Abstract
Chickpea, a widely cultivated legume, actively fix atmospheric nitrogen in root nodules through a symbiotic relationship with rhizobia bacteria. A recombinant inbred line (RIL) population, progressing from F2 to F7 generations, was developed in a short-period of 18 months using the Rapid Generation Advancement (RGA) protocol. The F7 RILs were evaluated during the 2020-21 and 2021-22 crop seasons under typical field conditions to quantify the effects of nodulation on seed yield (SY) and its associated traits. The analysis of variance revealed a highly significant difference (P < 0.01) among genotypes for seed yield and other agronomic traits, with no significant seasonal effect. In the pooled analysis, nodulating genotypes (NG) exhibited a substantial increase (P < 0.01) in SY (62.55%), 100-seed weight (SW100; 12.21%), harvest index (HI; 6.40%), number of pods per plant (NPPP; 39.55%), and number of seeds per plant (NSPP; 44.37%) compared to non-nodulating genotypes (NNG). Both NG and NNG exhibited a significant (P < 0.01) positive correlation between SY and NPPP (r = 0.64 and 0.63), NSPP (r = 0.66 and 0.61), HI (r = 0.27), and number of primary branches per plant (PBr) (r = 0.31), respectively. The top-performing genotypes for yield and related traits were predominantly nodulating. Genotype-trait bi-plot analysis identified nine nodulating genotypes as the most adaptable across the two seasons—six for SY, plant height, SW100, and three for days to first flowering and maturity. These findings underscore the critical role of nodulation in maximizing chickpea yields and the significant yield penalties associated with non-nodulation. To boost chickpea production, future breeding efforts should focus on developing genotypes with high compatibility with rhizobium strains.
Similar content being viewed by others
Introduction
Legumes have a specific mutualistic relationship with soil bacteria that fix the atmospheric nitrogen into plant-usable ammonical form in root nodules1, in contrast to other plants. Development of nodules and nitrogen fixation depends majorly on the legume cultivar and its specific rhizobial strain2,3,4 which will in turn improve the overall crop growth, productivity5,6 and soil health. Around 20 to 22 million tonnes of nitrogen per annum is fixed in the agricultural systems by the Leguminosae family members7 which contributes significantly towards reducing the global carbon footprint.
Chickpea (Cicer arietinum L.) is a highly nutritious, diploid (2n = 2x = 16) legume crop grown in an area of 14.2 million hectares across 56 countries globally8, of which, India is the largest producer and consumer. It is considered a storehouse of proteins, complex carbohydrates, vitamins, and micronutrients that are required for human nutrition9,10,11,12,13. Being a leguminous crop, chickpea fixes the atmospheric nitrogen by association with Rhizobium species14 which differentiates it from other cereal crops15. This ability to establish a symbiotic relationship is critical for legumes as it provides them with a readily available source of nitrogen, an essential macronutrient for plant growth and development. It stores the fixed nitrogen in the nodules present in its root system and converts it into ammonia that can be used by the plant16,17. It was estimated that about 70 kg of nitrogen per hectare is fixed annually by chickpea18, which helps in providing nitrogen not only to the host but also to the subsequent crops grown19 thereby, helping the farmers in reducing the cost of production.
Root nodulation is a complex process requiring the recognition of symbiotic bacteria, Nod-factor induced infection, and root growth20,21,22. Polyphenols named flavonoids released from the roots, stimulate Nod-factor production in rhizobia thereby initiating curling and colonization of root hair and the formation of root nodules23. Phytohormone signaling plays a major role during this process between the bacteria and the host24. The symbiotic nitrogen fixation (SNF) efficiency is dependent on the host, rhizobial strains, soil conditions25, availability of phosphorous26, and environmental conditions27. Under water stress, the biochemical activity in the nodules will get disrupted resulting in the senescence of nodules28,29,30 along with downgraded leg-hemoglobin content and nitrogenase activity31. The ability of the nodules to supply energy, transport, regulate oxygen molecules, and assimilate ammonia to the plants will help in increasing plant growth and yield32. However, the consequences of the chickpea roots being unable to make a symbiotic relationship with rhizobium are not well understood and documented.
Nitrogen is a key component of proteins, nucleic acids, and chlorophyll, all vital for plant function. Hence, effective nodulation directly impacts legume productivity and nutritional value. Previous studies in legumes have identified numerous genes and signaling pathways involved in nodulation, shedding light on the molecular basis of signaling between the plant and the bacteria33,34. Conversely, the genetic basis of non-nodulation in certain legume species has been linked to mutations in key nodulation genes or disruptions within the signaling cascades vital for establishing this symbiosis35. For example, studies in Medicago truncatula have revealed that mutations in the NFP (Nod Factor Perception) gene result in nodulation deficiency36. Unraveling the genetic mechanisms underlying both successful nodulation and its absence is crucial not only for enhancing nitrogen fixation in crops but also for broadening our understanding of the intricate plant-microbe interactions that underpin sustainable agriculture. Several studies reported in chickpea14,37,38,39 as well as in other leguminous crops viz., soybean40, cowpea30, lupin41, groundnut42, etc., were focused majorly on the external application of biochar, artificial fertilizers, growth hormones and the findings of strain-specific effect on nodulation and yield-related traits. However, the information on the impact of nodulation over non-nodulation on yield and yield-contributing traits was minimal. In this context, the current study aimed to (1) broaden the knowledge on the association between nodulation, yield, and its associated traits; and (2) quantify the value gain of the traits in a RIL population segregating for nodulation trait. In addition, we successfully showcased the utility of rapid generation advancement methods for developing mapping populations in a short period.
Results
Analysis of variance
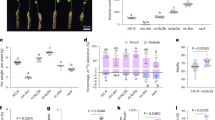
The results of the combined analysis of variance (ANOVA) revealed significant differences (Prob < 0.01) among the genotypes for the traits under study while for the factors, block and replication, no significant difference was observed (Table 1). For nodulating genotypes (NG), mean performance of agro-morphological and yield-related traits over the seasons was in the range of 42.74–47.37 (Days to first flowering- DF), 47.57–51.76 (Days to 50% flowering- DFF), 94.24-100.92 (Days to maturity- DM), 38.74–44.36 (Plant height- PH), 1.94–2.17 (Number of primary branches per plant- PBr), 6.03–8.48 (Number of secondary branches per plant- SBr), 293.76-343.36 (Number of pods per plant- NPPP), 349.08-395.66 (Number of seeds per plant- NSPP), 17.12–17.26 (100 seed weight- SW100), 34.50-36.85 (Harvest index- HI), 2472.78-2689.62 (Seed yield- SY) (Table 2). Similarly, non-nodulating genotypes (NNG) exhibited comparable trends over the seasons ranging from 44.58 to 50.60 (DF), 49.27–54.87 (DFF), 96.20-103.46 (DM), 34.86–39.28 (PH), 1.98–2.10 (PBr), 6.97–8.42 (SBr), 194.53-265.36 (NPPP), 214.48–30.23 (NSPP), 14.29–16.27 (SW100), 33.19–33.87 (HI) and 1638.78-1665.05 (SY) (Table 2). In addition to this, checks exhibited a range of 42.61–43.38 (DF), 47.50-48.53 (DFF), 93.26–98.52 (DM), 54.02–59.63 (PH), 1.99–2.07 (PBr), 5.06–7.81 (SBr), 296.37-321.63 (NPPP), 320.70–338.00 (NSPP), 21.55–21.67 (SW100), 31.68–34.34 (HI), 2644.09-2667.64 (SY) (Table 2) over the seasons.
The median values (black solid line) for agro-morphological and yield component traits across seasons were provided using Violin plot analysis (Fig. 1). All the traits exhibited almost consistent median values except for PH, PBr, and SW100 for both NG and NNG over the seasons, highlighting the importance of seasonal variations in the breeding selection process. Notably, the observed ranges of agro-morphological traits and yield components of NG, NNG, and checks, emphasize the inherent variability within the genotypes (nodulating and non-nodulating) across the seasons, which underscores the superiority of nodulating genotypes in terms of productivity.
Genetic parameters for the yield and agro-morphological traits
The genetic parameters for yield, yield components, and agro-morphological traits in NG and NNG chickpea are presented in Supplementary Tables 2 and 3, respectively. For NG (Supplementary Table 2), low Genotypic coefficient of variation (GCV) and Phenotypic coefficient of variation (PCV) were observed for DF, DFF, and DM; moderate to low for PH; low to moderate for PBr; and high to low for SBr. The genetic advance of mean (GAM) estimates was low for DM; low to moderate for DF and DFF; low to high for SBr; and moderate to high for PH and PBr in pooled data. Yield and yield component traits showed low PCV and GCV for HI, moderate to low for SW100, and high to moderate for NPPP, NSPP, and SY. GAM estimates were high for NPPP, NSPP, and SY, while SW100 and HI had moderate to low estimates in pooled data. For NNG (Supplementary Table 3), PCV and GCV estimates were low for DF, DFF, and DM; moderate to low for PH and PBr; and high to low for SBr. GAM estimates were low to moderate for DF and DFF; low for DM; moderate to high for PH; and low to high for PBr and SBr.
High estimates of PCV, GCV, heritability, and GAM for yield and yield components were observed for NPPP, NSPP, and SY. PCV and GCV were low to moderate for SW100 and HI, and high for SY. Promising NG and NNG genotypes were identified for yield and yield components (Table 3). Of the NG, NG-172 recorded the highest yield (3966.7 kg/ha) followed by NG-88 (3730.7 kg/ha), NG-188 (3584.7 kg/ha), NG-159 (3563.8 kg/ha), and NG-13 (3561.6 kg/ha). Despite the non-significant increase in SY, it was higher than the best checks, RVG 204 and Phule Vikram (Table 3). Among NNG, NNG-206 recorded the highest yield (3153.7 kg/ha) followed by NNG-182 (2881.3 kg/ha), NNG-163 (2676.4 kg/ha), NNG-80 (2661.9 kg/ha), and NNG-152 (2648.5 kg/ha), demonstrating comparable seed yield with the best checks.
Correlation of agro-morphological, yield, and yield component traits
Karl Pearson’s correlation coefficients were calculated for the agro-morphological, yield, and yield component traits of NG and NNG (Supplementary Tables 4 and 5). In NG under both seasons, SY showed a significant positive correlation with PH, NPPP (except 2020-21), NSPP, and SW100. Similarly, in pooled, SY was positively associated with PH, PBr, NPPP, NSPP, and HI. DF and DFF traits showed a significant negative correlation with DM and SW100 in both the crop seasons and pooled data. Whereas PH showed a significant positive correlation with SW100 in both the seasons and pooled. For NPPP, a positive significant correlation was recorded with NSPP and HI whereas NSPP recorded a significant positive correlation with HI alone (Fig. 2).
In NNG, the SY showed a significant negative correlation with NPPP in Rabi 2020-21; PH, NPPP, and NSPP in Rabi 2021-22 whereas it was significantly positively correlated with NPPP and NSPP in pooled. In both the cropping seasons and pooled season, a significant positive correlation was recorded between DF and DFF, PH and SW100, and NPPP and NSPP (Fig. 2).
Correlation of yield and yield components in nodulating and non-nodulating chickpea genotypes (DF-Days to first flowering, DFF-Days to 50% flowering, DM- Days to maturity, PH-Plant height, PBr- Number of primary branches per plant, SBR- Number of secondary branches per plant, NPPP- Number of pods per plant, NSPP- Number of seeds per plant, SW100-100 seed weight, HI-Harvest index, SY-Seed yield).
Scatter plot for yield and yield component traits in Chickpea RILs
The relationships between two numeric variables in the data set for the target traits and the performance of the genotypes in two environments were analyzed using a scatter plot (Fig. 3). The RILs that were in the 2nd and 4th coordinates are adaptable when considering the environment as the main factor while, those in the 1st and 3rd coordinates are stable across the environments. Specifically, genotypes situated in the 1st and 3rd coordinates were identified as NG and NNG, respectively.
The scatter plot analysis revealed distinct performance patterns for yield and yield component traits across different seasons in the NG and NNG (Fig. 3). In pooled (2nd and 3rd coordinate), RILs 172, 88, 188, 159, and 13 for seed yield; RILs 206, 156, 13, 172 and 17 for NPPP; the RILs 172, 40, 206, 13, 56 for NSPP; RILs 92, 199, 194, 17 and 63 for HI; RILs 163, 157, 194, 22, 68 for SW100 exhibited better performance, emphasizing the trait-specific adaptability of genotypes (Fig. 3). For Rabi 2020-21 (4th coordinate), the RILs 172, 88, 188, 178, 159 for SY; the RILs 172, 188, 178, 40, 159 NPPP; the RILs 172, 98, 188, 221, 13 for NSPP; the RILs 92, 199, 194, 17 and 63 for HI; RILs 163, 22, 207, 68, 157 for SW100 whereas for Rabi 2021-22 (2nd coordinate), the RILs 209, 101, 110, 151, 136 for SY; the RILs 156, 94, 4, 199, 196 for NPPP; RILs 156, 4, 94, 102, 175 for NSPP; the RILs 114, 22, 67, 199, 194 for HI; the RILs 163, 157, 169, 185, 208 for SW100 (Fig. 3) were scattered in their respective coordinates from other RILs depicting their better performance in their respective seasons, highlighting their potential for adaptability and yield enhancement.
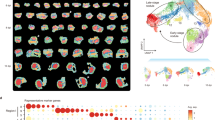
Genotype × trait (GT) bi-plot
The GT bi-plot analysis revealed that 53.92% of the trait variation could be explained, with PC1 and PC2 accounting for 40.72% and 13.20% of the total variance, respectively (Fig. 4). Apart from the checks, genotypes 68, 159, 13, 17, 187, and 123 demonstrated superior adaptability for the traits SY, PH, and SW100 across the seasons. Conversely, genotypes 213, 128, 106, 109, and 46 exhibited the highest adaptability for traits DFF and DM across the seasons (Fig. 4). Of the checks, PhuleVikram and RVG 204 were best adaptable for the traits SY, PH, and SW100 in both the seasons (Fig. 4). Notably, there were differences in genotype rankings for HI between seasons, as indicated by angles greater than 90° between vectors. Based on the vectors, the ranking of the genotypes for SY, PH, and SW100 can be observed as, PhuleVikram = RVG 204 > RILs 68 > 159 > 13 > 17 > 187 > 123. The polygon view depicted in Fig. 4 illustrated the distribution of genotypes in the trait bi-plot, highlighting important yield, yield components, and agro-morphological traits in chickpea.
Discussion
The formation of rhizobium nodulation is a key symbiotic mechanism in legume crops for their adaptation to marginal environments. The quantitative assessment of their impact on plant growth and economic yields is crucial for cultivar improvement and optimizing agricultural productivity. In this study, the selected parents are landraces, distinguishing one as a non-nodulating mutant (ICC 4918NN) derived from the other germplasm (ICC 4918). By employing the RGA approach, we successfully generated a RIL population within a mere 18 months, a testament to the efficiency and robustness of the methodology utilized43. While the potential of RGA has been acknowledged in other crops such as pea44, pigeon pea45, barley and wheat46, and canola46, its application in practical breeding programs remains largely unexplored. The variance analysis revealed a significant difference (P < 0.01) among the RILs for all the traits under the study. This indicates the presence of ample variability for the traits in the population (Table 1). Significant genetic variability among genotypes was observed in earlier studies evaluated for nodulation-related traits in chickpea47,48.
A notable finding in our study is the substantial increase in yield (62.55%) in NG compared to NNG. The improvement was majorly contributed by the increase in the NPPP (39.5%) and NSPP (44.4%). On a moderate level, nodulation has reduced the flowering time and maturity and enhanced the PH, SBr, SW100 and HI (Table 1). These results indicate that NGs were more efficient in synthesizing photosynthates, which led to produce more number of flowers for generating a large number of pods and seeds49,50 than NNG. In addition, the yield advantage reflects not only the inherent genetic potential of the legume plant but also the synergistic effects of the established symbiosis between roots and rhizobial bacteria. The result agrees with an earlier study in chickpea, which reported a 31% higher yield in NG compared to NNG counterparts51,52. The current study, with a more pronounced yield advantage, emphasizes the significance of rhizobial nodulation in optimizing chickpea productivity.
The poorer performance of NNG may be attributed to the deficiency of nitrogen fixation through nodulation, emphasizing its crucial role in chickpea productivity. The reliance on alternative nitrogen sources becomes crucial for NNG genotypes, and supplementing nitrogen in the form of fertilizers may be necessary to enhance yield53. This aligns with previous findings in chickpea and groundnut, where NNGs in nitrogen-rich soils could attain yields on par with NG’s54,55. Though the degree of reliance on nodulation for nitrogen fixation varies throughout legumes, it is perpetually present. For instance, in soybean, nodulation can contribute to a significant proportion of the plant’s nitrogen needs, with non-nodulating variants often displaying stunted growth and reduced yields25. Similarly, nodulation is critical for optimal growth in common beans, especially under nitrogen-deficient conditions56. Further investigations into the performance of NNG and NG under varying nitrogen availability can offer insights into their adaptability and yield potential across different soil conditions.
The present investigation aimed to bridge a crucial gap in the existing knowledge base by assessing the impact of nodulation on agro-morphological and yield traits in chickpea. Our results align with existing literature on the positive effects of rhizobial nodulation on various growth characteristics, emphasizing the importance of this symbiotic association. The observed higher values in PH, PBr, biomass, and yield traits in NG compared to NNG underscore the pivotal role of rhizobial nodulation in enhancing plant growth and productivity in chickpea (Table 2) and mobilization of insoluble nutrients in the soil, leading to improved nutrient uptake in other legumes57,58. In addition, the absence of rhizobium nodulation resulted in a significant reduction in various growth parameters which might be due to a deficit in (a) host-dependent strain fitness59, (b) up-regulated expression of nif genes related to flavonoid synthesis60, and (c) maintenance of plant Pi (Inorganic phosphate) levels61.
The estimation of genetic variability and inheritance through GCV, PCV and heritability allows the breeders to identify the traits with substantial genetic control and potential for selection in crop improvement programs. For phenological traits, the small difference between GCV and PCV values suggests a predominant influence of genetic factors on their variance. HI exhibited low estimates of both PCV and GCV, indicating a more substantial influence of environment (Supplementary Tables 2 and 3). This aligns with previous studies highlighting the major role of genetic components in the inheritance of flowering, maturity, and the environmental factors in determining HI62,63,64,65. For the traits NPPP, NSPP, HI, PBr, and SBr, a magnitude of low to high heritability was observed, suggesting several genetic factors controlling the inheritance of these traits. In particular, SY demonstrated a high magnitude of GCV, PCV, and heritability emphasizing its potential for genetic improvement through simple selection even in early generations efficiently63,65,66,70,71. This diverse heritability pattern underscores the importance of trait-specific breeding strategies to achieve improvements in chickpea agronomics64,65,72,73. High heritability coupled with high GAM was recorded for SY and SW100 across seasons (Supplementary Tables 2 and 3), indicating the predominance of additive gene action for these traits. Similar findings were reported under diverse genetic backgrounds in chickpea71,74,75, blackgram76 as well as in cowpea77,78.
Interestingly, the association among SY, NPPP, and NSPP was significantly positive in both NG and NNG (Fig. 2). This indicates the possibility of simultaneous improvement of multiple traits in chickpea genotypes and cowpea advanced breeding lines39,62,78,77. The ability to enhance multiple traits concurrently is crucial for developing improved chickpea varieties with enhanced agronomic performance. The scatter plot analysis (Fig. 3) demonstrated distinct performance patterns for yield and yield component traits across various seasons (Pooled, Rabi 2020-21 & 2021-22) in the NG and NNG indicating their superior performance in their respective seasons (Fig. 3). The use of scatter plots to differentiate genotypes based on their mean performance and its strategic application enhances the accuracy of genotype selection based on mean performance which is consistent with previous research conducted in chickpea66,67 and mungbean68,69. Further analysis of the genotypes for their yield and trait interactions using GT bi-plot identified the checks, Phule Vikram and RVG 204 on its vertex with high values for all the traits under the study which can be considered the best adaptable genotypes (Fig. 4). For SY, PH, and SW100, the NG #68, 159, 13, 17, 187, and 123 while 106, 109, and 46 for DFF and DM were best adaptable over the seasons. Comparable research of this kind in various crops79,80,81, provide a valuable insight for the selection of superior genotypes with enhanced adaptability and nodulating nature in the chickpea breeding programs.
The impact of rhizobium nodulation on grain yield and its association with yield-related traits was prominent in different legume crops. Earlier studies in chickpea emphasized the role of nodulation in enhancing seed yield and pod development62,71. Similarly, the seed yield was significantly improved by 40% in cowpea82, 33% in common bean56, and 45.6–50% in faba bean83,84 compared to control or non-inoculated treatments, indicating the critical role of symbiotic nitrogen fixation in achieving higher seed yields in legumes. Moreover, the crop-specific strains, soils, and weather factors are crucial in achieving yields. These shared trends highlight the conserved nature of the nodulation mechanism inherited historically for thriving legume crops under diverse agro-ecologies.
The comprehensive assessment of genetic variability, heritability, and genetic advance in this study provides a robust foundation for future chickpea breeding programs. The identification of traits with high heritability and substantial genetic advance, coupled with positive correlations among yield-related parameters, highlights avenues for effective genetic improvement. However, it’s crucial to recognize the crop-specific nature of these findings and to tailor breeding strategies accordingly. Future research should delve deeper into the molecular mechanisms underpinning nodulation in chickpea and explore comparative studies across leguminous crops to enhance our understanding of these complex interactions.
Materials and methods
Description of the environment
The present investigation was carried out at the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, India located at 17°30′ N latitude and 78°16′ E longitude with an altitude of 549 m above sea level during the normal crop season i.e., Rabi 2020-21 and 2021-22. The average rainfall, humidity, minimum, and maximum temperatures were recorded as 3.1 mm, 73.8%, 14.7 °C and 29.7 °C respectively during Rabi 2020-21 whereas in Rabi 2021-22 they were recorded as 8.0 mm, 63.3%, 15.7 °C and 30.0 °C respectively (http://icrisatintranet/nrmp/agroclimatology/weather.asp).
Development of the RIL population through rapid generation advancement (RGA)
The RIL population was developed using the Rapid Generation Advancement (RGA) method43 by crossing between two parents ICC 4918NN (female, non-nodulating type) and ICC 4918 (male, nodulating type). The hybridity confirmed F1 seeds were advanced to F2 in a glasshouse. From F2 the generations were advanced by selfing and single seed decent method. In 2019-20, 307 F2 seeds were harvested in the glasshouse and advanced to F3 through RGA where the immature pods were harvested 50 days after sowing (DAS) and sown on the same day for generation advancement. Four generations starting from F2 to F6 advanced in one year (Table 4). A maximum temperature of 25 ± 1 °C, 70–80% humidity, and artificial light with the help of incandescent bulbs was provided for a period of 12 h (from 18:00–6:00) during the RGA experiment. The F6 − 7 and F7 − 8 seeds were sown in the field in Rabi 2020-21 and 2021-22 for agronomic characterization (Fig. 5). A total of 230 RILs (130 nodulating genotypes, 94 non-nodulating genotypes), four checks (Phule Vikram, RVG 204, NBeG 47, and JG 14), and two parents (ICC 4918NN and ICC 4918) were used in our experiment. The parental genotypes were obtained from the gene bank of ICRISAT, India. The genotypes (RIL population) were categorized based on the presence or absence of nodules in Supplementary Table 1.
Field experiment
The experimental material was sown on Vertisols in Alpha Lattice Design with three replications. Each genotype was sown with a spacing of 60 × 15 cm (inter and intra) covering an area of 1.2 m2. Planting of the population was completed during the first week of November in Rabi, 2020, and in the second week of November in Rabi, 2021. All the standard agronomic practices were followed for better crop establishment. Two irrigations were provided, one at the initial stage of planting and the other at the podding stage to maintain sufficient moisture regime for better crop establishment. Data related to the agronomic traits such as DF, DFF, DM, PH, PBr, SBr were recorded in each plot. The genotypes were harvested when all the plants matured completely. The post-harvest data related to the NPPP, NSPP, SW100, HI and SY were recorded from their respective plots.
Screening for nodulation
To distinguish the nature of the genotypes in terms of nodulation, the RIL population was grown in semi-controlled glasshouse condition as per the protocol outlined in the previous studies of chickpea14. The experimental material was surface sterilized with sodium hypochlorite and treated individually with the rhizobial strains IC59 and IC76A, the predominant nodulating bacteria in south-central India54,85. The seeds of each genotype were sown in 9” pots filled with potting mixture composed of sterile soil, sand, and vermicompost (3:2:2). After germination, the seedlings were thinned to three and uprooted at 35 days after sowing86, to determine the presence or absence of nodules in the planting material. Each genotype was designated as NG and NNG based on the presence or absence of root nodules (Fig. 5).
Statistical analysis
A combined analysis of variance was carried out using the SAS MIXED procedure87, to test the significance of the main and interaction effects of seasons and genotypes, considering the season, genotype, replication as fixed, and block as random. Individual season variance was modeled into combined analysis with repeated statements using the REML (Restricted Maximum Likelihood) method. To analyze the fixed effects, BLUEs (Best Linear Unbiased Estimates) are estimated for both main and interaction effects from the pooled analysis. The significance of NG and NNG means were compared using t-statistics from the combined analysis. Karl Pearson’s correlation was performed among the agronomic traits by using the SAS CORR procedure87.
The magnitude of variation and its utility can be explained by genetic parameters like GCV, PCV, broad sense heritability, and GAM. These parameters were also estimated for the target traits using the SAS CORR procedure87. The estimates of PCV and GCV were classified as high (> 20%), moderate (10–20%) and low (< 10%)88; heritability as high (> 60%), moderate (30–60%), low (< 30%)89; GAM as high (> 20%), moderate (10–20%) and low (< 10%)89.
To visualize the associations among the traits and the trait profiles of the genotypes a Genotype × Trait (GT) bi-plot was generated. It is based on Single Value Decomposition (SVD) of trait-wise standardized data (Z ~ N (0,1)). The correlation between the traits was interpreted by using cosine angles of the vectors in between the traits. The angle < 90°, > 90°, and equal to 90° states the positive, negative, and no correlation between the traits, respectively. As the vector length of a particular trait explains the variation in genotypes, the longest vector explains more variation among the genotypes and the shortest vector depicts a very low level of variation among genotypes for a given trait.
Conclusion
The RIL mapping population was developed in less than two years, the first report in chickpea to show the practical application of the RGA protocol in developing breeding material. The influence of nodulation on yield was distinctly favorable in nodulating genotypes, characterized by increased SY, biomass, and PH compared to NNG. The stable performing genotypes with high yield and early flowering nature were mostly nodulating in nature depicting the beneficial effect of nodulation on the crop growth under moisture stress conditions. The promising genotypes identified can serve as donors for use in chickpea breeding programs. Furthermore, the results emphasize the critical role of compatible Rhizobium strain/s in achieving optimal yields. The study highlights that the absence of nodulation can lead to substantially lower yields in chickpea. Therefore, the distribution and presence of Rhizobium strains in cultivated fields emerge as influential factors affecting final yield levels. To enhance the understanding of nodulation in chickpea, further research on identifying the genomic regions/QTLs/markers associated with nodulation is in progress to enhance our knowledge and provide valuable insights into the molecular mechanisms controlling nodulation in chickpea, ultimately contributing to improved crop productivity and promoting sustainable agricultural practices.
Data availability
The data of this article supporting the conclusions will be made available by the corresponding author upon request.
Abbreviations
- RGA:
-
Rapid Generation Advancement
- RIL:
-
Recombinant Inbred Line
- SNF:
-
Symbiotic nitrogen fixation
- NG:
-
Nodulating genotypes
- NNG:
-
Non-nodulating genotypes
- DF:
-
Days to first flowering
- DFF:
-
Days to 50% flowering
- DM:
-
Days to maturity
- PH:
-
Plant height
- PBr:
-
Number of primary branches per plant
- SBr:
-
Number of secondary branches per plant
- NPPP:
-
Number of pods per plant
- NSPP:
-
Number of seeds per plant
- SY:
-
Seed yield
- SW100:
-
100-seed weight
- HI:
-
Harvest index
- ANOVA:
-
Analysis of variance
- GCV:
-
Genotypic coefficient of variation
- PCV:
-
Phenotypic coefficient of variation
- GAM:
-
Genetic advance of mean
- GT biplot:
-
Genotype-trait bi-plot analysis
References
McCauley, A. M. Nitrogen fixation by annual legume green manures in a semi-arid cropping system. Dissertation, Montana State University-Bozeman (2011).
Keneni, G. et al. Phenotypic diversity for symbio-agronomic characters in Ethiopian Chickpea (Cicer arietinum L.) germplasm accessions. Afr. J. Biotechnol. 11, 12634–12651 (2012).
Ouma, E. W., Asango, A. M., Maingi, J. & Njeru, M. Elucidating the potential of native rhizobial isolates to improve biological nitrogen fixation and growth of common bean and soybean in smallholder farming systems of Kenya. Int. J. Agron. 1–7 (2016).
Stajkovic, O. et al. Improvement of common bean growth by co-inoculation with Rhizobium and plant growth-promoting bacteria. Rom Biotechnol. Lett. 16, 5919–5926 (2011).
Huang, X. F. et al. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92, 267–275 (2014).
Perrig, D. et al. Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Appl. Microbiol. Biotechnol. 75, 1143–1150 (2007).
Herridge, D. F., Peoples, M. B. & Boddey, R. M. Global inputs of biological nitrogen fixation in agricultural systems. Plant. Soil. 311, 1–18 (2008).
Food and Agriculture Organization of the United Nations. FAOSTAT Statistical Database (FAO, 2022).
Diapari, M. et al. Genetic diversity and association mapping of iron and zinc concentrations in Chickpea (Cicer arietinum L). Genome 57, 459–468 (2014).
Diapari, M., Sindhu, A., Warkentin, T. D. & Bett, K. Tar’an, B. Population structure and marker-trait association studies of iron, zinc and selenium concentrations in seed of field pea (Pisum sativum L). Mol. Breed. 35, 1–14 (2015).
Gaur, P. M. et al. Inheritance of protein content and its relationships with seed size, grain yield and other traits in Chickpea. Euphytica 209, 253–260 (2016).
Jha, U. C. & Shil, S. A. N. D. I. P. Association analysis of yield contributing traits of Chickpea genotypes under high temperature condition. Trends Biosci. 8, 2335–2341 (2015).
Saha, S., Chakraborty, D., Sehgal, V. K. & Pal, M. Potential impact of rising atmospheric CO2 on quality of grains in Chickpea (Cicer arietinum L). Food Chem. 187, 431–436 (2015).
Gopalakrishnan, S., Srinivas, V., Vemula, A., Samineni, S. & Rathore, A. Influence of diazotrophic bacteria on nodulation, nitrogen fixation, growth promotion and yield traits in five cultivars of Chickpea. Biocatal. Agric. Biotechnol. 15, 35–42 (2018).
Wang, E. T., Chen, W. F., Tian, C. F., Young, J. P. W. & Chen, W. X. Ecology and Evolution of Rhizobia, Principles and Applications in Springer. 154–157 (2019).
Fox, A. R. et al. Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen fixing bacterium Pseudomonas protegens Pf5 X940. Environ. Microbiol. 18, 3522–3534 (2016).
Yang, Y. et al. Characterization of genetic basis on synergistic interactions between root architecture and biological nitrogen fixation in soybean. Front. Plant. Sci. 8, 1466 (2017).
Unkovich, M., Baldock, J. & Peoples, M. Prospects and problems of simple linear models for estimating symbiotic N2 fixation by crop and pasture legumes. Plant. Soil. 329, 75–89 (2010).
Lupwayi, N. Z., Clayton, G. W., Hanson, K. G., Rice, W. A. & Biederbeck, V. O. Endophytic rhizobia in barley, wheat and Canola roots. Can. J. Plant. Sci. 84, 37–45 (2004).
Limpens, E. et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Sci 302, 630–633 (2003).
Radutoiu, S. et al. Plant recognition of symbiotic bacteria requires two lysm receptor-like kinases. Nature 425, 585–592 (2003).
Sharma, V. et al. Molecular basis of root nodule symbiosis between Bradyrhizobium and ‘Crack-Entry’ legume groundnut (Arachis Hypogaea L). Plants 9, 276 (2020).
Mathesius, U. Comparative proteomic studies of root–microbe interactions. J. Proteome. 72, 353–366 (2009).
Buhian, W. P. & Bensmihen, S. Mini-review: Nod factor regulation of phytohormone signaling and homeostasis during rhizobia-legume symbiosis. Front. Plant. Sci. 9, 1247 (2018).
Yang, Z. W., Shen, Y. Y., Xie, T., Tan, G. & L. & Y. Biological nitrogen fixation efficiency in soybean under different levels of nitrogen supply. Acta Bot. Boreali Occident Sin. 29, 574–579 (2009).
Ward, A. Phosphorus Limitation of Soybean and Alfalfa Biological Nitrogen Fixation on Organic Dairy Farms. Dissertation, Nova Scotia Agricultural College (2011).
Liu, Y. Y., Wu, L. H., Baddeley, J. A. & Watson, C. A. Models of biological nitrogen fixation of legumes. A review. Agron. Sustain. Dev. 31, 155–172 (2011).
Ahmad, M. et al. Appraising endophyte–plant symbiosis for improved growth, nodulation, nitrogen fixation and abiotic stress tolerance: an experimental investigation with Chickpea (Cicer arietinum L). Agronomy 9, 621 (2019).
Ashraf, M. & Iram, A. Drought stress induced changes in some organic substances in nodules and other plant parts of two potential legumes differing in salt tolerance. Flora 200, 535–546 (2005).
Jemo, M. et al. Comparative analysis of the combined effects of different water and phosphate levels on growth and biological nitrogen fixation of nine Cowpea varieties. Front. Plant. Sci. 8, 2111 (2017).
Figueiredo, M. V. B., A, H. L., Burity., C., Martïnez, R. & Chanway, C. P. Alleviation of drought stress in the common bean (Phaseolus vulgaris L.) by co-inoculation with Paenibacillus polymyxa and Rhizobium tropici. Appl. Soil. Ecol. 40, 182–188 (2008).
Dutta, S. & Podile, A. R. Plant growth promoting rhizobacteria (PGPR): the Bugs to debug the root zone. Crit. Rev. Microbiol. 36, 232–244 (2010).
Oldroyd, G. E. D., Downie, J. A. & Bisseling, T. The nodule symbiosis. Nat. Rev. Genet. 12 (8), 566–580 (2011).
Jones, K. M., Kobayashi, H., Davies, D. P., Tiller, S. A. & Oldroyd, G. E. D. Ethylene perception regulates the development of nitrogen-fixing root nodules. Nature 446 (7132), 153–156 (2007).
Muehlbauer, F. J., Kraft, J. M. & Jha, U. C. Genetics of Chickpea. Crop Sci. 42 (1), 19–31 (2002).
Radutoiu, S. et al. NFP, the Medicago truncatula homolog of Rh5 and NORK, is a key factor in Nod factor perception and early steps of nodule development. EMBO J. 22 (19), 5126–5136 (2003).
Ditta, A. et al. Rock phosphate enriched organic fertilizer with phosphate solubilizing microorganisms improves nodulation, growth and yield of legumes. Commun. Soil. Sci. Plant. Anal. 49, 2715–2725 (2018a).
Ditta, A. et al. Application of rock phosphate enriched composts increases nodulation, growth and yield of Chickpea. Int. J. Recycl Org. Waste Agric. 7, 33–40 (2018b).
Rafique, M. et al. The combined effects of gibberellic acid and Rhizobium on growth, yield and nutritional status in Chickpea (Cicer arietinum L). Agron 11, 105 (2021).
Xia, X., Ma, C., Dong, S., Xu, Y. & Gong, Z. Effects of nitrogen concentrations on nodulation and nitrogenase activity in dual root systems of soybean plants. J. Soil. Sci. Plant. Nutr. 63, 1–13 (2017).
Bourebaba, Y. et al. Diversity of Bradyrhizobium strains nodulating Lupinus micranthus on both sides of the Western mediterranean: Algeria and Spain. Syst. Appl. Microbiol. 39, 266–274 (2016).
Asante, M., Ahiabor, B. D. K. & Atakora, W. K. Growth, nodulation, and yield responses of groundnut (Arachis Hypogaea L.) as influenced by combined application of rhizobium inoculant and phosphorus in the Guinea savanna zone of Ghana. Int. J. Agron. 1–7, 8691757 (2020).
Samineni, S., Sen, M., Sajja, S. B. & Gaur, P. M. Rapid generation advance (RGA) in Chickpea to produce up to seven generations per year and enable speed breeding. Crop J. 8, 164–169 (2020).
Ribalta, F. M. et al. Precocious floral initiation and identification of exact timing of embryo physiological maturity facilitate germination of immature seeds to truncate the lifecycle of pea. Plant. Growth Regul. 81, 345–353 (2017).
Saxena, K., Saxena, R. K. & Varshney, R. K. Use of immature seed germination and single seed descent for rapid genetic gains in Pigeonpea. Plant. Breed. 136, 954–957 (2017).
Watson, A. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants. 4, 23–29 (2018).
Gul, R., Khan, H., Khan, N. U. & Khan, F. Y. Characterization of Chickpea germplasm for nodulation and effect of Rhizobium inoculation on nodules number and seed yield. J. Anim. Plant. Sci. 24 (5), 1421–1429 (2014).
Gul, R., Khan, H., Khan, N. U., Latif, A. & Harada, K. Characterization for nodulation and detection of duplicate gene action of dominant epistasis controlling root nodulation in Chickpea (Cicer arietinum). Int. J. Agric. Biol. 20, 683–688 (2018).
Mallikarjuna, B. P., Viswanatha, K. P., Samineni, S. & Gaur, P. M. Association of flowering time with phenological and productivity traits in Chickpea. Euphytica 215, 1–8 (2019).
Siddique, K. H. M. & Khan, T. N. Early-flowering and high-yielding Chickpea lines from ICRISAT ready for release in Western Australia. Int. Chickpea Pigeonpea Newslett. 3, 22–24 (1996).
Rupela, O. P. & Johansen, C. Identification of non-nodulating, and low and high nodulating plants in Pigeonpea. Soil. Bio Biochem. 27, 539–544 (1995).
Rupela, O. P., Sharma, L. C. & Danso, S. K. A. Evaluation of N2 fixation by nodulation-variants of Chickpea in India. In improving yield and nitrogen fixation of grain legumes in the tropics and Sub-tropics of Asia. Int. At. Energy Agency. 1027, 99–119 (1997).
Koevoets, I. T., Venema, J. H., Elzegna, J. T. M. & Testerink, C. Roots withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant. Sci. 7, 1335 (2016).
Rupela, O. P. Natural occurrence and salient characters of Non nodulating Chickpea plants. Crop Sci. 32, 349–352 (1992).
Giller, K. E., Nambiar, P. T. C., Srinivasa Rao, B., Dart, P. J. & Day, J. M. A comparison of nitrogen fixation in genotypes of groundnut (Arachis Hypogaea L.) using 15N-isotope Dilution. Biol. Fertil. Soils. 5, 23–25 (1987).
Dos Santos Sousa, W. et al. Effects of Rhizobium inoculum compared with mineral nitrogen fertilizer on nodulation and seed yield of common bean. A meta-analysis. Agron. Sustain. Dev. 42, 52 (2022).
Ali, M. A., Naveed, M., Mustafa, A. & Abbas, A. The good, the bad, and the ugly of rhizosphere Microbiome in Probiotics and Plant Health. 253–290 (Springer, (2017).
Cakmakci, R., Donmez, M. F. & Erdogan, U. The effect of plant growth promoting rhizo bacteria on barley seedling growth, nutrient uptake, some soil properties, and bacterial counts. Turk. J. Agr For. 31, 189–199 (2007).
Burghardt, L. T., Epstein, B., Hoge, M., Trujillo, D. I. & Tiffin, P. Host-Associated rhizobial fitness: dependence on nitrogen, density, community complexity, and legume genotype. Appl. Environ. Microbiol. 88, e00526–e00522 (2022).
Xu, T. et al. Revealing the underlying mechanisms mediated by endophytic actinobacteria to enhance the rhizobia-chickpea (Cicer arietinum L.) symbiosis. Plant soil. 1–20 (2022).
Míguez-Montero, M. A., Valentine, A. & Pérez-Fernández, M. A. Regulatory effect of phosphorus and nitrogen on nodulation and plant performance of leguminous shrubs. AoB Plants. 12, 1–11 (2019).
Barmukh, R. et al. Construction of a high-density genetic map and QTL analysis for yield, yield components and agronomic traits in Chickpea (Cicer arietinum L). Plos One. 16 (5), e0251669 (2021).
Misra, G. et al. Baseline status and effect of genotype, environment and Genotype× environment interactions on iron and zinc content in Indian Chickpeas (Cicer arietinum L). Euphytica 216 (9), 1–16 (2020).
Paul, P. J. et al. Capturing genetic variability and selection of traits for heat tolerance in a Chickpea Recombinant inbred line (RIL) population under field conditions. Euphytica 214, 1–14 (2018).
Tsehaye, A., Fikre, A. & Bantayhu, M. Genetic variability and association analysis of Desi-type Chickpea (Cicer arietinum L.) advanced lines under potential environment in North Gondar, Ethiopia. Cogent Food Agri. 6 (1), 1806668 (2020).
Hussain, S. A. et al. Estimating genetic variability among diverse lentil collections through novel multivariate techniques. Plos One. 17 (6), e0269177 (2022).
Admas, S., Tesfaye, K., Haileselassie, T., Shiferaw, E. & Flynn, K. C. Genetic variability and population structure of Ethiopian Chickpea (Cicer arietinum L.) germplasm. Plos One. 16 (11), e0260651 (2021).
Samyuktha, S. M. et al. Delineation of Genotype× environment interaction for identification of stable genotypes to grain yield in Mungbean. Front. Agron. 2, 577911 (2020).
Ullah, H. et al. Selecting high yielding and stable Mungbean [Vigna radiata (L.) Wilczek] genotypes using GGE biplot techniques. Can. J. Plant. Sci. 92, 951–960 (2012).
Khan, M. M. H., Rafii, M. Y., Ramlee, S. I., Jusoh, M. A. & Mamun, M. Genetic analysis and selection of Bambara groundnut (Vigna subterranea [L.] Verdc.) landraces for high yield revealed by qualitative and quantitative traits. Sci. Rep. 11 (1), 1–21 (2021).
Kushwah, A. et al. Phenotypic evaluation of genetic variability and selection of yield contributing traits in Chickpea Recombinant inbred line population under high temperature stress. Physiol. Mol. Biol. Plants. 27, 747–767 (2021).
Gediya, L. N. et al. Phenotypic variability, path analysis and molecular diversity analysis in Chickpea (Cicer arietinum L). Vegetos 32, 167–180 (2019).
Mallu, T. S. et al. Assessment of genetic variation and heritability of agronomic traits in Chickpea (Cicer arietinum L). Int. J. Agron. Agric. Res. 5 (4), 76–88 (2014).
Parashi, V. S., Lad, D. B., Mahse, L. B., Kute, N. S. & Sonawane, C. J. Genetic diversity studies in Chickpea (Cicer arietinum L). BIOINFOLET 10 (1b), 337–341 (2013).
Rajasekhar, S. et al. Genetic variation for grain protein, Fe and Zn content traits in Chickpea reference set. J. Food Compos. Anal. p.104774 (2022).
Chauhan, S., Mittal, R. K., Sood, V. K. & Patial, R. Evaluation of genetic variability, heritability and genetic advance in blackgram [Vigna mungo (L.) Hepper]. Legum Res. 43 (4), 488–494 (2020).
Owusu, E. Y. et al. Genetic variability, heritability and correlation analysis among maturity and yield traits in Cowpea (Vigna unguiculata (L) Walp) in Northern Ghana. Heliyon 7(9), e07890 (2021).
Gerrano, A. S., van Jansen, W. S. & Kutu, F. R. Agronomic evaluation and identification of potential Cowpea (Vigna unguiculata L. Walp) genotypes in South Africa. Acta Agric. Scand. Sec- B Soil. Plant. Sci. 69, 295–303 (2019).
Bakhshi, B. et al. Trait profiling and genotype selection in oilseed rape using genotype by trait and genotype by yield* trait approaches. Food Sci. Nutr. 11 (6), 3083–3095 (2023).
Parissi, Z. et al. Analysis of genotypic and environmental effects on biomass yield, nutritional and anti nutritional factors in common Vetch. Agron 12, 1678 (2022).
Gayacharan et al. Understanding genetic variability in the Mungbean (Vigna radiata L.) genepool. Annals App Bio. 177 (3), 346–357 (2020).
Ahmet, E. K. Halime Özdamar Ünlü. Effect of rhizobium inoculation on yield and some quality properties of fresh Cowpea. Cogent Food Agric. 9, 2 (2023).
Gedamu, S. A., Tsegaye, E. A. & Beyene, T. F. Effect of rhizobial inoculants on yield and yield components of faba bean (Vicia fabae L.) on vertisol of wereillu district, South Wollo, Ethiopia. CABI Agric. Biosci. 2, 8 (2021).
Talaat, N. B. & Abdallah, A. M. Response of fababean (Vicia faba L.) to dual inoculation with Rhizobium and VA mycorrhiza under different levels of N and P fertilization. J. Appl. Sci. Res. 4, 1092–1102 (2008).
Gopalakrishnan, S. et al. Assessment of nodulation potential in mini-core genotypes and land races of Chickpea. J. Food Legum. 30, 65–72 (2017a).
Gopalakrishnan, S., Srinivas, V. & Samineni, S. Nitrogen fixation, plant growth and yield enhancements by diazotrophic growth-promoting bacteria in two cultivars of Chickpea (Cicer arietinum L). Biocatal. Agric. Biotechnol. 11, 116–123 (2017b).
SAS Institute Inc. SAS/STAT® 15.1 User’s Guide (SAS Institute Inc, 2018).
Sivasubramanian, S. & Madhavamenon, P. Genotypic and phenotypic variability in rice. Madras Agric. J. 60, 1093–1096 (1973).
Johnson, H. W., Robinson, H. F. & Comstock, R. E. Estimates of genetic and environmental variability in soybeans. Agron. J. 47, 314–318 (1955).
Acknowledgements
We, the authors, are thankful to the chickpea team members of the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) for providing research facilities and funds for the field trials, Acharya N. G. Ranga Agricultural University (ANGRAU) for providing fellowship during the research and International Center for Biosaline Agriculture (ICBA) for technical backstopping in development and submission of the manuscript.
Funding
The current research was supported by the Indian Council of Agricultural Research-International Crops Research Institute for the Semi-Arid Tropics (ICAR-ICRISAT) and The CGIAR Research Program on Grain Legumes and Dryland Cereals (CRP-GLDC) projects.
Author information
Authors and Affiliations
Contributions
S.R.N: Data collection and curation, Trial establishment, Software, Visualization, Writing-original draft preparation. M.D.M: Data curation, formal analysis, validation, writing the manuscript. U.C: Field layout, Intercultural operations, Data curation. S.S.G: Conceptualization of rhizobium inoculation, Methodology, Review, and editing the manuscript. S. R.V: Methodology, Review, and editing the manuscript. S.V: Rhizobium culture preparation and application, glasshouse evaluations. S.T: Conceptualization, Review, and editing of the manuscript. R.S: Data curation, Formal analysis, Editing of the manuscript. A.V: Statistical Analysis, Validation of Results, Visualization. H.K: Methodology and editing of the manuscript. S.S: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
International guidelines and legislation
This experimental research on chickpea (Cicer arietinum L.) crop adheres to international guidelines and legislation to ensure ethical, sustainable, and scientifically robust practices. The study complies with institutional, national, and international standards, including the Convention on Biological Diversity (CBD) and the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), ensuring the sustainable use and conservation of biodiversity.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nandigam, S., Mahendrakar, M.D., Srungarapu, R. et al. Rapid generation advancement of RIL population and assessing the impact of Rhizobium nodulation on crop yields in Chickpea. Sci Rep 15, 13945 (2025). https://doi.org/10.1038/s41598-025-98965-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98965-2
Keywords
This article is cited by
-
Advances in rhizobial technology: driving sustainable agriculture in the 21 st century
Archives of Microbiology (2026)