Abstract
Brain metastasis is emerging as the most serious concern for breast cancer patients. HER2-positive breast cancer is more prone to undergo brain metastasis than other subtypes; notably, there has been little improvement in the treatment of brain metastasis .Our study confirmed the relevance of HER2 status to brain metastasis risk via clinical data analysis and revealed that exerts GRB2 tumorigenic effects by regulating the Ras/MAPK pathway in vivo and in vitro. Both an in situ injection model and a direct cerebral injection model were used to explore the ability of GRB2 to promote the brain metastasis. Results indicated that HER2- positive is a risk factor for brain metastasis according to clinical data. GRB2 enhances proliferation, migration, and invasion while suppressing apoptosis in HER2-positive breast cancer cells in vitro, primarily by regulating phosphorylation and alternative splicing of key proteins within the Ras/MAPK pathway. Notably, tumor cells were able to cross the blood‒brain barrier in both models assessed in this study. Thus, GRB2 is an oncogenic factor that contributes to the malignancy of HER2-positive breast cancer, GRB2 and HER2 can synergistically promote tumor cell penetration of the blood‒brain barrier and induce metastasis.
Similar content being viewed by others
Introduction
The difficulties in treating brain metastasis (BMs) substantially impede progress in the treatment of breast cancer (BC)1,2. Approximately 10-30% of patients develop BMs, with 30-50% exhibiting the HER2 (ERBB2)-positive subtype3. The median survival time without treatment exceeds 2 months, whereas the overall survival period after treatment is only 13.1 months4,5. Radiation and surgery are the main methods, but their application is limited by strict criteria (tumor size ≤ 4 cm, N ≤ 3, PS 0–2) and some adverse effects (brain necrosis)1,6. The results of the DESTINY-Breast 01–037, PERMEATE8, and HER2CLIMB9 trials indicate that that tyrosine kinase inhibitors (TKIs)10and ADCs11can improve PFS or OS in patients with breast cancer with brain metastasis (BCBMs) by targeting HER2, encouraging further research into the mechanisms of BMs.
The ERBB receptor family consists of ERBB1-4 (HER1-4), which share 40-50% sequence similarity12. Neratinib and pyrotinib reduce the incidence of CNS symptoms by 85% and lower the risk of BMs by inhibiting HER1, HER2, and HER4 expression13,14. GRB2 (growth factor receptor-bound protein 2) plays a pro-oncogenic role in a variety of tumors, but it has received little attention in HER2-positive breast cancer15. The flexible protein GRB2 regulates tumor cell growth by interacting with HER1-4, and it is an essential component of the tyrosine kinase receptor cascade16. Our study aimed to explore the association between GRB2 and the occurrence of BCBMs in the HER2-positive BC subtype.
Results
HER2-positive is a risk factor for brain metastasis in patients with breast cancer according to analysis of clinical data
Among the 353 patients with BCBMs, 79 patients were in the HER2-positive/ HR-negative group, 105 patients were in the triple-negative group(TNBC), 115 patients were in the HER2-negative / HR- positive group, and 54 patients were in the HER2-positive/HR-positive group. The clinical data of these groups were analyzed by Cox Regression analysis, identifying size, ER, HER2, Ki-67, and LN as independent factors influencing TTBMs, ER and HER2 as common factors influencing TTBM, OSBM, and OS, as presented in Table 1.
We categorized all patients with BMs into four groups based on their ER and HER2 status: ERposHER2neg, ERnegHER2pos, ERnegHER2neg, and ERposHER2pos.The TTBM, OSBM, and OS demonstrated statistically significant differences among the four groups (Fig. 1A-C).Specifically, the median TTBM, OSBM, and OS of the ERposHER2neg group were higher than those of the ERposHER2pos group (Supplemental Fig. 1A-C) but lower than those of the ERnegHER2neg group (Supplemental Fig. 1D-F), these results suggest that ER and HER2 status are associated with brain metastasis in breast cancer, with HER2-positive being a more significant risk factor than ER-positive.Furthermore, no significant differences were observed between the ERnegHER2pos and ERposHER2pos groups in terms of median TTBM, OSBM, and OS (Supplemental Fig. 1G-I), indicating that, in the context of HER2-positive, ER status (positive or negative) does not influence the prognosis of breast cancer patients with brain metastasis. Similarly, no significant differences were found in the median TTBM and OSBM between the ERnegHER2pos and ERnegHER2neg groups (Supplemental Fig. 1J-L), suggesting that, in ER-negative patients, HER2 status (positive or negative) does not affect prognosis. However, the median OS of the ERnegHER2pos group was lower than that of the ERnegHER2neg group, indicating that HER2-positive is associated with poorer overall survival.
Association between GRB2/HER2 expression and prognosis in breast cancer. (A) Time to Brain Metastasis (TTBM) in breast cancer patients stratified by ER and HER2 Status. P-value was calculated by Mantel-Cox. **** denotes a P-value < 0.0001. (B) Overall Survival After Brain Metastasis (OSBM) in breast cancer patients stratified by ER and HER2 Status. (C) Overall Survival (OS) in breast cancer patients stratified by ER and HER2 Status. (D) GRB2 is a differentially expressed gene associated with brain metastasis in HER2-Positive breast cancer: Evidence from GSE43837. (E) Box plots illustrating differences in GRB2 expression levels between normal breast tissue(N) and breast cancer tissues(T). P-value < 0.001. (F) Overall Survival of breast cancer patients stratified by low or high GRB2 protein expression (lighter lines represent 95% confidence intervals). (G) Scatter plot of the correlation between GRB2 and HER2 expression. (H) Bubble plot of enriched pathways identified by GO: BP analysis. (I) Venn diagram illustrating the overlap between the MAPK pathway and the ERBB pathway. (J) Protein-protein interaction network analysis of key proteins.
These findings highlight that HER2 serves as a poor prognostic indicator, HER2-positive is associated with the incidence of brain metastasis in breast cancer and significantly influences both OSBM and OS in patients.
High expression of GRB2 in breast cancer is associated with a poor prognosis according to database analysis
We conducted bioinformatics analysis on GSE43837 and screened a total of 1607 DEGs, including 806 upregulated genes and 452 downregulated genes (Fig. 1D). In GEPIA 2.0, the expression of GRB2 was significantly elevated in tumor tissues (P < 0.001,Fig. 1E), and patients with high expression of GRB2 had worse survival outcomes (Fig. 1F). Finally, the coefficients for GRB2 and HER2 were R = 0.52 and P = 8.3e − 87 < 0.05, which means that these two proteins were positively correlated in breast cancer tissues (Fig. 1G). The top 15 enriched pathways were obtained by GO: BP analysis, and the MAPK pathway was the most obviously enriched pathway (Fig. 1H). A total of 14 overlapping proteins were obtained by cross-analysis of the MAPK pathway (KEGG pathway: map04010) and the ERBB pathway (KEGG pathway: map04012)17,18,19(Fig. 1I). PPI analysis of these 14 proteins revealed that GRB2 can bind to ERBB1-4 on the cell surface at the same time (Fig. 1J).
GRB2 expression is positively correlated with HER2 expression in vitro
Among the evaluated cell lines, MDA-MB-231 cells (triple-negative subtype: ER/PR/HER2-negative) exhibited the lowest basal expression of GRB2, with minimal HER2 protein detection (supplemental Fig. 2A-B). This cell line was consequently selected for model establishment. Systematic optimization identified the following conditions for stable transfection: HitransG P, MOI = 10, 2 µg/ml puromycin, 600 µg/ml geneticin, and dual lentiviral transduction with LV-HER2 plus LV-shRNA-GRB2-71 (Supplemental Fig. 2C-E).Comparative analysis revealed substantial GRB2 upregulation in both LV-HER2 and LV-HER2 + LV-shRNA-NC groups (P < 0.05 for both) relative to the control and LV-NC groups. Notably, GRB2 Silencing via LV-shRNA-GRB2-71 restored GRB2 expression to baseline levels, while demonstrating significant suppression compared to LV-HER2 and LV-HER2 + LV-shRNA-NC groups (P < 0.05) (Fig. 2A, supplemental table 2).HER2 expression patterns showed marked elevation in all HER2-transduced groups (LV-HER2, LV-HER2 + LV-shRNA-NC, and LV-HER2 + LV-shRNA-GRB2) compared to control and LV-NC groups (P < 0.05). Intriguingly, GRB2 knockdown in the LV-HER2 + LV-shRNA-GRB2 group resulted in concurrent HER2 downregulation relative to other experimental groups (P < 0.05, Fig. 2B, supplemental table 2), a finding corroborated by GEPIA2 database. These results collectively suggest a positive regulatory relationship between GRB2 and HER2 expression
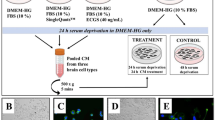
Results of cell proliferation, apoptosis, migration, and invasion across cell Lines. A-B. Expression levels of GRB2 and HER2.* denotes a P-value < 0.05. C-D. Differences in cell proliferation and apoptosis across experimental groups. E. Differences in cell migration and invasion capabilities.* denotes a P-value < 0.05.***denotes a P-value < 0.01. F. Microscopic examination of transwell migration and invasion assays. Blue staining indicates migrated or invaded cells, with a higher cell number reflecting stronger migration or invasion capability. G. Differential expression of proteins in the Ras/MAPK pathway by western blot. β-actin serves as the reference protein band, and the darkness of the target band indicates high expression of the target protein in this sample.
GRB2 promotes the proliferation, migration and invasion of HER2-positive breast cancer cells and inhibits their apoptosis in vitro
After the successful generation of stable strains, CCK-8 and flow cytometry assays were used to detect the proliferation and apoptosis ability of the tumor cells. After silencing GRB2, the proliferation rate of the tumor cells decreased (Fig. 2C and Supplemental Table 3) compare to the LV-HER2 and the LV-HER2 + LV-sh-NC group, and the apoptosis rate of the tumor cells significantly increased (Fig. 2D and Supplemental Table 3). The migration and invasion abilities of the tumor cells were detected via the Transwell assay (Fig. 2E, F, and Supplemental Table 3),the number of blue staining cells was significantly reduced following GRB2 silencing.Moreover, the expression levels of p-MEK1/2, p-ERK1/2, and Ras proteins were greater in the LV-HER2 group (Fig. 2G, Supplemental Table 4 ), and the expression levels were significantlyreduced following GRB2 silencing. In the LV-HER2 group and the LV-HER2 + LV-sh-NC group, tumor cell migration and invasion increased, whereas these abilities decreased following GRB2 silencing.
Silencing GRB2 reduces the tumorigenicity of HER2-positive breast cancer cells in vivo
In Model 1 (Fig. 3A), changes in body weight and tumor volume were recorded (Supplemental Fig. 3A), and fluorescence imaging was performed (Fig. 3B). After the mice were euthanized by means of isoflurane anesthesia, the tumors were removed (Fig. 3C). As shown in the LV-HER2 + LV- shRNA-GRB2 group, both the weight and volume of the isolated tumors were significantly reduced (Supplemental Fig. 3B), the fluorescence intensity clearly decreased, the apoptosis rate of the tumor cells was increased, as shown by the TUNEL assay (Supplemental Fig. 3C); the negative IR (inhibition rate) was significantly reduced in this group (Supplemental Fig. 3D), which indicated that tumor promotion was weakened (Supplemental Table 4) after the silencing of GRB2.
GRB2 promotes HER2-positive tumor cells to cross the blood brain barrier via the Ras/MAPK pathway in vivo. (A) In situ injection model (model 1). (B) Fluorescence intensity analysis using an in vivo animal imaging system.areas of high fluorescence intensity indicate active tumor sites or metastases. (C) Measurement of ex vivo tumor size and characteristics. (D) Results of Hematoxylin & Eosin (H&E) staining (×400), TUNEL assay, and Immunohistochemistry (IHC). H&E staining: Blue-purple represents nuclear staining, highlighting cell nuclei for morphological analysis. TUNEL assay: Yellow staining indicates apoptotic cells, reflecting DNA fragmentation during apoptosis. Immunohistochemistry: Brown represents a moderate positive signal for the target protein, while yellow indicates a weak positive signal, demonstrating varying levels of target protein expression. (E) H&E staining (×400) of Metastatic sites in the Brain, Lung, and Liver. (F) H-Score evaluation of GRB2 and HER2 protein expression levels. (G) Expression levels of key proteins in the Ras/MAPK pathway by western blot. (H) Western blot analysis of key proteins in the Ras/MAPK pathway.
The GRB2 and HER2 proteins synergistically promote tumor proliferation and inhibit tumor apoptosis in vivo
The majority of GRB2 protein expression was detected in the cytoplasm of tumor tissues via IHC (Fig. 3D), and positive cells were scattered in a single, dispersed distribution that was dark brown. The HER2 protein was also expressed in the cytoplasm, and the positive cells were widely distributed in lamellar or focal forms, which were light yellow. HE staining revealedthat tumor cells were present in all the groups (Fig. 3D-E), but the LV-HER2 group and the LV-HER2 + LV-sh-NC group had more pathological nuclear division. TUNEL staining revealed that the apoptosis rate did not significantly differ among the control group, the LV-NC group, and the LV-HER2 + LV-shRNA-GRB2 group, but the apoptosis rate was lower in the LV-HER2 group (Supplemental Fig. 3C). The H-scores for GRB2 and HER2 were equal in both groups, and with the silencing of GRB2, the H-score for HER2 decreased in the LV-HER2 + LV-shRNA-GRB2 group (Fig. 3F). As shown in supplemental Table 5, both GRB2 and HER2 were downregulated in the LV-HER2 + LV-shRNA- GRB2 group compared with the control group. The results of HE, IHC, TUNEL, and WB analyses revealed that the GRB2 and HER2 proteins work together to prevent tumor cells from dying and promote their growth.
Tumor cells can penetrate the blood‒brain barrier and cause metastasis
As required by ethics, the mice were sacrificed after 4 weeks. In Model 1, tumor cells were found in mouse lung tissue from the LV-HER2 + LV-shRNA-GRB2 group (Fig. 3E), and the other three groups presented signs of inflammation from neutrophils, eosinophils, and lymphocytes. In all the groups, tumor cells were found in their liver tissues. In the brain tissue, there was no clear tumor growth, but some neuronal cell edema and a low degree of microglial infiltration in the interstitium, especially more brain cell edema and inflammation, were observed in the LV-HER2 group. In Model 2 (Fig. 4A), tumor cells were found easily in the brain, liver, and lung tissues (Fig. 4B). These findings indicate that tumor cells can not only spread from breast tissues to the liver, lungs, and brain but also cross the BBB.
GRB2 and HER2 synergistically regulate the phosphorylation of key proteins in the Ras/MAPK pathway and promote the metastasis of breast cancer cells in vitro and in vivo
The expression levels of p-MEK1/2, p-ERK1/2, and Ras proteins were greater in the LV-HER2 group (Fig. 3G-H, Supplemental Table 5), and these levels were reduced after GRB2 silencing; the levels of MEK1/2 and ERK1/2 were not significantly different among all the groups after GRB2 silencing, which was in line with the results of the cell assay (Fig. 2G, Supplemental Table 4).
GRB2 binds and regulates the alternative splicing of KITLG in the Ras/MAPK pathway to promote the metastasis of HER2-positive breast cancer cells
According to the RNA-seq data, GRB2 knockdown led to related transcriptional changes (Fig. 5A): the expression of 5 Ras/MAPK pathway-related genes, FGFR3, EGF, FGFR4, PLA2G12A, and PLA2G4A was upregulated, and the expression of DUSP5, DUSP4, TGFB2, GRB2, TNF, RIN1, and PLA2G2F was downregulated. High-throughput transcriptome sequencing revealed 15 Ras/MAPK pathway-related genes with alternative splicing events in GRB2-knockdown cells. MAP3K3, CACNA1C, TRADD, GRB2, FLT3LG, DUSP8, MAP2K5, IL1RAP, PLA2G4B, KITLG, TAOK2, MAPK7, FOXO4, RASAL3, and GRIN1 are some of the genes involved (Fig. 5B). We performed an overlap analysis of genes whose expression was up- or downregulated (Fig. 5C) and the results of FRP-Seq (Fig. 5D) revealed that GRB2 and HER2 could work together to regulate the phosphorylation process of the Ras/MAPK pathway through EGF (epidermal growth factor) (Fig. 5E). Overlapping analysis of genes with alternative splicing and fRIP-seq data (Fig. 5F) revealed that GRB2 may regulate the alternative splicing of KITLG in the Ras/MAPK pathway.
Overlap analysis of RNA sequencing and fRIP-Seq results. (A) Heatmap visualization of GRB2 knockdown-induced up- or down-regulation of 5 Ras/MAPK pathway-related genes. (B) Heatmap reveals 16 Ras/MAPK signaling pathway-related genes with alternative splicing changes in GRB2 knockdown cells. (C) Venn diagram shows 1 overlapped gene between 11 Ras/MAPK pathway-related DEGs and 3196 GRB2-bound peaks genes from fRIP-seq data. (D) Venn diagram shows 1 overlapped gene between 16 Ras/MAPK pathway-related RASE and 40,709 GRB2-bound peaks genes from fRIP-seq data. (E) GRB2-binding peak genes of MESD. (F) GRB2-binding peak genes of KITLG.
Discussion
Many studies have demonstrated that HER2-positive is associated with a worse prognosis and increases the risk of BCBMs14,20,21. The clinical data from Xinjiang Tumor Hospital also confirmed that the proportion of the HER2-positive subtype was the highest (37.3%) among all BCBMs patients. GRB2, as a cross-protein of the ERBB and Ras/MAPK pathways and an RBP (RNA-binding protein) that possesses one SH2 domain and two SH3 domains15, mostly contributes to the synergistic procarcinogenic effect by initiating the phosphorylation of ERK1/2 and MEK1/2 or the alternative splicing process of KITLG in the Ras/MAPK pathway. Our research also revealed that GRB2 and HER2 might function cooperatively to promote the proliferation, invasion, and metastasis of breast cancer cells while suppressing their apoptosis.
In this study, we observed an intriguing finding: tumor cells were able to cross the blood‒brain barrier (BBB) in both the in situ injection model (Model 1) and the direct cerebral injection model (Model 2). Experiments with Model 1 demonstrated how HER2-positive cells spread from the breast tissue to the liver and lungs, followed by the observation of glial cells in brain tissues. Experiments with Model 2 revealed that tumor cells may not only proliferate in the brain but also reversibly cross the BBB and colonize the liver and lungs at a faster rate. Initially, we utilized human-derived tumor cells, which have a faster colonization rate in vivo or intracranially in mice, demonstrating that HER2-positive cells are highly malignant and easily proliferate in mice, whereas GRB2 overexpression can increase this ability. Second, we discovered that organ-specific colonization patterns22, such as liver and lung metastasis23, were present in both Model 1 and Model 2.While the brain metastasis pattern (brain-specific dissemination with minimal liver or lung involvement) remains statistically uncommon, our clinical observations identified a distinct cohort of HER2-positive patients presenting with isolated brain metastasis at initial diagnosis, notably devoid of detectable liver or lung lesions. Intriguingly, this subgroup demonstrated exceptional responsiveness to anti-HER2 targeted therapies, contrasting sharply with the typically poorer outcomes associated with visceral metastasis .Without timely treatment, the disease can deteriorate rapidly.Although brain metastasis may ultimately manifest in visceral organs (e.g., liver or lung), the latency period for brain-to-viscera dissemination is significantly prolonged compared to the reverse metastatic trajectory (viscera-to-brain) when targeted therapy exposure, potentially reflecting differential blood-brain barrier permeability and organ-specific metastatic niche compatibility. Furthermore, Stephen Paget’s “seed/soil hypothesis” suggests that the metastatic site is not random but rather architectural24,25; metastatic “organophilia” is influenced by the organ’s intrinsic structure, the cancer itself, and the organ’s specific microenvironment22. We found that the intracranial environment is favorable for the growth of tumor cells2,26, and we assume that Model 2’s pattern is comparable to that of gliomas27,28,29. Fourth, the metastatic cascade likely acts as a protective mechanism against metastasis, necessitating that tumor cells reach a sufficient concentration30 and virulence in vivo to overcome the cascade process. We also considered that as long as HER2 is not completely eliminated, GRB2 can assist the remaining “elite HER2 cells” in recovery31; therefore, anti-HER2 therapy32 should be followed by anti-GRB2 targeted therapy, but the current lack of clinically validated anti-GRB2 agents represents a critical translational gap in therapeutic development.
The greater degree of cell swelling and glial cell infiltration in the LV-HER2 group suggests that the BBB does not prevent HER2 and GRB2 from entering the brain, which is supported by the inflammatory response33,34. Alzheimer’s disease is characterized by chronic inflammation in the brain, and microglia contribute to the accumulation of amyloid β, which can trigger STAT3 activation on astrocytes, possibly resulting in acute injuries that promote brain metastasis35. Therefore, some inflammatory mediators may cause the immune system36 to react in a strange way and assist in tumor metastasis.
The BBB is a highly selective membrane that is composed mostly of glial cells and endothelial cells37,38. GRB2 and HER2 are both large molecules with molecular weights of 50.48 and 94.78 kDa, respectively, and are likely to penetrate the BBB via transcytosis39,40. Tumors developed faster in Model 2 than in Model 1, leading us to hypothesize that the BBB is a scale-like structure and that it is difficult to break through the barrier from the outside, but it is much easier for these cells to move from the inside of the brain to other sites via expansive growth.GLUT1 is highly expressed in blood-brain barrier endothelial cells, accounting for 10–15% of total proteins in cerebral capillaries.Brain metastatic tumor cells enhance glycolytic capacity through GLUT1 overexpression41, Ras/MAPK signaling can induce transcription factors that upregulate GLUT1 expression, the relationship between GLUT1 and GRB2 may play a role in the development of brain metastasis in HER2-positive breast cancer, which warrants further investigation in subsequent studies.
Finally, the mechanisms by which GRB2 and HER2 regulate selected proteins in the Ras/MAPK pathway were discovered. In particular, we found evidence that GRB2 aids in the movement of HER2-positive cells across the BBB, which leads to the spread of lesions inside and outside the brain. The treatment of HER2-positive breast cancer with a GRB2-targeting agent will be investigated in future studies. Although this study provides new insights into the mechanisms of brain metastasis in HER2-positive breast cancer, several limitations remain. First, the expression level of GRB2 in this study did not reach a significant level, which may limit its direct applicability as a therapeutic target. Second, GRB2 functions primarily indirectly, and its specific mechanisms have not yet been fully elucidated. Therefore, the current findings are insufficient to alter the clinical treatment strategies for brain metastasis. However, the potential importance of GRB2 in disease progression cannot be overlooked. Future research should further explore the interactions between GRB2 and downstream signaling proteins, as well as its role in other related pathways. For example, functional experiments could be conducted to validate the specific role of GRB2 in the ERBB2 pathway, or multiomics data could be integrated to comprehensively dissect its regulatory network. These studies help clarify the precise role of GRB2 in breast cancer brain metastasis and provide a theoretical foundation for the development of new therapeutic strategies.
Materials and methods
Clinical data collection
A total of 18,176 breast cancer patients who presented at Xinjiang Tumor Hospital between January 2010 and December 2020, including 353 patients who developed BMs, were included in the study. Age, tumor size, lymph node status, histological grade, vascular status, nerve invasion status, Ki-67 status, HER2 status, PR status, ER status, time of brain metastasis, and time of last follow-up were recorded at the first visit.
Bioinformatics analysis
The GEO dataset GSE43837 provide insight into the molecular basis of HER2-positive breast cancer outgrowth in the brain, which was used to compare the gene expression profiles of 19 HER2-positive human breast cancer brain metastasis and 19 HER2-positive nonmetastatic human breast tumors. Differentially expressed genes (DEGs) were screened with P < 0.05, LogFc≤-1.5, and LogFc ≥ 1.5 as the criteria. GO: BP pathway enrichment analysis was performed for the DEGs. STRING was subsequently used to analyze the protein‒protein interactions (PPIs) of the selected proteins. GEPIA2 was used to assess the expression and prognostic value of the target proteins.
Cell lines and cell culture
SK-BR3, MCF-7, and MDA-MB-231 cells were obtained from Procell Corporation in China. The cell line was cultured at 37 °C with 5% CO2, penicillin (1% 10 kU/ mL) / streptomycin (10 mg/mL) (PS, Procell, China), and 10% FBS (fetal bovine serum, Gibco, USA). The medium used for MDA-MB-231 cells was DMEM (high sugar, Gibco, USA), that used for SK-BR3 cells was McCoy’s 5 A (Gibco, USA), and that used for MCF-7 cells was MEM (Gibco, USA).
Plasmid construction and generation of stable cell lines
Four groups of MDA-MB-231 cells were used for the analyses: the control group: MDA-MB-231 cells cultured normally; LV-NC group: cells transfected with GV654 negative control lentivirus; the LV-HER2 group: cells transfected with the LV-HER2-positive lentivirus; The LV-HER2 + LV-sh-NC group: cells transfected with both the LV- HER2-positive lentivirus and GV654 negative control lentivirus; and the LV-HER2 + LV-shRNA-GRB2 group: cells transfected with both the LV-HER2-positive lentivirus and the LV-shRNA-GRB2-silenced lentivirus. Cells for the GRB2 silencing experiments were.
divided into three groups and transfected with the following shRNAs: LV-shRNA-GRB2-69: ccggCAGATATTCCTGCGGGACATActcgagTATGTCCCGCAGGAATATCTGtttttg; LV-shRNA-GRB2-70: ccggCGGCTTCATTCCCAAGAACTActcgagTAGTTCTTGGGAATGAAGCCGtttttg; LV-shRNA-GRB2-71: ccggGTCCAGGCCCTCTTTGACTTTctcgagAAAGTCAAAGAGGGCCTGGACtttttg. For siRNA experiments, the sequence of siNegative, a nontargeting control siRNA, was 5’-UUCUCCGAACGUGUCACGUTT-3’ (sense), and the sequence of the siRNA that targets GRB2 (siGRB2) was 5’-GGUGGAUUAUCACAGA UCUTT − 3’ (sense).
RNA extraction and qRT‒PCR
After 72 h, stable strains were generated, and the medium was removed, and 1 mL of TRIZOL was added to digest the cells. The culture flasks wereshaken until the cells were digested, and the total RNA was collected in 1.5 mL EP tubes. Tomeasure the levels of the GRB2 and HER2 genes via qRT‒PCR, 10 µl of BlasTaq 2×qPCR Master Mix, 0.5 µl of forward primer, 2 µl of cDNA, 0.5 µl of reverse primer, and up to 20 µl of RNase-free water were used for redenaturation (3 min, 95 °C, 1 cycle), denaturation (10 s, 95 °C, 40 cycles), and annealing/extension (30 s, 60 °C, 40 cycles). The primers used were as follows: GRB2-F, ATTCCTGCGGGACATAGA ACA; GRB2-R, GGTGACATAATTGCGGGGAAAC; HER2-F: TGTGACTGCCTGTCCCTACAA, HER2-R: CCAGACCATAGCACACACTCGGG.
Western blot analysis
After the stable strain was constructed, we collected 1 × 106 cells, rinsed them with cold PBS, centrifuged them, and removed the supernatant. Then, 100 µL of RIPA lysis buffer was added to the precipitate. BCA was used to determine the protein concentration, and the expression of Ras, MEK1/2, ERK1/2, GRB2, HER2, p-MEK1/2, and p-ERK1/2 was determined by western blot.
CCK-8 assay
Cells from each group were mixed with a cell suspension of 5 × 104 cells/mL and injected into a 96-well plate (100 µL per well, 5 duplicate wells). After 72 h, the medium was removed, 100 µL of 10% CCK-8 solution was added to each well, and the OD was measured at 450 nm with an enzyme marker 1 h later. This technique was designed to measure cell proliferation.
Flow cytometry assay
The culture medium from each set of cell samples was transferred to a centrifuge tube, which contained cells in suspension that had undergone apoptosis or necrosis. Adherent cells were washed twice with PBS, trypsinized, transferred to a tube and spun at 1000 rpm for 5 min, after which the supernatant was discarded. To resuspend the cells, 500 µL of 1× binding buffer was added, and the mixture was passed through a 200 mesh filter. Next, 10 µL of 7-AAD and 5 µL of Annexin were added to each tube, mixed slightly, and stored at 4 °C for 10 min in the dark. This procedure was completed in 30 min.
Transwell assay
The lower chamber of the transwell system (the bottom of the 24-well plate) was filled with 600 µL of medium, and the upper chamber was filled with 100 µL of cell suspension. For each group, three replicate wells were used, and the samples were incubated for 72 h. The small chambers were removed with tweezers and fixed in 4% formaldehyde at 25 °C for 20 min. After the upper chamber was stained with Giemsa, the cells were wiped away with a wet cotton swab, and cell migration was observed under a microscope.
Animal model
Forty-four SPF-grade NOD/SCID female mice weighing 18–20 g and aged 4–6 weeks were obtained from SiPeiFu Biotechnology Co. Ltd. (Suzhou, China). The mice were maintained at 22 ± 2 °C with a humidity of 60%~80% and provided free access to food and water for 1 week in an SPF animal laboratory (Hangzhou, China). The cells were prepared as single-cell suspensions, adjusted to 5*107/ml, and then injected with 200 µl (containing 5*106 cells and 100 µl of matrix gel) under the 4th mammary gland (Model 1) in the control, LV-NC, LV- HER2, and LV-HER2 + LV-shRNA-GRB2 groups (32 mice were randomly allocated into 4 groups, n = 8), and the tumor size was recorded every 3 days. In addition, the cells were prepared as single-cell suspensions, adjusted to 1*104/ml, and then injected directly into the brains (Model 2, bregma, 3 mm deep, right 2.5 mm lateral) of the LV-NC, LV-HER2, and LV-HER2 + LV-shRNA-GRB2 groups (12 mice were randomly allocated into 3 groups, n = 4). Mice were anesthetized with a mixture of 4–5% isoflurane (oxygen flow rate: 1–2 L/min) and subsequently euthanized by exsanguination.All animal experiments were conducted in the SPF animal laboratory of the Laboratory Animal Center of Hangzhou Medical College (SYXK, Zhejiang, 2019-0011).All experiments were performed with independent biological replicates .
Fluorescence in vivo imaging
After the two models were constructed, the remaining three groups received intraperitoneal injections of D-luciferin (100 µL) and 150 mg/kg luciferase substrate between weeks 4 and 5, and the whole body was subjected to data acquisition using an imaging system 10 min later. Then, the software of the imaging system was used to quantify the fluorescence intensity.
TUNEL assays
Antigenically fixed tissue slices were dewaxed and immersed in 3% H2O2 for 10 min before being digested with Proteinase K. Then, 1 µL of TdT and DIG-d-UTP was added to the buffer, the buffer solution was mixed thoroughly and added toot the samples in in a moist box; the samples were labeled for 2 h at 37 °C. The biotinylated anti-digoxin antibody and SABC were diluted 1:100 and incubated with the samples at 37 °C for 30 min. To assess apoptosis with fluorescence microscopy, the cells were stained with DAB (brown granular precipitate) for 3–10 min and then with hematoxylin for 3 min. The number of TUNEL-stained cells and the intensity of staining were also determined. The apoptosis rate was calculated as the number of TUNEL-positive cells/total number of cells × 100%.
HE staining and immunohistochemistry (IHC)
To preserve the brain, liver, lung, and breast cancer tissues from the mice, they were placed in a 10% formalin solution, wax blocks were made, and the samples were cut into 4 μm slices. The slices were then stained with Hematoxylin & Eosin (H&E) staining. To fix the antigens, the slices were dewaxed with xylene and then placed in 0.01 M citrate buffer for 10 min. After adding 50 µL of horseradish peroxidase-labeled secondary antibody, the slices were incubated for 20 min at room temperature. The sections were stained with DAB (a brown granular precipitate) for 3–10 min, after which the tissue sections were restained with hematoxylin for 3 min. We then examined the intensity and range of staining in the slices using a light microscopy. The antibodies used were a rabbit anti-GRB2 antibody (Bioss, China) and a rabbit anti-ERBB2 antibody (BOSTER, China). IHC was used to detect GRB2 and HER2 expression, and the H-scores were calculated. The total positive staining score was calculated by adding the sum of the staining strength score and the positive cell percentage score. Scores between 0 and 7 indicated low expression, and scores between 8 and 12 indicated high expression.
RNA sequencing and fRIP-seq
Following the downregulation of GRB2 in SK-BR3 cells by siGRB2 (5’-GGUGGAUUAUCAC AGAUCUTT-3’ ),high-throughput transcriptome sequencing was carried out to collect transcriptome data, the libraries for sequencing were generated and applied by the Illumina Novaseq Xplus instrument for 150nt paired-end sequencing. The date generated through functional RNA immunoprecipitation sequencing (fRIP-seq) and corresponding protein complex characterization via co-immunoprecipitation (Co-IP) are publicly available under GEO accession numbers GSE276962 and GSE276963. We studied how GRB2 binds to mRNAs and alternative splicing events at the molecular level in SK-BR3 cells by RNA sequencing and fRIP-seq methods.
Statistical analysis
All the data are expressed as M ± SD, and the statistical analysis was performed using SPSS 19.0 software. One-way analysis of variance was used, and P < 0.05 was considered to indicate statistical significance.
Data availability
The high-throughput transcriptome sequencing datasets mentioned in this study have been deposited in the Gene Expression Omnibus (GEO) repository under accession numbers GSE276962 and GSE276963.These datasets are available for research purposes upon formal request to the corresponding author.GSE43837 is derived from McMullin RP and is available from https:// www.ncbi. nlm.nih.gov/ geo/query/acc.cgi? acc=GSE43837.
References
Kingwell, K. Metabolic target for brain metastasis. Nat. Rev. Drug Discov. 20 (6), 426 (2021).
Wang, Y., Ye, F., Liang, Y. & Yang, Q. Breast cancer brain metastasis: insight into molecular mechanisms and therapeutic strategies. Br. J. Cancer. 125 (8), 1056–1067 (2021).
Corti, C. et al. Targeting brain metastases in breast cancer. Cancer Treat. Rev. 103, 102324 (2022).
Faure, C. et al. Allosteric Inhibition of HER2 by Moesin-Mimicking compounds targets HER2-Positive cancers and brain metastases. Cancer Res. 81 (21), 5464–5476 (2021).
Kuksis, M. et al. The incidence of brain metastases among patients with metastatic breast cancer: a systematic review and meta-analysis. Neuro Oncol. 23 (6), 894–904 (2021).
Boire, A., Brastianos, P. K., Garzia, L. & Valiente, M. Brain metastasis. Nat. Rev. Cancer. 20 (1), 4–11 (2020).
Hurvitz, S. A. et al. Trastuzumab Deruxtecan versus trastuzumab emtansine in patients with HER2-positive metastatic breast cancer: updated results from DESTINY-Breast03, a randomised, open-label, phase 3 trial. Lancet 401 (10371), 105–117 (2023).
Yan, M. et al. Pyrotinib plus capecitabine for patients with human epidermal growth factor receptor 2-positive breast cancer and brain metastases (PERMEATE): a multicentre, single-arm, two-cohort, phase 2 trial. Lancet Oncol. 23 (3), 353–361 (2022).
Curigliano, G. et al. Tucatinib versus placebo added to trastuzumab and capecitabine for patients with pretreated HER2 + metastatic breast cancer with and without brain metastases (HER2CLIMB): final overall survival analysis. Ann. Oncol. 33 (3), 321–329 (2022).
Anwar, M. et al. Pyrotinib treatment in patients with HER2-positive metastatic breast Cancer and brain metastasis: exploratory final analysis of Real-World, multicenter data. Clin. Cancer Res. 27 (16), 4634–4641 (2021).
Bartsch, R. et al. Trastuzumab Deruxtecan in HER2-positive breast cancer with brain metastases: a single-arm, phase 2 trial. Nat. Med. 28 (9), 1840–1847 (2022).
Kumagai, S., Koyama, S. & Nishikawa, H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer. 21 (3), 181–197 (2021).
Nagpal, A. et al. Neoadjuvant neratinib promotes ferroptosis and inhibits brain metastasis in a novel syngeneic model of spontaneous HER2(+ ve) breast cancer metastasis. Breast Cancer Res. 21 (1), 94 (2019).
He, Y. et al. Brain metastasis in de Novo stage IV breast cancer. Breast 71, 54–59 (2023).
Gril, B. et al. Grb2-SH3 ligand inhibits the growth of HER2 + cancer cells and has antitumor effects in human cancer xenografts alone and in combination with docetaxel. Int. J. Cancer. 121 (2), 407–415 (2007).
Lv, J. et al. Deubiquitinase PSMD14 enhances hepatocellular carcinoma growth and metastasis by stabilizing GRB2. Cancer Lett. 469, 22–34 (2020).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53 (D1), D672–D677 (2025).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28 (11), 1947–1951 (2019).
Liang, K. et al. Clinical features and outcomes of advanced HER2 + esophageal/GEJ cancer with brain metastasis. ESMO Open. 9 (1), 102199 (2024).
Karakaya, S., Karadag, I., Ates, O., Cakmak Oksuzoglu, O. B. & Demirci, U. Clinical outcomes and prognostic factors in HER-2 positive breast Cancer with brain metastasis: A Single-centre experience. J. Coll. Physicians Surg. Pak. 31 (2), 166–170 (2021).
Azubuike, U. F. & Tanner, K. Biophysical determinants of cancer organotropism. Trends Cancer. 9 (3), 188–197 (2023).
Kennecke, H. et al. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 28 (20), 3271–3277 (2010).
Fidler, I. J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 3 (6), 453–458 (2003).
Hu, M., Kenific, C. M., Boudreau, N. & Lyden, D. Tumor-derived nanoseeds condition the soil for metastatic organotropism. Semin Cancer Biol. 93, 70–82 (2023).
Gao, Y. et al. Metastasis organotropism: redefining the congenial soil. Dev. Cell. 49 (3), 375–391 (2019).
Yuzhalin, A. E. & Yu, D. Brain metastasis organotropism. Cold Spring Harb Perspect. Med. 10 (5), a037242 (2020).
Chen, W., Hoffmann, A. D., Liu, H. & Liu, X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis Oncol. 2 (1), 4 (2018).
He, K. et al. Metastasis organotropism in colorectal cancer: advancing toward innovative therapies. J. Transl Med. 21 (1), 612 (2023).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501 (7467), 338–345 (2013).
Xu, D. et al. Why does HER2-positive breast cancer metastasize to the brain and what can we do about it.
de Bono, J. S. & Rowinsky, E. K. The erbb receptor family: a therapeutic target for cancer. Trends Mol. Med. 8 (4 Suppl), S19–26 (2002).
Mamun, A. A. et al. Inflammation-targeted nanomedicine against brain cancer: from design strategies to future developments. Semin Cancer Biol. 86 (Pt 2), 101–116 (2022).
Adler, O. et al. Reciprocal interactions between innate immune cells and astrocytes facilitate neuroinflammation and brain metastasis via lipocalin-2. Nat. Cancer. 4 (3), 401–418 (2023).
Kleffman, K. et al. Melanoma-Secreted amyloid Beta suppresses neuroinflammation and promotes brain metastasis. Cancer Discov. 12 (5), 1314–1335 (2022).
Evans, K. T. et al. Microglia promote anti-tumour immunity and suppress breast cancer brain metastasis. Nat. Cell. Biol. 25 (12), 1848–1859 (2023).
Ahn, S. I. et al. Microengineered human blood-brain barrier platform for Understanding nanoparticle transport mechanisms. Nat. Commun. 11 (1), 175 (2020).
Profaci, C. P., Munji, R. N., Pulido, R. S. & Daneman, R. The blood-brain barrier in health and disease: important unanswered questions. J. Exp. Med. 217 (4), e20190062 (2020).
Xie, J., Shen, Z., Anraku, Y., Kataoka, K. & Chen, X. Nanomaterial-based blood-brain-barrier (BBB) crossing strategies. Biomaterials 224, 119491 (2019).
Sprowls, S. A. et al. Improving CNS delivery to brain metastases by Blood-Tumor barrier disruption. Trends Cancer. 5 (8), 495–505 (2019).
Johnston, S. N. et al. GLUT1 is redundant in hypoxic and glycolytic nucleus pulposus cells of the intervertebral disc. JCI Insight. 8 (8), e164883 (2023).
Funding
The research is supported by the Tianchi Talents - Young Doctor Project of Xinjiang Uygur Autonomous Region (2022TCYCLHY), the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2021D01C411),the Guiding Funds of Central Government for Supporting the Development of the Local Science and Technology(ZYYD2022), and the Tianshan Talents-Leading Talents in Science and Technology Innovation (2023TSLJ0038).
Author information
Authors and Affiliations
Contributions
Hongyu Li proposed the ideas and drafted the manuscript, Yalin Zhang and Xiao Han collaborated on the design and execution of the experiment, Bingyu Li and Dan Liu jointly made the figures and tables, Gang Sun supervised the procedure and performed the final modifications.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
All experiments involving human participants and/or human tissue samples were conducted in accordance with relevant guidelines and regulations, including the Declaration of Helsinki. All experimental protocols were reviewed and approved by the Ethics Committee of the Affiliated Cancer Hospital of Xinjiang Medical University (Approval ID: K-2024018).Informed consent was obtained from all participants and/or their legal guardian(s) prior to their inclusion in the study.All experiments involving live vertebrates were approved by the Animal Ethics Committee of Newlong Youshu Technology (Hangzhou) Co., Ltd (Approval ID: YS-m202501001). All procedures were performed in accordance with the Chinese Guidelines for Ethical Review of Laboratory Animal Welfare (GB/T 35892 − 2018) and international ethical guidelines for animal research. This study adheres to the ARRIVE guidelines 2.0 (https://arriveguidelines.org) to ensure comprehensive reporting of experimental methodology. Efforts were made to minimize animal suffering and reduce the number of animals used, following the principles of Replacement, Reduction, and Refinement (3Rs).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, H., Zhang, Y., Han, X. et al. GRB2 promotes brain metastasis in HER2-positive breast cancer by regulating the Ras/MAPK pathway. Sci Rep 15, 14736 (2025). https://doi.org/10.1038/s41598-025-99685-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-99685-3
Keywords
This article is cited by
-
Robust SH2 binding affinity indicates minimal SH3-to-SH2 communication in Grb2
Biology Direct (2026)