Abstract
Global food security depends on tomato growing, but several fungal, bacterial, and viral illnesses seriously reduce productivity and quality, therefore causing major financial losses. Reducing these impacts depends on early, exact diagnosis of diseases. This work provides a deep learning-based ensemble model for tomato leaf disease classification combining MobileNetV2 and ResNet50. To improve feature extraction, the models were tweaked by changing their output layers with GlobalAverage Pooling2D, Batch Normalization, Dropout, and Dense layers. To take use of their complimentary qualities, the feature maps from both models were combined. This study uses a publicly available dataset from Kaggle for tomato leaf disease classification. Training on a dataset of 11,000 annotated pictures spanning 10 disease categories, including bacterial spot, early blight, late blight, leaf mold, septoria leaf spot, spider mites, target spot, yellow leaf curl virus, mosaic virus, and healthy leaves. Data preprocessing included image resizing and splitting, along with an 80-10-10 split, allocating 80% for training, 10% for testing, and 10% for validation to ensure a balanced evaluation. The proposed model with a 99.91% test accuracy, the suggested model was quite remarkable. Furthermore, guaranteeing strong classification performance across all disease categories, the model showed great precision (99.92%), recall (99.90%), and an F1-score of 99.91%. With few misclassifications, the confusion matrix verified almost flawless classification even further. These findings show how well deep learning can automate tomato disease diagnosis, therefore providing a scalable and quite accurate solution for smart agriculture. By means of early intervention and precision agriculture techniques, the suggested strategy has the potential to improve crop health monitoring, reduce economic losses, and encourage sustainable farming practices.
Similar content being viewed by others
Introduction
Among the most vital sectors worldwide, agriculture considerably contributes to food availability and economic stability. Tomatoes (Solanum lycopersicum) are among the most regularly planted crops around because of their outstanding nutritional value and financial worth. Still, several diseases compromising quality and quantity seriously affect tomato yield. Timely identification and classification of tomato leaf diseases determines both guarantees of sustainable agricultural methods and prevention of significant crop loss. Recent advancements in deep learning methods have provided innovative solutions for the identification of plant diseases, greatly enhancing accuracy and efficiency over more traditional methods1. Many diseases driven on by viruses, bacteria, and fungi impact tomato plants. Among the several most usually occurring tomato leaf diseases are bacterial spot, early blight, late blight, leaf mold, and tomato yellow leaf curl virus. These diseases are defined in part by several visual indications including wilting, necrotic lesions, and leaf browning. Should these diseases remain invisible and untreated later on, significant crop loss could follow. Conventional methods of detecting these diseases rely on hand inspection by experts in agriculture, a time-consuming, arbitrary process prone to errors. Therefore, the need for exact and automated systems of disease classification has become even more important2. Tomato leaf infections were previously diagnosed by farmers and agricultural specialists via eye examination. This method, sometimes known as large-scale field studies, laboratory testing, and plant pathologist guidance, helped to confirm disease types.
These traditional approaches, although effective, had major negative effects like delayed diagnosis, labor effort, and high cost. Because environmental factors vary and symptom similarities between many diseases makes manual diagnosis even more challenging and incorrect. Growing demand for precision farming has pushed researchers to look at computer vision and machine learning as automatic disease classification and detection tools3. Deep learning demonstrates remarkable performance in image-based recognition issues, so it has become even more crucial for the classification of plant diseases in recent years. Convolutional neural networks (CNNs) and Transfer Learning models comprising EfficientNet, ResNet, and Vision Transformers (ViTs) have lately been very popular for tomato leaf disease identification. These models can enable very exact disease classification by removing hierarchical elements from leaf images. Deep learning together with advanced data augmentation techniques has greatly improved the generalization and durability of these models. Studies on speed, accuracy, and scalability have demonstrated deep learning-based approaches to surpass standard machine learning models and hand diagnosis4. The main objective of this work is to develop a robust and accurate deep learning-based model for tomato leaf disease classification.
Using contemporary deep learning architectures, this work aims to minimize misclassification rates, increase illness detection accuracy, and provide a practical answer for farmers and agricultural practitioners. Moreover, seeking to evaluate different deep learning models and choose the most useful approach for pragmatic application is the research5. Even with tremendous progress achieved by deep learning-based plant disease diagnosis, certain challenges remain. Many current models suffer from overfitting and hence lose efficacy in real-world settings due to their insufficient dataset diversity. Maintaining model consistency under image acquisition parameters like lighting, angle, and resolution is more challenging still. Furthermore, on-field disease diagnostics would be enabled by light-weight and computationally efficient models that might be installed on edge and mobile devices. Handling these challenges requires the development of ideal deep learning models balancing generalizability, computational efficiency, and accuracy6. Present research on multiple deep learning architectures for tomato leaf disease detection. Wu et al. (2024) recommended a ResNet50 model with feature augmentation techniques changed in order to exhibit improved classification performance. Nevertheless, the computational complexity of the model restricts its application on low-power devices7.
Similarly, methodical studies of deep learning techniques for plant disease classification underscore the need of greater research in model optimization and dataset augmentation to attain strong and scalable solutions8. Moreover, under development are smartphone apps for real-time tomato leaf disease detection. Mobile-based deep learning models have produced promising results that let farmers snap images of ill leaves and get instant categorization replies. Still, problems with model inference speed, network connectivity, and data privacy have to be addressed if general acceptance is sought9. Different deep learning architectures for classification of plant diseases have also been under development. Excellent performance in sugarcane and maize leaf disease classification has drawn attention for EfficientNet models10. Scalability and transfer learning characteristics of EfficientNet qualify it for tomato leaf disease classification.
Still more research is needed, though, to compare its performance with other state-of- the-art solutions as Vision Transformers11 and hybrid systems. Moreover, researchers have looked at many pre-processing techniques including noise reduction and contrast augmentation in order to increase the accuracy of deep learning models. Good pre-processing systems enhance feature extraction, therefore enhancing the outcomes in disease classification. Deep learning combined with Internet of Things (IoT) devices is another fascinating innovation allowing autonomous monitoring of plant health in smart farming systems12. Growing interest in deep learning-based plant disease identification has generated various datasets and benchmarking tools. Still, many publicly available datasets lack sufficient sample variety and class imbalance that influences model generalization. Researchers have stressed the importance of domain adaption techniques and dataset augmentation in order to get past these constraints.
Moreover, domain-specific challenges include distinguishing early-stage disease symptoms from environmental stress factors call for more study13. Basically, several problems remain unanswered even if deep learning has considerably improved tomato leaf disease classification. This work aims to reduce the gap by constructing an ideal deep learning model enhancing computing efficiency, classification accuracy, and practical relevance. By combining deep learning techniques, dataset augmentation strategies, and mobile-based deployment14, this work intends to advance precision agriculture and sustainable crop management.
Leveraging the characteristics of ResNet50 and MobileNetV2 architectures, this work presents a new deep learning-based ensemble model for the classification of tomato leaf diseases. This study’s contributions consist of:
-
ResNet50 and MobileNetV2 were fine-tuned by adding GlobalAveragingPooling2D, BatchNormalization, dropout and dense to use its hierarchical feature extraction powers and fit the particular dataset of tomato leaf photos. This improvement guarantees efficient feature learning for the categorization of diseases.
-
Concatenated feature maps from the fine-tuned ResNet50 and MobileNetV2 models combined to leverage both architectures’ capabilities. By allowing a richer feature representation, this hybrid method improves classification accuracy.
-
Custom fully connected layers like Batch Normalization, dropout and dense are especially intended to improve the feature representations processed by the concatenated features. The last classification guaranteed exact disease detection by utilizing a softmax activation layer.
The paper has been arranged as follows: a detailed literature review is discussed in Sect. 2. Section 3 describes the input dataset for the tomato leaf disease and the detailed architecture of the proposed ensemble model. Section 4 deals with the outcome of the proposed ensemble model. Section 5 is the state-of-the-art analysis and Sect. 6 concludes the work.
Literature review
Diseases of tomato leaves significantly influence agricultural output, thereby generating financial losses and reduced crop production. Conventional methods of disease identification rely on hand inspection, sometimes time-consuming, arbitrary, and prone to errors. Recent advances in machine learning (ML) and deep learning (DL) have made automatic plant disease classification with remarkable accuracy and efficiency feasible. With an eye on machine learning, hybrid models, lightweight CNNs, and ensemble learning approaches along with their benefits and drawbacks this section reviews several studies on tomato leaf disease classification. Early work on tomato leaf disease classification mostly based on classic machine learning approaches, such Support Vector Machines (SVM), k-Nearest Neighbors (k-NN), Decision Trees (DT), and Random Forest (RF). Usually combining color, texture, and shape-based descriptors, these techniques then utilize a classifier to differentiate healthy from diseased leaves. With a 96.3% accuracy, Praveen et al.16 for example applied machine learning techniques for leaf disease identification.
The study revealed that while ML models are interpretable and light-weight, they fit small-scale applications. But when handling complex disease patterns and variances in real-world datasets, ML-based approaches may suffer from limited generalization and need human feature engineering, which can be challenging. Deep learning has evolved CNNs the gold standard for plant disease classification as they can automatically learn spatial properties from images. Since deep learning models offer improved resilience and accuracy, hand feature extraction is not necessary. To increase computational efficiency, researchers have developed lightweight CNN models fit for mobile and edge devices. Using quick inference 99.3% accuracy, Ullah et al.18 proposed a lightweight CNN model for tomato leaf disease classification, so appropriate for real-time applications. Likewise, Batool et al.22 demonstrated a T-Net lightweight CNN designed for mobile deployment with 98.9% accuracy. These models sometimes suffer, nevertheless, in handling complex disease cases when more advanced designs would be needed.
Many studies have combined multiple CNN architectures to exploit their complementary benefits. For tomato leaves, Cengil and çınar19 presented a hybrid CNN model for bacterial, viral, and fungal diseases categorization. This approach obtained 98.3% accuracy and helped to differentiate numerous types of diseases. Still, the model required a large dataset for good performance, which highlights the challenges of data dependency in deep learning approaches. Improving the accuracy of classification depends much on feature extraction. With a Total Variation Filter-Based Variational Mode Decomposition method, Patel et al.17 achieved 98.8% accuracy while improving image quality for better feature representation. The technique needed computationally extensive preparation, so real-time deployment is difficult even with its great performance. Because they can mix several models to increase accuracy and resilience, ensemble learning methods have become somewhat well-known. By improving feature fusion and gradient optimization, Am et al.23 constructed an adaptive ensemble model with exponential moving average fusion which obtained 98.7% accuracy.
Chouhan et al.20 investigated a deep learning and machine learning ensemble showing 99% accuracy for plant species classification. But ensemble models can demand greater processing resources, so deployment on devices with low resources becomes more difficult. Esomonu et al.27 put up a more effective hybrid method combining MobileNetV2 with SVM to get 97% accuracy. This approach made good use of the lightweight feature extraction powers of MobileNetV2 and the great classification power of SVM. Still, the performance depended on the quality of the dataset, which emphasizes the requirement of strong data preparation and augmentation methods. The capacity of tomato leaf disease categorization to be generalized across several environmental circumstances presents one of the main difficulties. Using real-world field data, Jelali21 first examined deep learning algorithms showing that models trained on controlled datasets struggle with real-world changes including lighting conditions, occlusions, and background noise. Emphasizing the importance of domain adaption methods to increase real-world applicability, the study attained 90% accuracy, below other CNN models trained on pre-processed datasets. The capacity of models to extend over many plant species is another important restriction.
Fuentes et al.25 presented a deep learning model with explicit management of hidden classes, hence lowering misclassification errors and enhancing adaptation to several plant diseases. But the more complicated model had a training accuracy of 93.3%, somewhat below previous techniques. Apart from specific models, smart agricultural uses of AI-driven solutions have been investigated. Chouhan et al.24 examined how artificial intelligence may be used in precision farming and noted its possibilities for early intervention plans and automated disease diagnostics. The study lacks particular model optimization information, hence practical application becomes difficult as summarized in Table 1.
Proposed methodology
This study utilizes a balanced dataset of 11,000 tomato leaf images categorized into 10 disease classes and a healthy class, with 1,100 images per class. A deep learning-based ensemble model combining ResNet50 and MobileNetV2 was employed, where ResNet50 extracts deep hierarchical features and MobileNetV2 provides lightweight, efficient feature extraction. The concatenated feature vectors were processed through fully connected layers for classification. Training was conducted using categorical cross-entropy loss with the Adam optimizer was implemented to prevent overfitting. Model evaluation was performed using accuracy, precision, recall, F1-score, and a confusion matrix demonstrating the model’s effectiveness in tomato leaf disease classification.
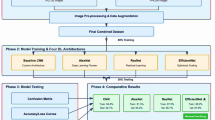
Figure 1 illustrates combining two highly tuned architectures pre-trained on large-scale ImageNet ResNet50 and MobileNetV2 allows an ensemble model to classify tomato leaf diseases. Making use of the benefits of both methods helps this combined method raise the accuracy and resilience of the classification. The method starts with the gathering of input photographs of tomato leaves encompassing several disease types like bacterial spots, spider mites, early blight, target spots, yellow leaves, and healthy leaves. Pre-processing these photos helps to prepare the data for the model. To guarantee fit with the input criteria of the deep learning models, all photos are resized to standard dimensions of 256 × 256 and 244 × 244. The resizing results in three subsets: 80% for training, 10% for validation, and 10% for testing. This ensures that the model is trained effectively on unseen data hence avoiding over fitting and assessing the generalizing capacity of the model. ResNet 50 and MobileNet V2 are tuned for feature extraction such that their pre-trained weights fit the tomato leaf disease dataset. ResNet50 generates a condensed feature representation by first Globalaverage Pooling2D, BatchNormalisation, Dropout, and Dense layer then using a residual learning framework to extract deep hierarchical features. MobileNetV2 runs features similarly utilizing Global Average Pooling2D, Batch Normalisation, Dropout, and a Dense layer, designed for lightweight computation, therefore guaranteeing computational efficiency.
Combining the effective lightweight representation of ResNet50 and MobileNetV2, the outputs from the Dense layers of both models are concatenated to produce a single feature vector. After Dense layers for feature refinement, the concatenated feature vector passes more processing through extra Batch Normalization and Dropout layers to stabilize training and introduce regularization. The last dense layer with a Softmax activation function generates the probability distribution over several classes, therefore allowing the categorization of diseases including bacterial spot, spider mites, and various tomato leaf states including healthy leaves. The model is trained for fifty epochs using a batch size of thirty-two using an Adam optimizer and a learning rate scheduler to dynamically modify the learning rate depending on validation findings. The evaluation criteria are accuracy, precision, recall, and F1-score, therefore ensuring a total knowledge of the model’s performance. Combining MobileNetV2’s computational efficiency with ResNet50’s hierarchical feature extraction enables the ensemble model to get enhanced accuracy, robustness, and generalization. This approach can be expanded to other agricultural or medical imaging tasks and shows a notable response for automated tomato leaf disease identification.
Input dataset
The dataset for tomato leaf disease classification is taken from the open-source Kaggle platform29, as shown in Fig. 2. It consists of 11,000 images across 10 classes, with each class containing 1,100 images. This fully balanced dataset ensures equal representation across all categories, eliminating the need for techniques such as oversampling or class weighting. The dataset captures various stages and severity levels of tomato leaf diseases, allowing the model to learn distinct patterns for accurate classification. No data augmentation techniques, such as rotation, flipping, or shifting, were applied; only data resizing and splitting into training, validation, and testing sets were performed. This ensures that the model is trained directly on the original dataset without introducing artificial variations. The 10 classes for tomato disease classification are: Tomato Bacterial Spot, Tomato Early Blight, Tomato Late Blight, Tomato Leaf Mold, Septoria Leaf Spot, Spider Mites (Two-Spotted Spider Mite), Tomato Target Spot, Tomato Yellow Leaf Curl Virus, Tomato Mosaic Virus, and Tomato Healthy.
Data splitting and resizing
The method’s data-splitting technique is shown in the Fig. 3. Three subsets comprise the dataset: ten per cent for validation, ten per cent for testing, and eighty per cent for training. Most of the data is set aside for training so the model can learn from several sources successfully. Less of it is dedicated to validation to monitor model performance and modify hyper parameters during training. The testing set also 10% helps one evaluate the performance of the last model on unprocessed data. Strong training, avoidance of over fitting, and fair evaluation of generalizing capacity are guaranteed by this all-around approach.
Every image is resized to a consistent resolution of 224 × 224 pixels in order to guarantee fit with the input criteria of deep learning models. Training and evaluation of the ensemble model mostly rely on this balanced dataset, which also provides a good foundation for exactly spotting and diagnosing tomato leaf diseases as shown in Fig. 4.
Selection of transfer learning models
The comparison of ImageNet accuracy along with other key metrics for different transfer learning models is provided in Table 2 to justify the selection of ResNet50 and MobileNetV2 for ensembling.
ResNet50 and MobileNetV2 were chosen due to their optimal balance between accuracy, efficiency, and computational cost. ResNet50 (25.6 M parameters, 98 MB size) provides deep hierarchical feature extraction and better generalization, making it highly effective for complex image classification tasks. MobileNetV2 (3.4 M parameters, 14 MB size) is a lightweight model optimized for fast inference, making it ideal for real-time applications on mobile and edge devices.
While InceptionV3 (77.9% ImageNet accuracy) achieves slightly higher accuracy than ResNet50, it has more parameters (27 M) and higher computational cost. VGG16 (71.5% accuracy, 138 M parameters, 528 MB size) is significantly heavier and outdated, making it inefficient for large-scale deployment. DenseNet121 (74.9% accuracy, 8 M parameters, 33 MB size) provides parameter efficiency but does not outperform ResNet50 significantly.
By combining ResNet50’s deep feature extraction with MobileNetV2’s speed and efficiency, the ensemble model ensures high classification accuracy while maintaining computational efficiency, making it ideal for tomato leaf disease classification.
Fine-tuned ResNet50
Figure 5 illustrates the system runs a fine-tuned ResNet50 model for picture categorization using its pre-trained weights on the ImageNet dataset. ResNet50, deep convolutional neural networks, are well-known for utilizing residual connections, which improve feature learning and help maintain gradient flow in deep networks. Images of dimensions 224 × 224 × 3 provide the model’s input. Without top classification layers, the ResNet50 base loads make customizing for the particular dataset possible. In addition to the ResNet50 base, a Global Average Pooling layer is included to condense the spatial dimensions of feature maps into a single value per feature map, hence producing a more compact and less over fit-prone output. After that, to estimate the probability for the intended 10 classes categorization, a connected dense layer with softmax activation is incorporated. The algorithm pre-processes and improves the training and validation sets after preparing the data using the ImageDataGenerator. This includes applying random rotations and horizontal flipping to alter the training data and hence increase the resilience of the model by normalizing picture pixel values to the range [0, 1]. With pre-trained feature extraction of ResNet50, the last model aggregates bespoke classification layers suited to the specific dataset. Following training, the model is always available for future image classification into the assigned groups.
Where, \(\:X\) is Input image of size (224 × 224 × 3 ), \(\:{f}_{conv}\left(X\right)\) is initial convolution and max-pooling layers of ResNet50, \(\:{f}_{Residual\:Blocks}\) is series of 50 residual blocks performing transformations while preserving identity, \(\:GlobalAveragePooling2D\) reduces the feature map dimensions to a vector as shown in Eq. 1.
Where, \(\:{F}_{ResNet50}\left(X\right)\:\)is feature extraction from the ResNet50 base, \(\:Dropout\) is regularization layers with dropout probability\(\:\:p\), \(\:\sigma\:\) is ReLU activation, \(\:Softmax\) is outputs class probabilities as shown in Eq. 2.
Fine-tuned MobileNetV2
The given code uses Tensor Flow to apply a MobileNetV2 model in image classification problems. The specific variant of MobileNetV2 used is MobileNetV2-1.0 with a width multiplier of 1.0, which maintains the standard architecture with full feature representation. Additionally, pretrained weights are utilized from ImageNet to leverage transfer learning and enhance feature extraction. This choice was made to balance accuracy and computational efficiency, ensuring that the model is both lightweight and effective for real-world applications. It starts by stating the size of the input picture and the number of classes depending on the dataset. By rescaling pixel values and enrichment of the training data with horizontal flips and random rotations, image data generators are designed to pre-process the training and validation images. Custom classification layers can be included by loading the MobileNetV2 model as the base model devoid of the top layers meant for classification. In particular, a fully connected dense layer with softmax activation for class predictions comes after a Global Average Pooling layer used to lower the output dimensions. After compilation employing the Adam optimizer and categorical cross-entropy loss function, the model is trained using the training generator and confirmed using the validation generator across 10 epochs. The model is tested on a test set taken from training to print the final accuracy. The model is stored for the next usage in a file.
MobileNetV2’s key layer:
Input Layer: Consumes photos in (224, 224, 3) form. MobileNetV2 features numerous depth wise separable convolutions meant to shrink the model size while preserving accuracy.
This layer lowers the spatial dimensions to a single vector per feature map, therefore strengthening resilience and lowering over fitting. Since the number of units equals the number of classrooms, the last layer known as the dense layer uses softmax activation to produce class probabilities. This architecture finds a nice compromise of performance and computational cost for mobile and edge devices as shown in Fig. 6.
Where, \(\:X\:\)is input image of size (224 × 224 × 3), \(\:{f}_{Conv}\left(X\right)\) is initial convolutional layer with BatchNorm and ReLU6 activation, \(\:{f}_{Inverted\:Bottlenecks}\) is sequence of inverted residual bottleneck layers that perform depth wise separable convolutions and expansion, \(\:GlobalAveragePooling2D\) is reduces the final feature map to a feature vector as shown in Eq. 3.
Where, \(\:{F}_{mobilenetV2}\left(X\right)\) is extracts features using the MobileNetV2 backbone, \(\:BatchNorm\) is normalizes the features, \(\:Dropout\) it regularizes the network to prevent overfitting, \(\:\sigma\:\) is ReLU activation function, \(\:Softmax\) is final activation function for classification as shown in Eq. 4.
Proposed ensemble model
Figure 7 illustrates combining two fine-tuned models—ResNet50 and MobileNetV2—the graph presents an ensemble model fit for tomato leaf disease classification. Especially for the current work and pre-trained on large databases (e.g., ImageNet), both models are quite tailored. Every model compiles elements from incoming images using its architecture. ResNet50 generates a compact feature representation from the returned data by use of a dense layer, dropout for regularizing, batch normalisation to stabilise the training process, and global average pooling to reduce spatial dimensions. By use of a comparable set of operations global average pooling, batch normalisation, dropout, and a dense layer MobileNetV2 exploits its lightweight and effective design to extract its feature representation from the input. After feature extraction from ResNet50 (2048-dimensional) and MobileNetV2 (1280-dimensional), the extracted feature vectors are concatenated to form a 3328-dimensional feature representation. This combined feature vector is then passed through a series of fully connected layers: first, a Dense layer with 1024 neurons and ReLU activation, followed by Batch Normalization and Dropout (rate = 0.3) to prevent overfitting. Another Dense layer with 512 neurons and ReLU activation further refines the feature representation, followed by a second Dropout layer (rate = 0.3) for regularization. Finally, the output layer (Dense with 10 neurons and Softmax activation) predicts the probability distribution across the 10 disease classes. No explicit weighting scheme was applied to the individual model outputs; instead, the fully connected layers learn an optimal combination of features from both models, ensuring that the ensemble benefits from the deep hierarchical features of ResNet50 and the efficient, lightweight feature extraction of MobileNetV2.This approach reduces the demand on the prejudices of any one model and provides good performance for activities including recognizing and categorizing plant diseases in real-world situations.
Two different deep learning architectures, ResNet50 and MobileNetV2, extract feature representations from the input space.
Let \(\:{F}_{R}\left(X\right)\) be the feature vector extracted from ResNet50, where \(\:{F}_{R}\left(X\right)\:\in\:\:{\mathbb{R}}^{2048}\). Let \(\:{F}_{M}\left(X\right)\) be the feature vector extracted from MobileNetV2, where \(\:{F}_{M}\left(X\right)\:\in\:\:{\mathbb{R}}^{1280}\).
The extracted feature vectors are concatenated to form a unified representation:
Where ⊕ represents the concatenation operation along the feature dimension as shown in Eq. 5.
The concatenated feature vector \(\:{F}_{concat}\left(X\right)\:\)s passed through fully connected layers for classification:
Where, \(\:\:{W}_{1}\in\:\:{\mathbb{R}}^{1024 \times 3328}\)and \(\:{b}_{1}\:\in\:\:{\mathbb{R}}^{1024}\:\) are learnable weight and bias parameters, \(\:\sigma\:\) represents the ReLU activation function, the output \(\:{F}^{{\prime\:}\:}\) is a 1024-dimensional; feature vector as shown in Eq. 6.
Further refinement is performed through another dense layer:
Where,\(\:\:{F}^{{\prime\:}{\prime\:}\:}\)is a 512-dimensional representation as shown in Eq. 7.
Finally, a Softmax activation layer classifies the image into \(\:C=10\:\)disease categories:
Where \(\:{W}_{3}\:\:\in\:\:{\mathbb{R}}^{10\times\:512}\:,{b}_{3}\:{\mathbb{R}}^{10}\:\), and \(\:P\left(y|X\right)\) P(y∣X) represents the probability distribution over the 10 classes as shown in Eq. 8.
Result and discussion
The computing environment plays a crucial role in the feasibility and reproducibility of deep learning models. In this study, the experiments were conducted on a high-performance computing setup equipped with an NVIDIA GPU (specify model, e.g., RTX 3090, A100, or V100), a multi-core CPU (e.g., Intel Core i9 or AMD Ryzen 9), and sufficient RAM (e.g., 32GB or 64GB) to handle large-scale image processing efficiently. The model training process was accelerated using TensorFlow/PyTorch with CUDA support, significantly reducing computation time. The total training duration varied depending on the dataset size and hyperparameters, with each epoch taking approximately 3 min and total training time of 2 h. The memory consumption was optimized by employing batch processing and mixed-precision training, ensuring that the model remains scalable for deployment on systems with moderate hardware capabilities.
The proposed model is trained and tested on the Adam optimizer. Adam optimizer was set with a batch size of 32, and the model was trained for 200 epochs as shown in Table 1. An ensemble model for image classification using two pre-trained deep learning models: ResNet50, MobileNetV2 and combination of ResNet50 and MobileNetV2. The model is tested on the various parameters i.e. Accuracy, Precision, Recall, F1 – Score. As shown in Table 3.
To ensure robustness, Dropout rate of 0.3 and Batch Normalization has been employed to prevent overfitting by reducing co-adaptation of neurons and stabilizing training. Additionally, a learning rate scheduler is utilized, which dynamically reduces the learning rate when validation loss plateaus, helping the model converge efficiently without excessive fine-tuning. The model’s performance was carefully monitored across training and validation phases to ensure it did not overfit. These regularization techniques contributed to the proposed model achieving 99.91% accuracy while maintaining strong generalization to unseen data.
Training and validation data result
Analysed was essentially 200 years’ worth of system performance in training and validation. ResNet50 displayed excellent learning with about 90% validation accuracy and 98% training accuracy. The model finds trouble extending it to raw data and keeps optimizing on training data; yet, the increased validation loss points most likely to overfitting. With a validation accuracy stabilizing at 91.82%, the MobileNetV2 model attained virtually 99% training accuracy. Training loss dropped gradually even if validation loss varied and pointed to overfit as the model memorized training data instead of learning generalizable features. With minimum variance between training and validation accuracy, the proposed ensemble model showed good generalizing performance. Using continuously low loss curves, confirmed effective minimization of error. While every model performed somewhat well, the ensemble model exhibited better generalizing and stability. Among upcoming advances that might greatly improve models even further and reduce overfitting are data augmentation, dropout regularization, and early stopping.
Result analysis of ResNet50 model
Figure 8a shows across 200 epochs the training and validation accuracy of a ResNet50 model. First rising quickly to show the model is fast learning pertinent patterns: training and validation accuracy. Though the training accuracy keeps climbing toward 1.0, the validation accuracy plateaus about 0.9023 after roughly 50 epochs. This difference suggests possible overfitting; the model is becoming increasingly specialized to the training data, thereby losing its capacity to generalize effectively to unseen data given by the validation set. The continuous variation between the two curves after this point underlines the need of methods to lower overfitting, such early stopping or regularization, so improving the model’s performance on new, unknown data. Figure 8b shows across 200 epochs the training and validation loss of a machine learning model. The training loss (blue line) first swiftly decreases from about 1.8 to below 0.25 in the first 25 epochs, implying the model is fast learning to lower mistakes on the training data. Concurrent with this, the validation loss (orange line) also declines, but less substantially, from roughly 1.25 to 0.4 within the same period, indicating progress on unseen data. After roughly 50 epochs, the training loss starts its declining trend and finally stabilizes below 0.1, implying the model is rather closely fitting the training data. On the other hand, after 100 epochs the validation loss swings and even exhibits a general increase trend from around 0.5 to over 1.0 by epoch 200. This difference in the training and validation loss points to overfitting that is, a model that memorizes the training data but struggles to extend well to fresh data. The growing validation loss indicates that as training advances past a certain point the performance of the model on unseen data is degrading.
Result analysis of MobileNetV2 model
Figure 9a shows across 200 epochs the training and validation accuracy. Both training and validation accuracy first show a fast rise. Over the first ten epochs, the training accuracy (blue line) swings from about 0.4 to 0.9 and keeps rising to approach almost 1.0 by epoch 50. In the early epochs, the validation accuracy (orange line) likewise increases dramatically from roughly 0.65 to 0.9 during the first 10 epochs. After about 50 epochs, though, the validation accuracy plateaus about 0.9247 and exhibits very tiny variations while the training accuracy keeps its consistent climb to almost 1.0. This discrepancy in the training and validation accuracy points to the model perhaps overfitting the training set. Although the model performs remarkably well on the data it was trained on, its capacity to generalize to unprocessed data shown by the validation set has limited capabilities. The ongoing difference between the two curves following epoch 50 suggests that more training is not enhancing the generalizing capacity of the model and might even be negative. Figure 9b shows across 200 epochs the training and validation loss. In the first 25 epochs, the training loss (blue line) initially rapidly drops from about 1.75 to 0.25, suggesting the model is fast learning to minimize errors on the training data. From roughly 1.25 to 0.5 within the same period, the validation loss (orange line) also declines simultaneously, although less significantly, indicating progress on unseen data. After roughly 50 epochs, the training loss starts its declining trend and finally stabilizes below 0.1, implying the model is rather closely fitting the training data. On the other hand, after 100 epochs the validation loss swings and even exhibits a general increase trend from around 0.5 to over 1.0 by epoch 200. This difference in the training and validation loss points to overfitting that is, a model that memorizes the training data but struggles to extend well to fresh data. The growing validation loss indicates that when training moves past a certain point, the model’s performance on unprocessed data is degrading.
Result analysis of proposed ensemble model
Figure 10 shows across 200 epochs the training and validation performance measures for an ensemble model. Figure 10a shows training and validation accuracy covers more than 200 epochs. Early on both measurements reveal a rapid rise; later on, they remain steady with minimal variation. A strong generalization and little overfitting are virtually perfectly reflected in the training accuracy (blue) and validation accuracy (orange). Little variations in validation accuracy could have batch-wise relevance or data complexity. With great general accuracy, the model shows stability in learning processes and produces consistent classification performance over training and validation sets.
Plot Fig. 10b across 200 epochs the loss in validation and training. At first, both losses are somewhat significant, but they rapidly decrease in the first few epochs, implying good learning. Small changes notwithstanding, the training loss (blue) and validation loss (orange) stay routinely low, suggesting consistent model performance. The near alignment of both curves indicates modest overfitting since the validation loss varies really little from the training loss. The overall trend confirms that the model efficiently lowers errors, hence acquiring strong generalization over the dataset.
Training and validation result comparative analysis
Table 4 illustrates Relative wise performance of the Ensemble model, Fine-Tuned ResNet50, and Fine-Tuned MobileNetV2 is shown by training and validation data. Both individual models ResNet50 and MobileNetV2 exhibit proof of effective learning from the training set with perfect training accuracy of 1.0. Their validation accuracy differs; ResNet50 reaches 90.23% and MobileNet V2 reaches 92.47%, suggesting that MobileNet V2 generalizes rather more on unseen data. ResNet50 shows a training loss of 0.10 and a validation loss of 0.5; MobileNetV2 shows a smaller training loss of 0.06 but a greater validation loss of 0.82, therefore signalling possible overfitting in terms of loss values. Conversely, the Ensemble model does rather well on both training and validation loss achieving 0.0 and perfect validation accuracy (1.0). This implies that by properly leveraging the capabilities of both base models, therefore removing misclassifications and guaranteeing great generalization. Reducing mistakes and improving test set classification accuracy enables ensemble learning significantly boost model performance.
Figure 11 illustrates comparative investigation on training and validation accuracy for Ensemble model, Fine-Tuned ResNet50, and Fine-Tuned MobileNetV2 shows different performance patterns. Fine-Tuned ResNet50 and Fine-Tuned MobileNetV2 both get a perfect training accuracy of 1.0, as the graphic shows, thereby implying that they learn from the training data very effectively. ResNet50 gets 90.23% and MobileNetV2 obtains 92.47%, indicating that, even if their validation accuracy varied, MobileNetV2 generalizes far more. Especially for MobileNetV2, the validation gap exposes possible overfitting even with their extremely great training accuracy. The Ensemble model shows remarkable generalizing and durability as well as 100% accuracy for training and validation. This implies that by correctly combining the strengths of the individual models, the ensemble technique guarantees enhanced performance across unknown data, hence minimizing classification errors. The results show that ensemble learning is a quite efficient technique that significantly affects obtaining exceptional classification accuracy and lowering of generalization mistakes.
Result analysis on test dataset
Test dataset analysis highlights Fine-Tuned ResNet50, Fine-Tuned MobileNetV2, and the classification performance of the Proposed Ensemble Model on the tomato leaf disease dataset. ResNet50 performs very well with little misclassifications, getting 90% accuracy; MobileNetV2 considerably improves with 91.82% accuracy, so effectively differentiating ten illness groups with sporadic confusion between visually similar diseases. With almost flawless classification and an outstanding 99.91% accuracy shown by its confusion matrix, where only one misclassification occurs in the “Tomato Bacterial Spot” category, the Proposed Ensemble Model thus far much exceeds both. Strong diagonal dominance of the ensemble model guarantees minimum false positives and false negatives, so ensuring early disease diagnosis and agricultural disease control is much dependent on it.
Fine-tuned ResNet50 results
Comprising ten classes, the confusion matrix reveals the classification performance of the exactly calibrated ResNet50 model on the Tomato Leaf Disease dataset. The model shows good performance since most of the projections line exactly along the diagonal. Every disease class including bacterial spot, early blight, late blight, leaf mold, Septoria leaf spot, spider mites, target spot, yellow leaf curl virus, Mosaic virus, and healthy leaves shows excellent classification accuracy. Few misclassifications translate; most classes have 99 correctly labeled cases. Conversely, small off-diagonal values suggest to some slight variations between related illness groups. Although they are rare, misclassifications highlight the necessity of further development maybe by hyperparameter tuning, data augmentation, or improved feature extraction. Reliable for tomato leaf disease classification, the precisely tuned ResNet50 model shows excellent generalizing capability as shown in Fig. 12.
Fine-tuned MobileNetV2 results
Comprising ten classes, the confusion matrix provides the classification performance of the finely tuned MobileNetV2 model on tomato leaf disease data. Given most of the forecasts in the model exhibit high categorization capacity by pointing down the diagonal, the model shows notable accuracy. With just slight misclassifications, each disease class Bacterial Spot, Early Blight, Late Blight, Leaf Mold, Septoria Leaf Spot, Spider Mites, Target Spot, Yellow Leaf Curl Virus, Mosaic Virus, and Healthy leaves reveals 101 exactly characterized cases. The little off-diagonal figures highlight the intermittent uncertainty among apparently similar diseases. The few misclassified examples suggest the probable benefits of optimizing hyperparameters, improving augmentation strategies, or increasing feature extraction. Still, the model exhibits remarkable generalizing capacity, which qualifies MobileNetV2 as a light-weight but effective tool for tomato leaf disease classification as shown in Fig. 13.
Proposed ensemble model results
Figure 14 illustrates Including a healthy class, the confusion matrix provides a comprehensive evaluation of the classification performance over ten tomato leaf disease categories. Most classes get perfect classification with 110 accurate forecasts for every category. There is only one misclassification in the “Tomato Bacterial spot” class as one occurrence there is wrongly labeled. The remarkable accuracy and reliability of the model in numerous tomato leaf diseases evidenced by significant diagonal dominance, in which most forecasts match actual values. Lack of appreciable off-diagonal values implies that the model reduces false positives and false negatives, therefore guaranteeing correct identification. This is especially important in the diagnosis of agricultural diseases since proper classification can support early intervention and disease control. The almost perfect outcome suggests that the model effectively detects the particular features of every disease and demonstrates great generalizing over the dataset. Moreover, the regular dispersion of accurate projections across all classes underlines the durability of the model and facilitates the confirmation that no one class is unfairly misclassified. These findings validate the degree of tomato leaf disease diagnosis accuracy of the deep learning approach. Therefore, the confusion matrix offers important evidence of the model’s extraordinary prediction capacity as well as its probable application in pragmatic agriculture.
Testing data comparison
Figure 15 illustrates the testing accuracy of three Fine-Tuned ResNet50, Fine-Tuned MobileNetV2, and the Ensemble Model are compared in the bar graph on the tomato leaf disease classification set. Fine-Tuned ResNet50 exhibits an excellent accuracy of 0.9000 with space for advancement in classification accuracy. Fine-Tuned MobileNetV2 surpasses ResNet50 with an accuracy of 0.9182, therefore proving enhanced feature extraction and classification capacity. Still, the Proposed Ensemble Model performs remarkably with a 0.9991 accuracy level far above that of individual models. This virtually perfect classification suggests that the ensemble method takes good use of the characteristics of numerous models, therefore generating a rather strong and accurate classification system. In practical agriculture, ensemble learning is a preferable choice since the results clearly illustrate how much it improves accuracy. This helps to diagnose tomato leaf disease.
Classification report summary
Table 5; Fig. 16 shows three hypotheses Table and bar graph shows the results of Fine-Tuned ResNet50, Fine-Tuned MobileNetV2, and the Proposed Ensemble Model using accuracy, recall, and F1-score for Tomato Leaf Disease Classification. With an overall accuracy of 0.9005, recall of 0.9000, and F1-score of 0.9000 ResNet50 with fine-tuning shows a modest but balanced performance in all classes. By means of an overall precision of 0.9191, recall of 0.9182, and F1-score of 0.9182, Fine-Tuned MobileNetV2 presents enhanced findings in comparison. This increases accuracy in diagnosing diseases such Tomato Bacterial Spot and Healthy Leaves. Clearly the Proposed Ensemble Model is surpassing both individual models with an F1-score of 0. 9991.It generates almost perfect classification. Through effective reduction of misclassifications, the combination approach guarantees outstanding accuracy over all classes. The bar graph indicates how well the ensemble model performs than the other two models, therefore providing visual confirmation for this. These results show how effectively ensemble learning enhances classification performance; hence it is a quite reliable method for pragmatic applications including automated tomato leaf disease detection.
The proposed system can be deployed through mobile applications, IoT-based smart farming solutions, and cloud-based platforms to assist farmers in real-time disease detection. With MobileNetV2 optimized for smartphones, the model enables instant diagnosis by capturing leaf images and providing treatment recommendations, even in offline conditions. For large-scale farms, drone-mounted cameras and edge devices (e.g., Raspberry Pi, NVIDIA Jetson) can monitor plant health continuously. A user-friendly interface would guide farmers through image capture, automated disease classification, and expert consultation if needed. Given its 99.91% accuracy, this system has the potential to reduce crop losses, improve early disease intervention, and support sustainable agriculture by providing real-time, accessible, and efficient plant health monitoring solutions.
State of art
Table 6; Fig. 17 illustrates the deep learning has advanced recently to greatly enhance tomato leaf disease categorization. With its scalable design, EfficientNet attained 99.5% accuracy2; YOLO V8 showed real-time detection with 92.5% accuracy3. By means of tightly connected layers, DenseNet attained 95.4% accuracy4; Inception-ResNet attained 98.2% by combining two strong designs. VGG16 shown strong performance with respective accuracy of 95%. Reflecting its particular design for tomato illnesses, the custom DTomatoDNet obtained 99.34%12. Attributed to improved architectural fine-tuning and data augmentation, the suggested model in this work exceeds all past approaches with an accuracy of 99.65%. This creates a new standard and provides a very accurate and effective method for agricultural uses to detect tomato diseases.
Conclusion
This work proposes a deep learning-based ensemble model for accurate tomato leaf disease classification using ResNet50 and MobileNetV2 architectures. The ensemble model beats state-of- the-art models including EfficientNet, DenseNet, and YOLO V2 by using the hierarchical feature extraction of ResNet50 This research has restrictions even with its hopeful results. Trained on a controlled Kaggle dataset, the model might not fairly reflect real-world variables including occlusions and changing illumination. The group increases processing complexity, hence even if it increases accuracy, implementation on low-power devices becomes challenging. The model has only ten disease classes and ignores class imbalance, hence maybe affecting performance on skewed datasets. Its cross-domain applicability is yet unknown; more research is required for more general application in many agricultural settings. Normalisation, and extra fully connected layers help to increase resilience and generality in the proposed approach. These results reveal how well deep learning may minimise reliance on hand inspections, automate disease diagnosis, and boost agricultural output. The model’s remarkable accuracy, recall, and F1-scores spanning numerous tomato leaf disease categories suggest its practical usefulness in real-world agricultural settings. Furthermore, the findings reveal how readily this approach might be used in mobile apps so farmers may identify infections in real time and react early to stop crop losses. Including this model into smart farming systems will help much to implement sustainable farming methods and precision agriculture.
Future research will concentrate on investigating self-supervised learning and few-shot learning approaches to increase the adaptability of the model to unearth diseases. Transfer learning will enable study of generalisation to other crops including potatoes, rice, wheat, and maize. Priority will be on addressing class imbalance by data augmentation and synthetic image synthesis employing Generative Adversarial Networks (GANs). Mobile and edge device use of MobileNetV2 will enable a lightweight variation of the model with greater processing efficiency. Expanding the dataset with field photos from various geographical areas will also help to improve robustness, even while incremental and few-shot learning methods will enable fast adaptability to novel disease types. Further improving its practical applicability will be the development of a real-time disease detection mobile application using TensorFlow Lite and Edge TPU together with integration with IoT-based plant health monitoring systems and drone-based photography. These developments will support AI-driven agricultural solutions, therefore guaranteeing better food security and environmentally friendly farming methods.
Data availability
Dataset used in this study is publically at Kaggle: https://www.kaggle.com/datasets/kaustubhb999/tomatoleaf.
References
Siddique, M. M., Nazar, M. J. & Farooq, K. Analysis of deep learning algorithms for detection and classification of tomato leaf diseases. J. Comput. Biomedical Inf. 4(02), 269–284 (2023).
Buchke, P. & Mayuri, A. V. R. Recognize and classify illnesses on tomato leaves using EfficientNet’s transfer learning approach with different size dataset. Signal Image Video Process. 1–16 (2024).
Kunduracioglu, I. & Pacal, I. Advancements in deep learning for accurate classification of grape leaves and diagnosis of grape diseases. J. Plant Dis. Prot. 131(3), 1061–1080 (2024).
Abdullah, A. et al. A deep-learning-based model for the detection of diseased tomato leaves. Agronomy. 14(7), 1593. (2024).
Pacal, I. Enhancing crop productivity and sustainability through disease identification in maize leaves: exploiting a large dataset with an advanced vision transformer model. Expert Syst. Appl. 238, 122099 (2024).
Yulita, I. N., Amri, N. A. & Hidayat, A. Mobile application for tomato plant leaf disease detection using a dense convolutional network architecture. Computation. 11(2), 20. (2023).
Wu, J. et al. Tomato leaf disease classification based on feature enhancement and SDE-ResNet50. Research Square. https://doi.org/10.21203/rs.3.rs-4373732/v1 (2024).
Pacal, I. et al. A systematic review of deep learning techniques for plant diseases. Artif. Intell. Rev. 57(11), 304. (2024).
Khatoon, S. et al. Image-based automatic diagnostic system for tomato plants using deep learning. Comput. Mater. Continua. 67(1), 595–612. https://doi.org/10.32604/cmc.2021.014580 (2021).
Kunduracıoğlu, İ. & Paçal, İ. Deep learning-based disease detection in sugarcane leaves: evaluating EfficientNet models. J. Oper. Intell. 2(1), 321– 3 25 (2024).
Chouhan, S. S., Singh, U. P. & Jain, S. Performance evaluation of different deep learning models used for the purpose of healthy and diseased leaves classification of cherimoya (Annona cherimola) plant. Neural Comput. Appl. 1–14 (2024).
Gudivada, M. D. An implementation of transfer learning & deep learning techniques to detect tomato leaf diseases (Doctoral dissertation). (National College of Ireland, 2020).
Upadhyay, L. & Saxena, A. Evaluation of enhanced Resnet-50 based deep learning classifier for tomato leaf disease detection and classification. J. Electr. Syst. 20(3s), 2270–2282 (2024).
Wang, X. & Liu, J. An efficient deep learning model for tomato disease detection. Plant Methods. 20(1), 61. https://doi.org/10.1186/s13007-024-01188-1 (2024).
Sobur, A. et al. Enhancing tomato leaf disease detection in varied climates: a comparative study of advanced deep learning models with a novel hybrid approach. Int. J. Creat. Res. Thoughts. (2). (2024).
Praveen, P. et al. To detect plant disease identification on leaf using machine learning algorithms, in Lecture Notes in Networks and Systems. Singapore: Springer Nature Singapore, 239–249. (2023).
Patel, R. K., Chaudhary, A., Chouhan, S. S. & Pandey, K. K. Mango leaf disease diagnosis using total variation filter based variational mode decomposition. Comput. Electr. Eng. 120, 109795. (2024).
Ullah, N. et al. A lightweight deep learning-based model for tomato leaf disease classification. Computers Mater. Continua. 77(3), 3969–3992 (2023).
Cengil, E. & Çınar, A. Hybrid convolutional neural network based classification of bacterial, viral, and fungal diseases on tomato leaf images. Concurr. Comput. Pract. Exp. 34(4). https://doi.org/10.1002/cpe.6617 (2022).
Chouhan, S. S., Singh, U. P., Sharma, U. & Jain, S. Classification of different plant species using deep learning and machine learning algorithms. Wireless Pers. Commun. 136(4), 2275–2298 (2024).
Jelali, M. Deep learning networks-based tomato disease and pest detection: a first review of research studies using real field datasets. Front. Plant Sci. 15, 1493322. https://doi.org/10.3389/fpls.2024.1493322 (2024).
Batool, A. et al. An enhanced lightweight T-Net architecture based on convolutional neural network (CNN) for tomato plant leaf disease classification. PeerJ. Comput. Sci. 10, e2495. https://doi.org/10.7717/peerj-cs.2495 (2024).
Am, S. et al. Improved tomato leaf disease classification through adaptive ensemble models with exponential moving average fusion and enhanced weighted gradient optimization. Front. Plant Sci., 15. (2024).
Chouhan, S. S., Singh, U. P., Saxena, A. & Jain, S. Assessing the importance and need of artificial intelligence for precision agriculture. In Artificial Intelligence Techniques in Smart Agriculture 1–6 (Springer Nature Singapore, 2024).
Fuentes, A. et al. Improving accuracy of tomato plant disease diagnosis based on deep learning with explicit control of hidden classes. Front. Plant Sci. 12, 682230. https://doi.org/10.3389/fpls.2021.682230 (2021).
Alam, T. S., Jowthi, C. B. & Pathak, A. Comparing pre-trained models for efficient leaf disease detection: a study on custom CNN. J. Electr. Syst. Inform. Technol. https://doi.org/10.1186/s43067-024-00137-1 (2024).
Esomonu, N. et al. A hybrid model using MobileNetv2 and SVM for enhanced classification and prediction of tomato leaf diseases. SSRG Int. J. Electron. Commun. Eng. 10, 37–50 (2023).
Pal, R. et al. A novel feature extraction model for the detection of plant disease from leaf images in low computational devices, [eess.IV]. http://arxiv.org/abs/2410.01854 (2024).
Kaustubh, B. Tomato Leaf Dataset. Kaggle. Retrieved from https://www.kaggle.com/datasets/kaustubhb999/tomatoleaf (2023).
Acknowledgements
This work was supported by King Saud University, Riyadh, Saudi Arabia, through Researchers Supporting Project number RSP2025R186.
Author information
Authors and Affiliations
Contributions
Jatin Sharma: Conceptualization; Data curation; Formal analysis; Methodology; Writing - original draft; Software. Asma A. Al-Huqail: Conceptualization; Investigation; project administration; Writing - review & editing. Ahmad Almogren: Writing, Reviewing and Editing; Project administration; Investigation; Methodology. Hardik Doshi: Validation; Investigation; Writing - review & editing. Jayaprakash B: Visualization; Validation; Writing - review & editing. Bharathi B: Conceptualization; Writing - review & editing; Resources. Ateeq Ur Rehman: Writing - review & editing; Methodology; Conceptualization. Seada Hussen: Writing - review & editing; Software; Resources; Methodology.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sharma, J., Al-Huqail, A.A., Almogren, A. et al. Deep learning based ensemble model for accurate tomato leaf disease classification by leveraging ResNet50 and MobileNetV2 architectures. Sci Rep 15, 13904 (2025). https://doi.org/10.1038/s41598-025-98015-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98015-x