Abstract
TCP genes are plant-specific transcription factors that play essential roles in plant growth, development, metabolism, and responses to biotic and abiotic stresses. However, the roles of TCP genes in Cucurbitaceae species remain unknown. In this study, 111 and 119 TCP genes were identified in the Benincaseae (C. melo, C. sativus, C. lanatus and L. siceraria) and Cucurbiteae (C. maxima, C. moschata and C. pepo) tribes, respectively, and were analyzed. Segmental duplication, tandem duplication, and whole-genome duplication (WGD) were identified as the major driving factors in the expansion of TCP genes in Cucurbitaceae species, with the majority of TCP genes undergoing purifying selection. Using the melon genome as a reference, an integrated map containing 29 loci across nine chromosomes was constructed, 28 of which were shared by seven Cucurbitaceae species. Gene structure analysis revealed that their function was conserved. The result of promoter sequence analysis indicated that TCP genes have many phytohormone-related cis-regulatory elements. GO term enrichment analysis showed that TCP genes were the major regulators of many downstream transcriptional networks and primarily functioned in the nucleus. Transcriptome analysis of different tissues and developmental stages of the Cucurbiteae tribe revealed tissue-specific spatial and temporal expression patterns of TCP genes, suggesting that TCP genes play an important role in the growth and development of Cucurbitaceae. Gene expression profiling demonstrated that TCP genes are involved in the responses of plants to abiotic and biotic stresses. In conclusion, this is the first systematic analysis of TCP genes in Cucurbitaceae, which provides deeper insights into their evolutionary dynamics and functional properties, which may be crucial for the genetic improvement of Cucurbitaceae.
Similar content being viewed by others
Introduction
Transcription factors (TFs), also known as transacting factors, contain two functional structural domains are involved in DNA binding or the activation/repression of gene expression in response to stimuli, which forms the genetic basis of phenotypic evolution1,2,3. TFs play important roles in the growth, metabolism, and responses to environmental stresses in higher plants, and their variation (expansion and diversification) plays a central role in the evolution of key phenotypes4,5. Identifying and deciphering the molecular functions of DNA-binding domains, transcriptional regulatory domains, oligomerization sites, and nuclear localization signals of key TFs enhances our understanding of signal transduction pathways and plant responses to stress. TF families regulate specific target genes involved in various processes that are crucial for plant development6,7. TFs can be categorized into various families, including WRKYs (WRKYGQ), SPLs (SQUAMOSA promoter binding protein-like), NACs (NAM, ATAF, and CUC), AP 2/ERF (Apetala 2/ethylene response factor), and TCPs (TEOSINTE BRANCHED 1/CYCLO BRANCHIDEA/ PCF) based on their structural domain. The TCP TF family is plant-specific8,9.
TCP genes are named after the four initially characterized members: TB1 (TEOSINTE BRANCHED 1) in maize, CYC (Cycloidea) in snapdragons, and PCF 1/2 (proliferating cell nuclear antigenic factors 1 and 2)10. TCP TFs contain a basic helix-loop-helix (bHLH) structure comprising approximately 59 amino acids at the N-terminus that form, a conserved structural domain that is essential for DNA binding and is involved in protein-protein interactions and protein localization11,12. TCP genes can be categorized into two classes: the PCF class (TCP-P class) and TCP-C class, referred to as Class I and Class II, respectively10. In addition to the different binding sequences, four amino acids that are present in the Class II structural domain are absent from the Class I structural domain. Class I can bind to the GTGGNCCC sequences and Class II binds to the GGNCCCAC sequences13. Class II TCPs have a unique R structural domain and ECE motif, which allow them to be subdivided further into CIN and CYC/TB1 subclasses, which may be involved in protein-protein interactions14,15.
TCP genes are involved in several plant growth and development processes, such as seed germination, leaf morphogenesis, flower development, lateral branch formation, circadian rhythms, hormone biosynthesis and signal transduction9,11,16,17,18,19. AtTCP14 can activate seed embryo growth and break dormancy in seeds16. Activation of AtTCP16 regulation of microspore development using transgenic technology, affects pollen development20. SlTCP12/15/18 play an important role in the regulation of fruit development and ripening in tomatoes21. Overexpression of GhCYC2 leads to the acquisition of a ray-flower-like morphology in disk flowers, which is involved in defining the complex inflorescence structure of Asteraceae22. TCP genes are involved in the regulation of plant secondary metabolism, which plays an essential role in responses to environmental stresses, such as, salt, drought, and low temperature stress23,24,25. The overexpression of OsPCF2 in rice using transgenic technology can activate responses to salt stress and PEG-induced drought stress26. Similarly, PeTCP10 overexpression increases drought resistance in bamboo26. Overexpression of DgTCP1 could increase cold resistance in chrysanthemums12. In addition, overexpression of miR319 decreases the expression of Class II TCP TFs, thereby increasing salt tolerance in grasses11. Furthermore, GbTCP3/4/7 are involved in flavonoid biosynthesis in Ginkgo biloba27.
Cucurbitaceae is one of the most genetically diverse plant families, and most of its members are vital vegetable or medicinal plants and are widely distributed globally28. Members of the Cucurbitaceae family include many popular vegetables and fruits, such as melon (C. melo), cucumber (C. sativus), watermelon (C. lanatus), bottle gourd (L. siceraria), zucchini (C. pepo), and pumpkin (C. maxima or C. moschata)29. Cucurbitaceae is commonly used as model for understanding the molecular regulation of plant tendrils and the biosynthesis of bitter compounds30,31. In recent years, genome sequencing and bioinformatics, whole genome sequencing, and data sharing have developed rapidly. The first cucurbit species to be sequenced was cucumber, and with the generation of chromosome conformation capture technology and novel computational methods, the genomes of more cucurbit species have been sequenced and updated, such as Cucumis melo, Cucumis sativus, Citrullus lanatus, Lagenaria siceraria, Cucurbita pepo, Cucurbita moschata, and Cucurbita maxima32,33,34,35,36,37,38,39. The assembly and updating of high-quality genomes enable researchers to explore gene family members and determine their biological functions.
Considering the critical role of TCP genes in different biological processes, TCP homolog genes have been explored in various plants. TCP genes have been identified in many species, including 24 AtTCP genes in Arabidopsis. Moreover, Parapunova et al. detected 30 SlTCP genes in tomatoes, and 27 ClTCP and 27 CsTCP genes in watermelon and cucumber, respectively11,40,41. Moreover, 66 TCP genes in Triticum aestivum, and 74 TCP genes in Gossypium raimondii were identified42,43. TCP genes emerged early in the history of land plants and expanded rapidly during the evolution of higher plants9. TCP genes in the same class or evolutionary branch generally show highly conserved gene/protein structures and expression profiles in terrestrial plants, suggesting a common origin and functional conservation. CmTCP1 is reportedly involved in development of tendrils from lateral shoots in melon40. TEN encodes a TCP transcription factor conserved within the cucurbits, and regulates tendril-less phenotype9. Interaction network predictions have indicated that this gene family tends to form protein complexes that are involved in different biological processes, such as phytohormone pathways, the cell cycle, and environmental responses9.
Due to their wide variation among species, the evolutionary relationships of TCP genes among cucurbit species are not well characterized. In this study, to improve our understanding of the evolutionary history of the TCP genes in Cucurbitaceae species, we used seven genomic and multiple transcriptomic datasets to reconstruct the evolutionary relationships between TCP genes. A total of 230 TCP genes were identified in cucurbit species, and their chromosomal distribution, gene structure, motif and evolutionary relationships, and promoter cis-regulator elements were analyzed. We found that whole-genome duplications (WGDs) play crucial roles in increasing biodiversity and function in both growth process and responses to environmental stress29. To study the duplication events of inter- and intraspecific TCP genes in cucurbit species using gene covariance analysis, we calculated synonymous substitution (Ks) and non-synonymous substitution rates (Ka) and their ratios (Ka/Ks) to determine the time of differentiation of TCP gene pairs and the main forces promoting differentiation in cucurbit species. In addition, we analyzed transcriptomic datasets of TCP genes in Cucurbita species from different tissues and in response to various environmental stresses, and we observed that TCP genes were involved in plant growth, development, and responses to biotic and abiotic stresses. This study reviews the identification of TCP genes in Cucurbitaceae and their molecular function analysis, laying a foundation for the molecular regulation of major traits in cucurbit plants, quality improvement and plant breeding.
Materials and methods
Identification of TCP genes in cucurbitaceae
In order to explore the evolutionary relationships of TCP genes in cucurbit crops, the whole genome sequences of seven representative Cucurbitaceae species (Cucumis melo, Cucumis sativus, Citrullus lanatus, Lagenaria siceraria, Cucurbita pepo, Cucurbita moschata, and Cucurbita maxima) were obtained (http://cucurbitgenomics.org/). Based on the TCP protein sequences of the model plants (PF03634) (http://pfam.sanger.ac.uk/), the HMMER software with default parameters was used to establish a profile HMM with an E-value cutoff of 1e− 10 to search the gene libraries of the seven Cucurbitaceae species and obtain candidate TCP genes. The candidate gene sequences were subjected to multiple homology comparison, redundant gene sequences were removed, and structural domains were detected using the online tools CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and SMART (http://smart.embl-heidelberg.de/) to obtain TCP genes in Cucurbitaceae. Basic information on TCP genes (e.g., amino acid number, molecular weight, instability index, fat index, and the grand average of hydropathicity [GRAVY] values) was obtained from cucurbit crops, using the ExPASy Proteomics Server online tool in standard mode (http://expasy. Org).
Collinearity analysis and chromosomal location of TCP genes
To analyze the type and relative order of genes conserved among different species that diverged from a uniform ancestral type, seven Cucurbitaceae species (C. melo, C. sativus, C. lanatus, L. siceraria, C. maxima, C. moschata and C. pepo) were subjected to covariance analysis using TBtools software44. To measure collinearity among these species of Cucurbitaceae and between melon and other Cucurbitaceae species (C. sativus, C. lanatus, L. siceraria, C. maxima, C. moschata and C. pepo), Circos and collinearity plots were constructed using the default parameters of the TBtools multiple synteny plots. In addition, tandem and segmental duplication events among TCP genes were detected using TBtools and MCScanX with the e-value set to 1.0 × 10[− 10 44,45. Using Mapchat software under standard model conditions, the TCP gene location information for the seven species (Cucumis melo L. cv. DHL92, Cucumis. sativus L. var. sativus var. 9930, Citrullus lanatus subsp. vulgaris cv. 97103, Lagenaria siceraria var. USVL1VR-Ls, Cucurbita pepo subsp. pepo, Cucurbita moschata var. Rifu, Cucurbita maxima var. Rimu), obtained from the Cucurbitaceae Genome Website (http://cucurbitgenomics.org/), was visualized.
Phylogeny tree construction and multiple sequence alignment analysis
Multiple sequence comparisons of the full-length TCP protein sequences from Cucumis melo, Cucumis sativus, Citrullus lanatus, Lagenaria siceraria, Cucurbita pepo, Cucurbita moschata and Cucurbita maxima were performed in MEGA 11 using ClustalW with default pairwise settings. A phylogenetic tree was constructed using the Maximum Likelihood (ML) method in the MEGA 11 software with 1000 bootstrap replicates. Evolutionary trees were generated using the iTOL (https://itol.embl.de/) website46. TCP genes were categorized into groups based on the TCP conserved structural domains belonging to Cucurbitaceae species. TCP conserved structural domains were analyzed by amino acid sequence comparison using DNAMAN software, under standard conditions.
Gene structure analysis of TCP genes
TCP gene sequences were extracted from the location information of genes in the genome, and the TCP gene structures in Cucurbitaceae species were analyzed using the Gene Structure Display Server (GSDS) (https://gsds.gao-lab.org/index.php) online tool47. The Basic-Helix I-Loop-Helix II structures of the TCP genes were analyzed in MEGA 11 using ClustalW.
TCP gene pairs and divergence time Estimation
TCP gene collinearity analysis and visualization were performed using the Tbtools software44. Briefly, collinearity, CTL and .gff files were generated using the One Step MCScanX-Super Fast module, gene pair files were generated from the covariance files using the file merge module of MCScanX, TCP gene pairs were generated using the Text Block Extraction and Filtering module. The protein sequences and open reading frames (ORFs) of the TCP gene pairs were compared using ClustalW in the MEGA 11 software. The CODEML program was used to calculate the synonymous substitution rate (Ks) and non-synonymous substitution rates (Ka) of PAL2NAL (http://www.bork.embl.de/pal2nal/).
Gene ontology and promoter cis-regulator element analysis
GO (Gene Ontology) enrichment analysis of TCP genes in the Benincaseae tribe (C. melo, C. sativus, C. lanatus and L. siceraria) and Cucurbiteae tribe (C. maxima, C. moschata and C. pepo) was performed using the agriGo online tool (http://systemsbiology.cau.edu.cn/agriGOv2/) with default parameters, and the terms are presented using a Venn plot (https://jvenn.toulouse.inrae.fr/app/example.html). Sequence similarity was used to predict function, sequence alignment was used to predict homology, E-values and bit scores were used to filter low-quality homology matches, GO annotation terms associated with proteins involved in well-known biological processes were used to classify function, and GO terms with P values ≤ 0.05 were considered highly representative. Sequences within 2000 bp upstream of the ATG promoter in TCP genes were examined and searched for in the Cucurbitaceae genome. Cis-regulator elements were identified using the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare), and cis-regulator elements were statistically analyzed and visualized using heat maps constructed in R.
Transcript expression analysis of TCP TFs
Using published RNA-seq data, the expression patterns of CmTCP genes were analyzed in fruit, root, female flower, male flower and leaf in melon48. Expression patterns of CsTCP genes were analyzed in the root, stem, leaf, tendril base, tendril, female flower, male flower, ovary, fertilized ovary and unfertilized ovary in cucumber49. Expression profiling of ClTCP genes was conducted in multiple watermelon tissues, including root, leaf, FF10 (Fruit flesh 10 days after pollination), FF18, FF26, FF34, FR10 (Fruit rind at 10 days after pollination), FR18, FR26 and FR34 in watermelon23,29. The expression patterns of LsTCP genes were analyzed in root, flower, leaf, fruit and stem in bottle gourd50. Expression profiling of TCP genes was conducted in C. maxima and C. moschata tissues, including leaf, root, fruit and stem38. The expression patterns of CpTCP genes were analyzed in leaf, fruit at 5d (days after pollination), fruit at 10d, fruit at 15d, fruit at 20d, fruit at 40d, seed at 5d, seed at 10d, seed at 15d, seed at 20d, and seed at 40d in C. pepo38,39. TCP gene expression data analysis was performed using the free online platform OmicShare Tools (https://www.omicshare.com/tools). Dynamic mountain range maps were used to visualize gene tissue expression specificity, with the height of the mountain range indicating the number of genes, and the degree of smooth extension indicating gene expression.
Transcriptome data of Cucurbitaceae TCP gene expression in response to different environmental stressors were obtained from publicly available transcriptome databases, downloaded from the Cucurbit Expression Atlas (http://cucurbitgenomics.org/rnaseq/home), and analyzed to identify genome-wide differentially expressed genes (DEGs) before and after cold14,51, salt51,52, PM (Powdery mildew)53,54,55, and DM (Downy mildew)56,57 stress. The FDR (or P-value) and Log2 (Fold-change) values published in the literature were used to identify DEGs. If the gene ID was inconsistent with that of the reference genome, the TCP gene sequence was aligned to the reference genome to determine the gene ID. The ComplexHeatmap showing the expression of TCP genes was plotted using the R language software. The transcript abundance of TCP genes was calculated as the number of exon model reads per kilobase per million mapped reads (RPKM). For transcriptome analyses of TCP genes in response to abiotic and biotic stresses, a threshold of FDR values (or P-value) ≤ 0.05 and │Log2 (Fold-change)│ ≥ 1 were used to define DEGs.
Results
Identification and basic information of TCP genes in cucurbitaceae species
TCP genes were identified in seven cucurbit genomes using HMMER, including C. melo, C. sativus, C. lanatus, L. siceraria, C. pepo, C. moschata and C. maxima (Supplemental Table 1). A total of 230 TCP genes were identified and predicted using the CDD and SMART online tools, which verified the existence of the following numbers of unique TCP domains in each species: 29 in C. melo, 27 in C. sativus, 27 in C. lanatus, 28 in L. siceraria, 42 in C. maxima, 42 in C. moschata, and 35 in C. pepo (Supplemental Table 1). All identified TCP genes were renamed according to their locations on the chromosome (Supplemental Fig. 1 and Supplemental Table 2). The TCP genes were widely distributed throughout the genome, but were not uniformly distributed between chromosomes. (Supplemental Fig. 1). Basic information on the TCP genes is summarized in Table 1. The results showed that the lengths of the TCP proteins ranged from 61 (CmTCP4) to 631 (CpTCP25) amino acid residues. The molecular weights of TCP proteins ranged from 7.11 (CmTCP4) to 69.83 (CpTCP25) kDa, and the PIs ranged from 5.27 (LsTCP26) to 10.07 (CmoTCP11) in Cucurbitaceae species (Supplemental Table 2). The theoretical PI was < 7 for acidic proteins and > 7 for basic proteins, there were four acidic proteins in C. melo, seven in C. sativus, six in C. lanatus, nine in L. siceraria, ten in C. maxima, six in C. moschata, and four in C. pepo (Supplemental Table 2). The instability index was less than 40 for stable proteins, and most TCP genes encode stable proteins, except for two TCP genes in C. melo, two in C. sativus, one in C. lanatus, three in C. maxima, and one in C. moschata. The higher the aliphatic index, the better the stability of the protein, and the more suitable they are to function in different environments. GRAVY score was negative for hydrophilic proteins, and all TCP genes encoded hydrophilic proteins (Supplemental Table 2).
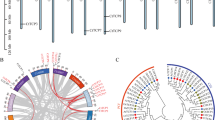
Collinearity analysis of segmental repeats of TCP genes in seven representative species. The red lines represent 15 segmental duplicates of 29 CmTCP genes. The orange lines represent 13 segmental duplicates of 27 CsTCP genes. The yellow lines represent 12 segmental duplicates of 27 ClTCP genes. The green lines represent 15 segmental duplicates of 28 LsTCP genes. The blue lines represent 36 segmental duplicates of 42 CmaTCP genes. The purple lines represent 38 segmental duplicates of 42 CmoTCP genes. The black lines represent 26 segmental duplicates of 35 CpTCP genes. The detailed information is listed in Supplemental table 3.
Gene structure and collinearity analysis of TCP genes
To further validate the 230 TCP genes found in the Cucurbitaceae species, we performed multiple sequence alignments on the TCP domains of all TCP genes to comprehend the phylogenetic relationships of the TCP genes in each species. TCP domain comparison and phylogenetic analysis showed that TCP proteins were divided into two classes: Class I (blue) and Class II (red and orange), Class II was further divided into the subclasses CYC/TB 1 and CIN (Supplemental Fig. 2). TCP genes belong to a protein superfamily, which distinguishes them from other gene families. Although most TCP genes have an integral basic helix-loop-helix structure, a partial absence of the basic structural sequence was present in CsTCP1, LsTCP5/6, CmaTCP24, and CpTCP14, which was also found in other species. The results of the gene structure analysis showed that most TCP genes in the Benincaseae tribe contained one or two CDS regions, and a few contained three CDS regions, including CsTCP4/18/27 and LsTCP3/4/13/24. Although most TCP genes in the Cucurbiteae tribe contained one or two CDS regions, others contained more CDS regions; for example, CmaTCP41 and CpTCP21 have seven CDS regions, CmoTCP33 had six, and CpTCP7 have five (Supplemental Fig. 2).
Comprehensive map of TCP loci in Cucurbitaceae species. TCP genes were localized on 12 chromosomes in melons. The different species are represented by different squares, in the following order: C. melo, C. sativus, C. lanatus, L. siceraria, C. maxima, C. moschata, and C. pepo. The color of the squares indicates the number of genes.
To further explore the relationships among the TCP genes across different species, duplication events in the seven species (C. melo, C. sativus, C. lanatus, L. siceraria, C. pepo, C. moschata, and C. maxima) were investigated (Fig. 1). The results revealed 55 segmental duplication events in the Benincaseae (C. melo, C. sativus, C. lanatus, and L. siceraria) (Fig. 1A and Supplemental Table 3). Furthermore, 15, 13, 12, and 15 collinear gene pairs were identified in C. melo, C. sativus, C. lanatus, and L. siceraria, respectively (Fig. 1A and Supplemental Table 3). More segmental duplication events were identified in the Cucurbiteae tribe (C. pepo, C. moschata and C. maxima), and 36, 38, and 26 collinear gene pairs were observed in C. maxima, C. moschata and C. pepo, most of which were observed between sub-genomes (Fig. 1B and Supplemental Table 3). Our findings suggest that segmental duplication plays a crucial role in the expansion of TCP genes.
Construction of the TCP gene integration map
Melon, cucumber, and watermelon are horticultural crops that are grown worldwide. Using the melon genome as a reference, collinearity analysis revealed a similar number of collinear regions in both genomes (C sativus and C lanatus) (Supplemental Table 4). Watermelon and bottle gourds were closely related, and collinearity analysis revealed approximately 53 collinear gene pairs between melon and bottle gourds (Supplemental Table 4). More collinear genes were detected in the Cucurbita tribe when using melon as the reference genome. In total, 81, 77, and 61 collinear gene pairs were detected in C maxima, C moschata, and C pepo, respectively (Supplemental Table 4).
Cucurbit species have 20 chromosomes, 19 of which can be divided into two sub-genomes, A and B, except for chromosome 4, which consists of two sub-genome A chromosomes and one sub-genome B chromosomes38. The number of TCP genes retained in both subgenomes was similar, which was consistent with similar evolutionary proportions of genes lost or gained in both subgenomes after polyploidization in cucurbit species (Fig. 2). Modern structural studies of chromosomes in Cucurbitaceae species indicate that their genome is an ancestral Cucurbitaceae karyotype consisting of 12 protochromosomes37. To investigate the conserved loci of TCP genes on chromosomes, we constructed an integrated genetic map containing 29 TCP loci, using the melon genome as a reference (Fig. 2). Most of the loci were common to all species, except for the eighth locus at the end of chromosome 6 (Fig. 2). Interestingly, some loci were shared by different genes of the same species; for example, the first locus contained one CmTCP, three CsTCP, three ClTCP, three LsTCP, four CmaTCP, three CmoTCP, and four CpTCP (Supplemental Fig. 3).
Phylogenetic tree of the TCP gene family was constructed based on the amino acid sequences of the TCP structural domains in Cucurbitaceae species. Maximum likelihood (ML) tree of the 230 TCP proteins categorized into Class I and II. Class II was subdivided into the clades CIN and CYC. Colored lines represent different classes, and colored dashed lines represent the TCP genes in different species.
Analysis of phyletic evolution and duplication events in TCP genes
To explore the evolutionary relationships among the 230 TCP proteins in Cucurbitaceae, we constructed an ML evolutionary tree based on TCP structural domain comparisons using MEGA11. The 230 TCP proteins were categorized into Class I and Class II, and no species-specific branching was observed, suggesting that the classification and function of the TCP genes were conserved (Fig. 3). The number of proteins in Class II (129 members) was slightly larger than that in Class I (101 members), and Class II was further divided into CIN and CY/TB 1 branches, with the CIN branch (76 members) being larger than the CY/TB 1 branch (53 members). All investigated cucurbits had CIN branch TCP genes, and TCP genes in the CYC branch were also present in the seven cucurbit species, but at a lower frequency than the other branch (Fig. 3). In our phylogenetic tree, the TCP proteins from each species were clustered in a single sub-branch, indicating that they expanded after diverging from a common ancestor. For example, TCP proteins from melons were clustered in Class I with 13 members and in the CIN branch with 10 members (Fig. 3). Phylogenetic analyses suggested that each gene classification in the family was conservatively distributed during cucurbit evolution, and that TCP genes in the Class I and CIN branches might have been ancient.
To further study the duplication events among TCP genes, we analyzed the collinearity of TCP genes at the genomic level using the TBtools software. Among the 230 TCP genes, 379 collinearity events were found between melons and other cucurbit crops, suggesting duplication events experienced by the TCP genes. To understand the divergence of the gene pairs, we calculated the synonymous substitution rate (Ks/Ka), with most members of the same subfamily experiencing duplicate events (Supplemental Tables 5 and 6). Combined with the results of the phylogenetic tree, these findings suggest a more distant evolutionary relationship, which makes discovering divergence more challenging. Ka/Ks indicates the method of gene selection based on experience. Ka/Ks < 1, indicates that the gene underwent purifying selection, indicating that the selection process neutralized the mutation to maintain protein stability. In contrast, Ka/Ks > 1, suggests that the gene underwent positive selection, indicating that the gene has undergone a large mutation that ultimately leads to a change in the encoded protein. Our identified TCP gene pairs had Ka/Ks values ranging from 0.05 to 0.44, therefore all of these genes underwent purifying selection, suggesting that the TCP genes have experienced little to no severe mutational disruption (Supplemental Table 5).
GO enrichment and Cis-regulator element analysis of TCP genes
To fully explore TCP gene function, GO enrichment analysis was performed for the 230 TCP genes in Cucurbitaceae species. A total of 114 GO terms (P-value < 0.05) were identified, of which 95 belonged to biological process (BP), 10 belonged to cellular components (CC), and nine belonged to molecular function (MF). Regarding the BP-related terms, five GO terms were enriched in the Benincaseae tribe, including ‘regulation of cellular metabolic processes’ (GO:0031323), ‘regulation of metabolic processes’ (GO:0019222), ‘regulation of cellular processes’ (GO:0050794), ‘regulation of biological processes’ (GO:0050789), and ‘biological regulation’ (GO:0065007) (Fig. 4 and Supplemental Table 7). Moreover, 72 GO terms were co-enriched in Cucurbiteae tribe, except for ‘organic cyclic compound binding’ (GO:0097159). The findings are consistent with the fact that TCP genes are a major regulator and suggest that TCP genes are involved in many downstream transcriptional networks and functions primarily in the nucleus of Cucurbitaceae species. Several other terms, such as ‘nucleic acid-templated transcription’ (GO:0097659) and ‘response to endogenous stimulus’ (GO:0009719) were enriched, which are consistent with the proposed function of TCP genes (Fig. 4 and Supplemental Table 7).
Regions that are 2000 bp upstream of the C. melo, C. sativus, C. lanatus, L. siceraria, C. maxima, C. moschata, and C. pepo TCP genes were extracted to identify cis-regulatory elements and explore promoter region functions. In total, 619, 607, 508, 537, 1084, 873, and 849 cis-regulatory elements were identified in C. melo, C. sativus, C. lanatus, L. siceraria, C. maxima, C. moschata, and C. pepo, respectively. The cis-regulators included phytohormone responsive elements (ABA, auxin, gibberellin, MeJA, and SA responsive elements), stress responsive elements (defense and stress, drought, light, and wound responsive elements) and growth and development elements (anaerobic induction, circadian control, and meristem expression) (Fig. 5). Light-responsive elements were the most prevalent functional elements of TCP gene promoters in each species, indicating that light plays a critical role in regulating TCP gene function throughout plant growth and development (Supplemental Fig. 4). The second and third predominant cis-regulator elements were ABA-responsive- and MeJA-responsive elements, which were also widely distributed in TCP gene promoter regions of Cucurbitaceae species (Supplemental Fig. 4). These results suggest that these elements may control the actions of the two phytohormones. Furthermore, we identified components involved in meristem expression and anaerobic induction, which supports the critical function of TCP genes in preserving meristem homeostasis (Fig. 5).
Cis-regulator elements in the promoter regions of TCP genes. The detailed information is listed in Supplemental Figure 4. The colored bar shows the number of cis-regulator elements. The horizontal axis of the heatmap indicates the class of cis-regulator elements. The ordinate represents TCP genes of different species, from smallest to largest.
Expression profiles of TCP genes in different tissues
To better understand the expression patterns of TCP genes, their expression patterns in different tissues from the seven cucurbit species were analyzed using published RNA-seq data. The expression of 29 CmTCP genes in the fruit, roots, female flowers, male flowers, and leaves of melons were analyzed (Fig. 6). In addition, TCP genes expression in different tissues of other cucurbit species was analyzed. The dynamic volcano plots were constructed to visualize tissue-specific expression of TCP genes in cucurbit species, with the height of each peak indicating the number of genes, and the degree of smoothing indicating the amount of gene expression (Fig. 6). The result showed that the expression profiles of TCP genes were different among tissues, suggesting that TCP genes have different functions in different plant development stages. In all cucurbit species, TCP genes showed moderate expression levels in the leaves, suggesting that TCP genes play a role in plant growth (Fig. 6). CmTCP, CsTCP, and LsTCP were highly expressed in flowers, indicating the potential significance of these TCP genes in driving tissue differentiation. CmaTCP, CmoTCP, and CpTCP were also highly expressed in the stems, indicating that these TCP genes play an important role in plant development. The results suggest that different TCP genes play different roles in plant growth and development.
Expression analysis of TCP genes in different tissues under normal growth conditions in the seven Cucurbitaceae species5,38,48,56. Transcript levels of TCP genes were normalized to Reads Per Kilobase per Million mapped (RPKMs) and is displayed as the mean of three biological repeats. In the volcano plot, dashed lines above the peaks represent the median line, and the dashed lines below the peaks represent TCP genes in each species. FF, fruit flesh. FR, fruit rind.
Activation of TCP gene transcription in response to abiotic and biotic stressors
To determine whether TCP genes are involved in abiotic stress, we analyzed the expression of CmTCP and ClTCP after cold stress in melons and watermelons. As shown in Fig. 7A, TCP genes exhibited diverse expression patterns after cold stress, which varied among the cucurbit species. After cold stress, TCP21 expression was significantly upregulated in melons, and the expression of TCP21, which has the same gene ID, was significantly upregulated in watermelons (Fig. 7A). These results suggest that TCP21 may be involved in the regulating responses to cold stress in cucurbit species. To further understand the role of TCP genes in adaptations to environmental stress, we analyzed the expression of TCP genes in the leaves, leaf mesophyll, and leaf veins following salt stress. Compared to that under normal conditions, the expression of CsTCP6/7/13/20/26 was significantly increased after salt stress treatment, and the expression of 12 CmoTCP genes (CmoTCP4/5/7/13/21/27/28/29/34/36/38/41) was significantly altered (Fig. 7B). After salt stress, CmoTCP2/10 expression was significantly upregulated in the leaf mesophyll, as well as CmaTCP2 expression (Fig. 7B). After salt stress, CmoTCP31 and CmaTCP2/15/32 expression was significantly upregulated in the leaf veins (Fig. 7B). These results indicate that TCP genes may be involved in the response to salt stress in cucurbit species.
To predict the possible functions of TCP genes in environmental adaptation, we investigated the transcriptional profiles of TCP genes under biotic stress conditions, including powdery and downy mildew stress. CmTCP29 expression was downregulated after powdery mildew infection in resistant (MR-1) and susceptible melons (top mark) (Fig. 8A). Notably CmTCP18 expression was significantly elevated after powdery mildew infection with MR-1. Compared with that under normal conditions, the expression of seven CmTCP genes (CmTCP3/9/13/18/21/28/29) was upregulated at 48 h post-infection (hpi) (Fig. 8A). After powdery mildew infection, CsTCP13/19 expression was significantly upregulated in SSL508-28 cells, whilst CsTCP14 expression was significantly upregulated on D8 (Fig. 8A). CsTCP1/25 expression showed a gradual increase after downy mildew infection, whereas CsTCP23/31 and CsTCP18 expression showed a gradual decrease in resistant (PI197088) and susceptible melons (Vlaspik), respectively (Fig. 8B). In addition, we analyzed TCP genes expression in the early stages of Pseudoperonospora cubensis infection (0/6/24 hpi) and found that CsTCP1/5/12/15/16/19 expression increased gradually, whilst CsTCP13/24/25 expression decreased gradually (Fig. 8B). Overall, the findings indicate that the transcript levels of TCP genes differ under different stresses, suggesting that TCP genes play an important role in response to abiotic and biotic stresses in cucurbit species.
TCP gene expression patterns under different abiotic stresses. Heatmap of TCP genes in response to cold sress in C. melo and C. lanatus (A) and salt sress in C. maxima, C. moschata, and C. sativus (B). RNA-seq data obtained from CuGenDB (http://cucurbitgenomics.org/) and public data were used14,34,52. Color coding was based on the scale given. Boxes filled with dots indicate no detection in the species. Transcript levels were calculated as log2 transformed Reads Per Kilobase of exon model per Million mapped reads (RPKM).
Expression profile of TCP genes in response to biotic stresses. (A) Expression profiles of TCP genes in response to PM infection in C. melo (cultivars: Top Mark and MR-1, SBA number: PRJNA358655; cultivars: Rochet, SBA number: PRJNA434538) and C. sativus (cultivars: SSL508-28 and D8, SBA number: PRJNA321023). PM, powdery mildew. (B) Expression profiles of CsTCP genes in response to DM infection in cucumbers (cultivars: Vlaspik and PI 197088, SBA number: PNJNA285071; cultivars: PI 197088, SBA number: PNJNA388548). DM, Downy mildew. The axis shows the logarithmic normalized RPKM. hpi, hours after post-pathogen infection. dpi, days after post-pathogen infection.
Discussion
The plant-specific TF TCP plays various roles in processes related to plant growth, development, and stress responses. With the extensive sequencing of plant genomes, genome-wide identification of TCP genes has been performed in a variety of plants, including rice, maize, Arabidopsis, tomatoes, and potatoes41,58,59,60. Across species, TCP gene sequences are generally conserved, with most members belonging to two classes37,42. Although TCP gene analyses have been conducted for individual Cucurbitaceae species, there is a lack of comprehensive summaries24. In this study, TCP genes from seven species of Cucurbitaceae were recharacterized and analyzed, and 230 members were identified, of which 101 TCP genes belonged to Class I and 129 members belonged to Class II. Most plants have relatively similar numbers of TCP genes, for example there are 22 in rice, 29 in maize, 24 in Arabidopsis, and 30 in tomatoes, which is similar to the number of TCP genes in the seven cucurbit species. Notably, there is some variation in the size of these plant genomes; for example, the melon genome is 375 Mb, the cucumber genome is 243.5 Mb, and the C. maxima, C. moschata and C. pepo genomes are 269.9, 271.4, and 263 Mb, respectively28. Therefore, it can be inferred that cucurbits contain a similar number of TCP genes, which are not necessarily related to the genome size of the species. In addition, according to the PI range of TCP genes in Cucurbita species, most of TCP genes are basic and hydrophilic proteins that are mainly distributed in the nucleus and may act in alkaline environments, suggesting that TCP proteins might play a role in regulating nuclear genes, which is in agreement with the results of our GO enrichment analysis. The results of the above analyses indicate that TCP genes in Cucurbitaceae are largely evolutionarily and functionally conserved.
Analyzing the evolutionary relationship of TCP genes aids the prediction of their functions. By analyzing CrTCP3 as a homologous protein of AtTCP11, previous authors deduced that this gene may regulate leaf and stem development61. Phylogenetic and structural domain analyses of TCP genes in different species revealed that TCP genes in Cucurbitaceae species are divided into Classes I and II, and Class II is further divided into the CIN and CYC/TB1 subclasses62. All cucurbit TCP proteins contain a typical TCP structural domain. Phylogenetic analyses showed that TCP genes from the same species tended to cluster together in both Class I and Class II analyses, which is consistent with the findings of previous studies on other species and demonstrates that genomic differences emerged during the evolution of cucurbit species9,62. TCP genes play key regulatory roles in flower development. PeCIN8/10 play an important role in orchid ovule development by regulating cell division63. Moreover, CIN-type genes may play a key role in petal formation and are highly expressed in the ovule development and pistil initiation stages64. This is consistent with the findings of the present study that CIN-type genes in cucurbit species play an important role in the development of female floral organs. Among the Class I-type genes, CmTCP14 suppresses organ size and prolongs flowering time in chrysanthemums30, and AtTCP11 and AtTCP16 are involved in early pollen development20,61. Additionally, CIN genes play important regulatory roles in plant growth and development, particularly in the development of the lateral organs10.
The main causes of gene amplification are segmental duplication, tandem duplication, and WGD. Tandem duplication genes have multiple functions in plants65. Tandem duplication of biosynthetic genes drives the diversity of plant secondary metabolites, which might be related to the evolution of certain unique traits66. The rye genome contains 7,077 tandemly duplicated genes, which correlate with the abundance of secondary metabolites in rye67, including ScTCP genes identified with three tandem duplicate gene pairs7. After gene duplication, most redundant copies of genes are exposed of during the long evolutionary process, but some duplicates are retained depending on the level of adaptation to the environment, which explains the differences in copy numbers between species62. Both Class I and Class II TCP genes have undergone continuous expansion, developing from a few members in algae and bryophytes to dozens of members in angiosperms, with some species having more than 50 members, such as soybeans, which have 55 TCP genes9,62. The TCP gene family has gradually expanded and diversified as green plants have diversified, and similar changes have occurred in Cucurbitaceae species. Gene duplication and collinearity analyses showed that most TCP genes (155/230) arose from gene duplication events; however, the expansion of TCP genes may be caused by different mechanisms in different Cucurbitaceae species. Based on the evolutionary relationship among Cucurbitaceae, both the Benincaseae and Cucurbiteae tribes experienced an initial WGD event, with species belonging to the Benincaseae and Cucurbiteae tribes forming the sister clades28. In our study, the number of TCP genes in the Cucurbiteae tribe was higher than that in the Benincaseae tribe, which may be because the Cucurbiteae tribe underwent another WGD event during its evolution. Both ancient and recent WGD events occurred in most angiosperms, which led to the expansion of the TCP gene family; this observation also supported our findings. Therefore, we conclude that the expansion of the TCP gene family in Cucurbitaceae was mainly driven by dispersed duplication and segmental duplications/WGD. In addition, collinearity analysis revealed the same high number of fragment repeats, and the purification selection was positive. Functional annotation analysis showed that TCP genes were not only involved in ‘regulation of cellular metabolic processes’, ‘regulation of metabolic processes’, ‘regulation of cellular processes’, ‘regulation of biological processes’, ‘biological regulation’, but also in response to light and hormones and tissue development. TFs regulate gene expression; therefore, TCPs have diverse functions, which is consistent with the findings of previous studies on the function of TCP genes7,68.
TFs regulate gene expression at the transcriptional level by binding to cis-regulator elements69,70. In this study, a series of cis-regulator elements were identified in the promoter regions of TCP genes, including growth-associated, hormone-associated, and stress response-associated cis-regulator elements71. The presence of different types of cis-regulator elements in the promoter regions, indicates that Cucurbitaceae TCP genes might be associated with a wide range of stress responses. The active components of secondary metabolites play an important role in plant adaptation to unfavorable environments and some environmental stressors can be used as inducers72. Previously, positive effects of UV-B radiation and MeJA application on TIA induction were confirmed15. In Ginkgo, GbTCP6/11/13 were significantly up-regulated following exogenous MeJA administration27. In the present study, light-responsive elements were the most prevalent in the promoters of TCP genes across species, suggesting that light is a key regulator of TCP gene function during plant growth and development. The second and third predominant cis-regulatory elements were ABA- and MeJA-responsive elements. Furthermore, these elements are widely distributed in the promoter region of TCP in cucurbit species. These results suggest that TCP genes may be involved in hormone signaling in cucurbit species. In addition, we identified other components involved in phloem organization and anerobic induction, indicating the critical function of TCP gene in phloem homeostasis9.
TCP genes play an important role in the adaptation of plants to a wide range of biotic and abiotic stresses10,71. Furthermore, TCP genes may have an evolutionarily conserved function in gene regulation, and comparative analyses analyzing upstream and downstream TCP signaling pathways indicate that TCP regulatory pathways are conserved among species. Our analysis of the transcriptional profiling libraries of TCP genes under abiotic (cold and salt stress) and biotic (powdery mildew and downy mildew) conditions showed that TCP genes were generally similar among different species of Cucurbitaceae. Preliminary studies have demonstrated the broad functional conservation of TCP genes in Rosaceae species, which is consistent with the results of this study15. However, further investigation is required to confirm our preliminary findings. Overall, analyzing the biological functions of these TCP genes will provide an important theoretical basis for improving the response of melons to environmental stresses.
Data availability
Publicly available datasets were analyzed in this study. The C. melo, C. sativus, C. lanatus, L. siceraria, C. maxima, C. moschata and C. pepo genome data presented in this study are openly available in CuGenDB at http://cucurbitgenomics.org/download. The RNAseq data presented in this study are openly available in https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA80169, PRJNA285071, PRJNA321023, PRJNA328189, PRJNA358655, PRJNA383830, PRJNA388584, PRJNA434538, PRJNA437579, PRJNA464060 and SRP012849.
References
Wray, G. A. et al. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419. https://doi.org/10.1093/molbev/msg140 (2003).
Amoutzias, G. D. et al. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol. Biol. Evol. 24, 827–835. https://doi.org/10.1093/molbev/msl211 (2007).
Abbas, F. et al. Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 12, 623742. https://doi.org/10.3389/fpls.2021.623742 (2021).
Ambawat, S., Sharma, P., Yadav, N. R. & Yadav, R. C. MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biology Plants: Int. J. Funct. Plant. Biology. 19, 307–321. https://doi.org/10.1007/s12298-013-0179-1 (2013).
Lai, X., Chahtane, H., Martin-Arevalillo, R., Zubieta, C. & Parcy, F. Contrasted evolutionary trajectories of plant transcription factors. Curr. Opin. Plant. Biol. 54, 101–107. https://doi.org/10.1016/j.pbi.2020.03.002 (2020).
Cao, Y., Li, K., Li, Y., Zhao, X. & Wang, L. MYB transcription factors as regulators of secondary metabolism in plants. Biology 9 https://doi.org/10.3390/biology9030061 (2020).
Zhan, W., Cui, L., Guo, G. & Zhang, Y. Genome-wide identification and functional analysis of the TCP gene family in Rye (Secale cereale L). Gene 854, 147104. https://doi.org/10.1016/j.gene.2022.147104 (2023).
Lehti-Shiu, M. D., Panchy, N., Wang, P., Uygun, S. & Shiu, S. H. Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochim. Et Biophys. Acta Gene Regul. Mech. 1860, 3–20. https://doi.org/10.1016/j.bbagrm.2016.08.005 (2017).
Liu, M. M. et al. Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20143591 (2019).
Li, Y. et al. Genome-wide identification and analysis of TCP gene family among three Dendrobium species. Plants (Basel Switzerland). 12 https://doi.org/10.3390/plants12183201 (2023).
Schommer, C., Debernardi, J. M., Bresso, E. G., Rodriguez, R. E. & Palatnik, J. F. Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant. 7, 1533–1544. https://doi.org/10.1093/mp/ssu084 (2014).
Li, X. et al. A natural antisense RNA improves chrysanthemum cold tolerance by regulating the transcription factor DgTCP1. Plant Physiol. 190, 605–620. https://doi.org/10.1093/plphys/kiac267 (2022).
Aggarwal, P. et al. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant. Cell. 22, 1174–1189. https://doi.org/10.1105/tpc.109.066647 (2010).
Diao, Q., Cao, Y., Fan, H. & Zhang, Y. J. B. P. Transcriptome analysis Deciphers the mechanisms of exogenous nitric oxide action on the response of melon leaves to chilling stress. 64, 465–472 (2020).
Hao, J. et al. Genome-wide identification and expression analysis of TCP family genes in Catharanthus roseus. Front. Plant Sci. 14, 1161534. https://doi.org/10.3389/fpls.2023.1161534 (2023).
Tatematsu, K., Nakabayashi, K., Kamiya, Y. & Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant. Journal: Cell. Mol. Biology. 53, 42–52. https://doi.org/10.1111/j.1365-313X.2007.03308.x (2008).
Kieffer, M., Master, V., Waites, R. & Davies, B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant. Journal: Cell. Mol. Biology. 68, 147–158. https://doi.org/10.1111/j.1365-313X.2011.04674.x (2011).
Aguilar-Martínez, J. A., Poza-Carrión, C. & Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant. Cell. 19, 458–472. https://doi.org/10.1105/tpc.106.048934 (2007).
Giraud, E. et al. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant. Cell. 22, 3921–3934. https://doi.org/10.1105/tpc.110.074518 (2010).
Takeda, T. et al. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol. Biol. 61, 165–177. https://doi.org/10.1007/s11103-006-6265-9 (2006).
Guo, Z. H. et al. Expression analysis of TCP genes in Peach reveals an involvement of PpTCP.A2 in ethylene biosynthesis during fruit ripening. Plant. Mol. Biology Report. 36, 588–595. https://doi.org/10.1007/s11105-018-1105-z (2018).
Broholm, S. K. et al. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U.S.A. 105, 9117–9122. https://doi.org/10.1073/pnas.0801359105 (2008).
Ling, L. et al. Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genom. 20, 537–550. https://doi.org/10.1007/s10142-020-00733-0 (2020).
Wen, H. et al. Genome-Wide identification and characterization of the TCP gene family in cucumber (Cucumis sativus L.) and their transcriptional responses to different treatments. Genes 11 https://doi.org/10.3390/genes11111379 (2020).
Li, D. et al. Comparative genomic investigation of TCP gene family in eggplant (Solanum melongena L.) and expression analysis under divergent treatments. Plant Cell Rep. 41, 2213–2228. https://doi.org/10.1007/s00299-022-02918-2 (2022).
Almeida, D. M., Gregorio, G. B., Oliveira, M. M. & Saibo, N. J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 93, 61–77. https://doi.org/10.1007/s11103-016-0547-7 (2017).
Yu, L. et al. Genome-wide identification and expression pattern analysis of the TCP transcription factor family in Ginkgo biloba. Plant Signal. Behav. 17 https://doi.org/10.1080/15592324.2021.1994248 (2022).
Ma, L. et al. Cucurbitaceae genome evolution, gene function and molecular breeding. Hortic. Res. 9 https://doi.org/10.1093/hr/uhab057 (2022).
Guo, J. et al. Phylotranscriptomics in Cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Mol. Plant. 13, 1117–1133. https://doi.org/10.1016/j.molp.2020.05.011 (2020).
Zhou, Y. et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat. Plants. 2, 16183. https://doi.org/10.1038/nplants.2016.183 (2016).
Wang, J. et al. An overlooked paleotetraploidization in Cucurbitaceae. Mol. Biol. Evol. 35, 16–26. https://doi.org/10.1093/molbev/msx242 (2018).
Castanera, R., Ruggieri, V., Pujol, M., Garcia-Mas, J. & Casacuberta, J. M. An improved melon reference genome with single-molecule sequencing uncovers a recent burst of transposable elements with potential impact on genes. Front. Plant Sci. 10, 1815. https://doi.org/10.3389/fpls.2019.01815 (2019).
Garcia-Mas, J. et al. The genome of Melon (Cucumis Melo L). Proc. Natl. Acad. Sci. U.S.A. 109, 11872–11877. https://doi.org/10.1073/pnas.1205415109 (2012).
Huang, S. et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. https://doi.org/10.1038/ng.475 (2009).
Osipowski, P. et al. A high-quality cucumber genome assembly enhances computational comparative genomics. Mol. Genet. Genomics: MGG. 295, 177–193. https://doi.org/10.1007/s00438-019-01614-3 (2020).
Guo, S. et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 45, 51–58. https://doi.org/10.1038/ng.2470 (2013).
Wu, S. et al. The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant. Journal: Cell. Mol. Biology. 92, 963–975. https://doi.org/10.1111/tpj.13722 (2017).
Sun, H. et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant. 10, 1293–1306. https://doi.org/10.1016/j.molp.2017.09.003 (2017).
Montero-Pau, J. et al. De Novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 16, 1161–1171. https://doi.org/10.1111/pbi.12860 (2018).
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D. & Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. https://doi.org/10.1007/s00239-006-0174-z (2007).
Parapunova, V. et al. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14 https://doi.org/10.1186/1471-2229-14-157 (2014).
Ma, J. et al. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4, 6645. https://doi.org/10.1038/srep06645 (2014).
Zhao, J. et al. Genome-wide identification and expression profiling of the TCP family genes in Spike and grain development of wheat (Triticum aestivum L). Front. Plant Sci. 9, 1282. https://doi.org/10.3389/fpls.2018.01282 (2018).
Chen, C. et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 (2020).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (e49). https://doi.org/10.1093/nar/gkr1293 (2012).
Letunic, I. & Bork, P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinf. (Oxford England). 23, 127–128. https://doi.org/10.1093/bioinformatics/btl529 (2007).
Hu, B. et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinf. (Oxford England). 31, 1296–1297. https://doi.org/10.1093/bioinformatics/btu817 (2015).
Latrasse, D. et al. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics Chromatin. 10 https://doi.org/10.1186/s13072-017-0132-6 (2017).
Li, Z. et al. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 12, 540. https://doi.org/10.1186/1471-2164-12-540 (2011).
Xu, P. et al. Long-read genome assembly and genetic architecture of fruit shape in the bottle gourd. Plant. Journal: Cell. Mol. Biology. 107, 956–968. https://doi.org/10.1111/tpj.15358 (2021).
Huang, Y. et al. Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae. J. Exp. Bot. 70, 5879–5893. https://doi.org/10.1093/jxb/erz328 (2019).
Niu, M. et al. An early ABA-induced stomatal closure, Na + sequestration in leaf vein and K + retention in mesophyll confer salt tissue tolerance in Cucurbita species. J. Exp. Bot. 69, 4945–4960. https://doi.org/10.1093/jxb/ery251 (2018).
Vaughn, J. N. et al. Graph-based pangenomics maximizes genotyping density and reveals structural impacts on fungal resistance in melon. Nat. Commun. 13, 7897. https://doi.org/10.1038/s41467-022-35621-7 (2022).
Polonio, Á. et al. RNA-seq analysis and fluorescence imaging of melon powdery mildew disease reveal an orchestrated reprogramming of host physiology. Sci. Rep. 9, 7978. https://doi.org/10.1038/s41598-019-44443-5 (2019).
Yang, X. et al. Comparative ribosome profiling reveals distinct translational landscapes of salt-sensitive and -tolerant rice. BMC Genom. 22, 612. https://doi.org/10.1186/s12864-021-07922-6 (2021).
Liu, X. et al. Identification of novel loci and candidate genes for cucumber downy mildew resistance using GWAS. Plants (Basel Switzerland). 9 https://doi.org/10.3390/plants9121659 (2020).
Ma, S. et al. Genome-wide identification, structural, and gene expression analysis of BRI1-EMS-Suppressor 1 transcription factor family in Cucumis sativus. Front. Genet. 11, 583996. https://doi.org/10.3389/fgene.2020.583996 (2020).
Chai, W., Jiang, P., Huang, G., Jiang, H. & Li, X. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biology Plants: Int. J. Funct. Plant. Biology. 23, 779–791. https://doi.org/10.1007/s12298-017-0476-1 (2017).
Danisman, S. et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. https://doi.org/10.1104/pp.112.200303 (2012).
Bao, S., Zhang, Z., Lian, Q., Sun, Q. & Zhang, R. Evolution and expression of genes encoding TCP transcription factors in Solanum tuberosum reveal the involvement of StTCP23 in plant defence. BMC Genet. 20, 91. https://doi.org/10.1186/s12863-019-0793-1 (2019).
Viola, I. L., Manassero, U., Ripoll, N. G., Gonzalez, D. H. & R. & The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 435, 143–155. https://doi.org/10.1042/bj20101019 (2011).
Wang, J. L., Wang, H. W., Cao, Y. N., Kan, S. L. & Liu, Y. Y. Comprehensive evolutionary analysis of the TCP gene family: further insights for its origin, expansion, and diversification. Front. Plant Sci. 13, 994567. https://doi.org/10.3389/fpls.2022.994567 (2022).
Lin, Y. F. et al. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 67, 5051–5066. https://doi.org/10.1093/jxb/erw273 (2016).
Zhou, Y. et al. Genome-wide identification, characterization and expression analysis of the TCP gene family in Prunus mume. Front. Plant Sci. 7, 1301. https://doi.org/10.3389/fpls.2016.01301 (2016).
Huang, Y. et al. The impact of tandem duplication on gene evolution in Solanaceae species. J. Integr. Agric. 21, 1004–1014. https://doi.org/10.1016/S2095-3119(21)63698-5 (2022).
Wang, Y. et al. Deletion and tandem duplications of biosynthetic genes drive the diversity of triterpenoids in Aralia elata. Nat. Commun. 13, 2224. https://doi.org/10.1038/s41467-022-29908-y (2022).
Li, G. et al. A high-quality genome assembly highlights Rye genomic characteristics and agronomically important genes. Nat. Genet. 53, 574–584. https://doi.org/10.1038/s41588-021-00808-z (2021).
Fang, Y. et al. Roles of miR319-regulated TCPs in plant development and response to abiotic stress. Crop J. 9, 17–28. https://doi.org/10.1016/j.cj.2020.07.007 (2021).
Yu, C. et al. Role of female-predominant MYB39-bHLH13 complex in sexually dimorphic accumulation of taxol in Taxus media. Hortic. Res. 9, uhac062. https://doi.org/10.1093/hr/uhac062 (2022).
Feng, S. et al. Investigation of the role of TmMYB16/123 and their targets (TmMTP1/11) in the tolerance of Taxus media to cadmium. Tree Physiol. 43, 1009–1022. https://doi.org/10.1093/treephys/tpad019 (2023).
Feng, Z. J. et al. Soybean TCP transcription factors: evolution, classification, protein interaction and stress and hormone responsiveness. Plant. Physiol. Biochemistry: PPB. 127, 129–142. https://doi.org/10.1016/j.plaphy.2018.03.020 (2018).
Hao, J. et al. Ultraviolet-B irradiation increases antioxidant capacity of Pakchoi (Brassica Rapa L.) by inducing flavonoid biosynthesis. Plants (Basel Switzerland). 11 https://doi.org/10.3390/plants11060766 (2022).
Funding
This work was supported by Natural Science Foundation of Shandong Province (ZR2023QC248), Liaocheng University, China (grant numbers 318052244, 318052290 and 31946221226), and the Key Research and Development Program of Liaocheng (grant number 2022YDNY11). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
HJ, and SSW designed the experiments, supervised the study, and managed the projects. SSW and WLL performed most of the research and drafted manuscripts. SSW performed bioinformatics analysis and charting. HJ analyzed and discussed the results. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, S., Li, W. & Jin, H. Evolution and comparison of the expression of TCP genes in the benincaseae and cucurbiteae tribes. Sci Rep 15, 15470 (2025). https://doi.org/10.1038/s41598-025-99296-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-99296-y










