. The median overall survival (OS) of patients with pancreatic or colorectal cancer was assessed using Kaplan–Meier analysis. The skeletal muscle index (SMI = CSA muscle/(body height [m])2) was calculated2. For each entity, the association of SMI with OS was analyzed using Harrell’s C-index and receiver operating characteristic (ROC) analysis to identify patients with above-median OS, as well as Kaplan–Meier survival analyses. Hazard ratios (HR) with 95% confidence intervals were calculated using Cox proportional hazards regression. All statistical analyses were performed using R version 4.4.0. The significance level was set at 0.05, and all tests were conducted two-sided.
Results
Patient characteristics
This study included 337 patients who underwent oncological surgery. Among these patients, 174/337 (51.6%) had metastatic colorectal cancer, 150/337 (44.5%) had pancreatic ductal adenocarcinoma, 7/337 (2.1%) had carcinoma of the papilla of vater, and 6/337 (1.8%) had extrahepatic cholangiocarcinoma. Of the included patients, 140/337 (41.5%) were female. The age of the patients ranged between 21 and 85 years, with a mean age of 64.5 years (standard deviation, 10.8 years).
CT scans were performed a median of 15 days prior to surgery (interquartile range [IQR], 5 to 29 days). Slice thickness ranged from 0.5 to 8 mm, with a median slice thickness of 5 mm (IQR, 3 to 5 mm). The median number of slices per CT scan was 129 (IQR, 90 to 195).
Deep learning-based vs. manual measurements
At the level of L3, Bland–Altman analysis indicated that the CSA of SMpsoas and VAT was lower when measured using the pipeline compared to manual measurements (Fig. 2a,d). In contrast, no relevant systematic difference was observed between manual and automated measurements for the CSA of SAT (Fig. 2g). All measurements correlated strongly or very strongly (SMpsoas r = 0.776, 95% CI 0.730 to 0.815; P < 0.001; VAT r = 0.993, 95% CI 0.991 to 0.994; P < 0.001; SAT r = 0.984, 95% CI 0.981 to 0.987; P < 0.001) (Fig. 2b,e,h). Exemplary segmentations are shown in Fig. 2c,f,i.
Comparison of manual and automated measurements: Bland–Altman plots (a, d, g) showing the differences between cross-sectional area (CSA) measurements for the psoas muscle tissue (a, b, c), visceral adipose tissue (d, e, f), and subcutaneous adipose tissue (g, h, i), with outliers marked in red; Scatter plots (b, e, h) illustrating the respective correlations; Example images of the respective segmentations (c, f, i) with manual segmentations (red), deep learning-based segmentations (blue), and intersections (purple).
Causes of measurement discrepancies
In a subset of patients who underwent pancreatic surgery, the levels identified as the center of L3 by the manual reader and the pipeline were compared. In 150/163 (92.0%) patients, the same or directly neighboring slices were identified. In 4 cases (2.4%), the level differed by two or three slices but was still within the L3 vertebral level. In 8 cases (4.9%) the deep learning-based segmentations included errors, with some errors associated with vertebral anomalies (e.g., sacralization of lumbar vertebrae or lumbarization of sacral vertebrae, Fig. 3a), and others occurring independently of any anomalies (Fig. 3b) (see Online Resource 3, Causes of Measurement Differences).
Examples of deep learning-based segmentation errors: segmentation errors of the vertebral bodies (a, b), caused by vertebral anomalies (a, lumbarization of S1) or by factors not associated with anomalies (b); Segmentation errors of tissue (d–f), including inaccurate segmentation of specific muscle groups (c, d), included extremities (e, corrected), and noisy CT scans (f); skeletal muscle tissue delineated in red, visceral adipose tissue in green, subcutaneous adipose tissue in yellow, and intermuscular adipose tissue in pink.
The CSA of VAT measured using the pipeline was lower compared to manual measurements. This discrepancy was primarily due to the overestimation that occurred when voxels within the abdominal cavity were selected as VAT based solely on HU thresholds, leading to the misclassification of structures such as bowel contents (Fig. 2f). SMpsoas measurements showed greater variability between manual and automated measurements, whereas SAT and VAT demonstrated more consistent correlations (Fig. 2). In the analysis of all outliers, causes of deviations included mislabeling of the quadratus lumborum muscle (Fig. 3c), the abdominal wall (Fig. 3c) or the psoas muscle (Fig. 3d), inclusion of extremities in the analyzed slice (Fig. 3e), or noisy CT scans (Fig. 3f).
Influence of the imaging plane on measurements
To assess the impact of the selected slice, the automatically determined CSA per slice was compared to reference measurements obtained at the center of L3. CSA of SMpsoas and SAT were larger in the lower lumbar spine (Fig. 4a), whereas no similar trend was observed for SMtotal and VAT. The extent of deviation for all measurements progressively increased with the distance from the reference level at the center of L3. Measurements taken just one slice above or below the reference level, but still within the L3 vertebra, exhibited deviations of up to 50% in some cases. Notably, SMtotal showed greater stability across different measurement levels (Fig. 4a).
Cross-sectional areas (CSA) per slice relative to the reference measurement obtained at the center of L3 (a) for the psoas muscle tissue, total skeletal muscle tissue, subcutaneous adipose tissue, and visceral adipose tissue with negative and positive values indicating measurements taken below and above the center of L3, respectively; mean CSA of the respective tissue type (b) across different vertebral levels (L2, L3, and L4) relative to L3.
Reducing measurement variability
To address the variability of measurements across different slices, we computed the mean CSA per vertebral level and compared the values at L2 and L4 relative to L3 (Fig. 4b). SMpsoas and SAT were consistently greater at lower levels, whereas no systematic trend was observed for the SMtotal and VAT. The influence of vertebral level on measurement variability was explored using a linear mixed-effects model. For SMpsoas, the fixed effect estimates showed a significantly lower CSA at L2, and a significantly higher CSA at L4 compared to L3. A similar, but less pronounced trend was observed for SAT. For SMtotal, the CSA was lower at both L2 and L4 compared to L3, with smaller relative differences (Table 2).
Association with overall survival
We compared the prognostic relevance of the SMI derived from manual psoas muscle measurements SMpsoas at the L3 center and automated total skeletal muscle measurements SMtotal averaged across L3: In patients with pancreatic cancer, the SMtotal-based SMI demonstrated significantly higher prognostic accuracy than SMpsoas-based SMI (C-index 0.598 vs. 0.494; P < 0.001). Additionally, the AUC in ROC analysis for predicting above-median OS was higher for automated SMtotal-based SMI compared to manual SMpsoas-based SMI (Fig. 5a). In patients with colorectal cancer, prognostic accuracy was comparable between the two methods, with no significant difference in C-Index (0.579 vs. 0.626; P = 0.111). The AUC for automated SMtotal-based SMI was slightly lower than for manual SMpsoas-based SMI (Fig. 5b). Kaplan–Meier survival analyses further illustrated the differences in OS between high- and low-SMI groups, with similar variations depending on the measurement method and cancer type (Fig. 6).
ROC analysis of skeletal muscle index (SMI) predicting above-median overall survival (OS) for (a) patients with pancreatic cancer (> 19.9 months [95% CI, 16.0 to 23.0 months]), and (b) patients with metastatic colorectal cancer (> 47.9 months [95% CI, 40.7 to 61.0 months]). The red line represents SMI derived from manual psoas muscle measurements (SMpsoas) at the center of L3, while the blue line represents SMI based on total skeletal muscle tissue (SMtotal) averaged across all levels of L3 as determined by the pipeline.
Kaplan–Meier survival curves illustrating overall survival (OS) in patients with pancreatic cancer (a, b) and metastatic colorectal cancer (c, d), stratified by skeletal muscle index (SMI) derived from manual psoas muscle measurements (SMpsoas) at the center of L3 (a, c) or based on pipeline-derived total skeletal muscle tissue (SMtotal) averaged across L3 (b, d). Patients were grouped based on the median SMI within each cohort and measurement method. Hazard ratios (HR) and p-values were determined using Cox proportional hazard regression analysis.
Discussion
In this study, we developed a software pipeline that integrates pre- and postprocessing steps with open-access segmentation models to automate the measurement of body composition parameters from routine CT scans. In a cohort of 337 surgical-oncological patients, we assessed the concordance of automated measurements with manual measurements originating from earlier, unpublished analyses, explored potential causes of discrepancies, and investigated strategies to compensate for measurement errors.
We found a strong correlation between manual and automated measurements of subcutaneous and visceral adipose tissue (SAT, VAT) at the level of L3, with no relevant systematic differences. However, the concordance between manual and automated measurements of the psoas muscle tissue (SMpsoas) was less consistent. Errors related to vertebral anomalies, such as lumbarization of S1 or sacralization of L5, occasionally resulted in incorrect selection of L3 by the pipeline29, or muscle tissue was not correctly segmented. Additionally, noisy CT scans and the proximity of tissue to structures with similar HU values complicated threshold-based sub-segmentations21.
We observed that the CSA of SMpsoas and SAT varied significantly depending on the slice selected for tissue segmentation. In contrast, SMtotal remained more stable across vertebral levels, making it a more robust and reproducible marker for skeletal muscle mass. By averaging SMtotal across the entire L3 vertebra, segmentation inconsistencies are mitigated, and measurement reliability is improved. In this study, the mean SMtotal across the entire L3 vertebra, as determined by the pipeline, showed comparable predictive value for above-median OS compared to manually determined SMpsoas at the center of L3.
Other groups have reported on individual deep learning-based tissue segmentation tools, but common issues include small datasets, unavailable training labels, restricted model weights, and a lack of clinical validation14,15,18. A recent study claimed that their model outperformed TotalSegmentator in tissue segmentation; however, labels or model weights were not provided21. Blankemeier et al. developed a pipeline that includes tissue segmentation, but the model is based on a 2D nnU-Net and was not evaluated clinically16. The Body and Organ Analysis (BOA) tool offers a comprehensive pipeline, including model weights30 and the underlying training labels31, and the association of its measurements with OS has been documented32. However, BOA uses the vertebral levels predicted by TotalSegmentator, and thus some limitations (and mitigation strategies) described in this study apply to BOA as well.
It is important to note that manual segmentation methods are not without errors. For instance, in the subset of patients who underwent pancreatic surgery, 2 of 13 discrepancies between manual and automated selection of the axial slice were caused by human errors. Moreover, a purely HU-based segmentation approach can lead to additional inaccuracies, particularly in noisy CT scans. Manual segmentation is prone to observer fatigue, particularly in repetitive tasks, whereas deep learning models offer scalability and can further improve by addressing identified weaknesses, for instance with refined training and postprocessing adjustments.
One limitation of this study is the absence of a pixel-by-pixel ground truth approximation. Since manual segmentations were not systematically stored in a format enabling voxel-wise comparisons, a Dice similarity coefficient could not be calculated. Although the Dice score is a widely used metric for segmentation accuracy, it does not necessarily correlate with clinical relevance15, and we therefore focused on evaluating whether body composition measures derived from the deep learning-based pipeline are comparable to manually obtained measures. While this study discusses issues arising from segmentation errors, it does not provide improved training labels or propose new models. Future work could build on openly available datasets to improve labels, particularly for the segmentation of vertebral bodies with anatomical anomalies. This could enhance the accuracy and reliability of automated segmentation tools, addressing some of the limitations we identified.
The prognostic value of the SMI was limited in this study, particularly in the ROC-analysis of patients with metastatic colorectal cancer. This limitation could stem from the multifactorial nature of oncological patient outcomes, which the SMI alone may not fully capture. Additionally, using above-median OS as an evaluation metric might not be ideal for patients with longer survival, as sarcopenia at the time of diagnosis could more significantly impact short-term outcomes33.
Although this study focused on pancreatic and metastasized colorectal malignancies due to data availability, the published pipeline itself is disease-agnostic and can be applied across oncological and non-oncological conditions. Future studies should explore the prognostic value of different measurement techniques across various patient groups and cancer types. A deeper understanding of body composition and its changes could be used to optimize the combination, timing, and intensity of multimodal therapy for individual cancer patients.
In summary, our open-access pipeline integrates established deep learning segmentation models with postprocessing steps to extract body composition measures from routine CT scans. Automated measurements showed strong correlation with manual measurements and were prognostic for overall survival. Averaging CSA across the entire L3 vertebra minimized measurement variability, reinforcing SMtotal as a robust and reliable marker for sarcopenia assessment. Our study highlights the clinical potential of automated segmentation tools while identifying key areas for future improvement and validation.
Data availability
The clinical data and CT scans analyzed this study cannot be shared due to data protection laws. The source code and a docker image of the pipeline are available under Apache 2.0 license at https://github.com/fohofmann/BodyComposition/releases/tag/v0.1. The segmentation tasks are performed using TotalSegmentator, available at https://github.com/wasserth/TotalSegmentator under Apache 2.0 license27. The weights for the segmentation models were available under free or non-commercial usage licenses from https://github.com/wasserth/TotalSegmentator. TotalSegmentator uses the nnU-Net framework34.
References
Surov, A., Strobel, A., Borggrefe, J. & Wienke, A. Low skeletal muscle mass predicts treatment response in oncology: A meta-analysis. Eur. Radiol. 33, 6426–6437 (2023).
Prado, C. M. M. et al. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: A population-based study. Lancet Oncol. 9, 629–635 (2008).
Engelke, K., Museyko, O., Wang, L. & Laredo, J.-D. Quantitative analysis of skeletal muscle by computed tomography imaging—state of the art. J. Orthop. Transl. 15, 91–103 (2018).
Mourtzakis, M. et al. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl. Physiol. Nutr. Metab. 33, 997–1006 (2008).
Surov, A. & Wienke, A. Prevalence of sarcopenia in patients with solid tumors: A meta-analysis based on 81,814 patients. J. Parenter. Enter. Nutr. 46, 1761–1768 (2022).
Martin, L. et al. Cancer cachexia in the age of obesity: Skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J. Clin. Oncol. 31, 1539–1547 (2013).
Blauwhoff-Buskermolen, S. et al. Loss of muscle mass during chemotherapy is predictive for poor survival of patients with metastatic colorectal cancer. J. Clin. Oncol. 34, 1339–1344 (2016).
Park, B. et al. Consensus-defined sarcopenia predicts adverse outcomes after elective abdominal surgery: Meta-analysis. BJS Open 7, zrad065 (2023).
Simonsen, C. et al. Sarcopenia and postoperative complication risk in gastrointestinal surgical oncology: A meta-analysis. Ann. Surg. 268, 58–69 (2018).
Su, H., Ruan, J., Chen, T., Lin, E. & Shi, L. CT-assessed sarcopenia is a predictive factor for both long-term and short-term outcomes in gastrointestinal oncology patients: A systematic review and meta-analysis. Cancer Imaging 19, 82 (2019).
Bundred, J., Kamarajah, S. K. & Roberts, K. J. Body composition assessment and sarcopenia in patients with pancreatic cancer: A systematic review and meta-analysis. HPB 21, 1603–1612 (2019).
Koh, F. H. et al. Paradigm shift in gastrointestinal surgery−combating sarcopenia with prehabilitation: Multimodal review of clinical and scientific data. World J. Gastrointest. Surg. 13, 734–755 (2021).
Shen, W. et al. Total body skeletal muscle and adipose tissue volumes: estimation from a single abdominal cross-sectional image. J. Appl. Physiol. 97, 2333–2338 (2004).
Lee, Y. S. et al. Deep neural network for automatic volumetric segmentation of whole-body CT images for body composition assessment. Clin. Nutr. 40, 5038–5046 (2021).
Mai, D. V. C. et al. A systematic review of automated segmentation of 3D computed-tomography scans for volumetric body composition analysis. J. Cachexia Sarcopenia Muscle 14, 1973–1986 (2023).
Blankemeier, L. et al. Comp2Comp: Open-source body composition assessment on computed tomography. Preprint at http://arxiv.org/abs/2302.06568 (2023).
Koitka, S., Kroll, L., Malamutmann, E., Oezcelik, A. & Nensa, F. Fully automated body composition analysis in routine CT imaging using 3D semantic segmentation convolutional neural networks. Eur. Radiol. 31, 1795–1804 (2021).
Weston, A. D. et al. Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology 290, 669–679 (2019).
Ahmad, N. et al. Automatic segmentation of large-scale CT image datasets for detailed body composition analysis. BMC Bioinform. 24, 346 (2023).
Islam, S. et al. Fully automated deep-learning section-based muscle segmentation from CT images for sarcopenia assessment. Clin. Radiol. 77, e363–e371 (2022).
Hou, B., Mathai, T. S., Liu, J., Parnell, C. & Summers, R. M. Enhanced muscle and fat segmentation for CT-based body composition analysis: A comparative study. Int. J. Comput. Assist. Radiol. Surg. https://doi.org/10.1007/s11548-024-03167-2 (2024).
Cruz-Jentoft, A. J. et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 48, 16–31 (2019).
Jones, K., Gordon-Weeks, A., Coleman, C. & Silva, M. Radiologically determined sarcopenia predicts morbidity and mortality following abdominal surgery: A systematic review and meta-analysis. World J. Surg. 41, 2266–2279 (2017).
Waduud, M. A. et al. Volumetric versus single slice measurements of core abdominal muscle for sarcopenia. Br. J. Radiol. 92, 20180434 (2019).
Flaris, A. N. & Konstantinidis, T. Letter to the editor: Is total psoas muscle area at L3 truly representative of its volume?. Br. J. Radiol. 95, 20210137 (2022).
Pigneur, F. et al. Psoas muscle index is not representative of skeletal muscle index for evaluating cancer sarcopenia. J. Cachexia Sarcopenia Muscle 14, 1613–1620 (2023).
Wasserthal, J. et al. TotalSegmentator: Robust segmentation of 104 anatomic structures in CT images. Radiol. Artif. Intell. 5, e230024 (2023).
Fedorov, A. et al. 3D Slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30, 1323–1341 (2012).
Bacon-A. Inaccurate Vertebrae Segmentations. github, wasserth/TotalSegmentator https://github.com/wasserth/TotalSegmentator/issues/268 (2024).
Haubold, J. et al. BOA: A CT-based body and organ analysis for radiologists at the point of care. Invest. Radiol. 59, 433–441 (2024).
Koitka, S. et al. SAROS: A dataset for whole-body region and organ segmentation in CT imaging. Sci. Data 11, 483 (2024).
Keyl, J. et al. Prognostic value of deep learning-derived body composition in advanced pancreatic cancer—a retrospective multicenter study. ESMO Open 9, 102219 (2024).
Fumagalli, I. A. et al. Automated CT analysis of body composition as a frailty biomarker in abdominal surgery. JAMA Surg. https://doi.org/10.1001/jamasurg.2024.0628 (2024).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Acknowledgements
The authors gratefully acknowledge LMU Klinikum for providing computing resources through the Clinical Open Research Engine (CORE).
Funding
Open Access funding enabled and organized by Projekt DEAL.
FOH received research grants sponsored by the Bavarian Centre for Cancer Research (BZKF), Munich, Germany, by the Munich Clinician Scientist Program MCSP FoeFoLe + , LMU Munich, Germany and by the CORTEX fellowship program, German Cancer Consortium (DKTK), Heidelberg, Germany.
Author information
Authors and Affiliations
Contributions
Conceptualization: FOH, CH, TSS, HN, MI; Methodology: FOH, CH; Formal analysis and investigation: FOH; CH, TT, SJ, LAA, UA, ABG; Writing—original draft preparation: FOH, CH; Writing—review and editing: TSS, HN, MI, JW, BWR; Funding acquisition: FOH; Resources: FOH, TSS, HN, MI, JW, BWR; Supervision: ABG, MI, JW, BWR.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hofmann, F.O., Heiliger, C., Tschaidse, T. et al. Validation of body composition parameters extracted via deep learning-based segmentation from routine computed tomographies. Sci Rep 15, 11909 (2025). https://doi.org/10.1038/s41598-025-96238-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-96238-6