Abstract
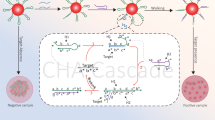
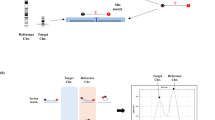
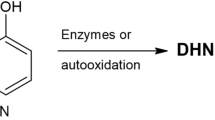
Melanin is a potent inhibitor of PCR, which interferes with forensic DNA typing by binding to Taq polymerase and disrupting its function, leading to allele dropout and decreased peak heights. Traditional mitigation measures, such as dilution and replacement of the polymerase, often result in the loss of DNA and are not suitable for low-template forensic samples. In this study, melanin-induced inhibition was explored using molecular docking and dynamics simulation, which identified stable interaction with catalytic residues TYR671 and PHE667 (Kd = 31.76 ± 0.02 µM), thereby interfering with polymerase function. STR typing of the inhibitor-treated sample showed a total allelic loss of SE33 and Penta E, dropout at D12S391, and substantially reduced peak heights. The overall peak height was 443,409.3 RFU. Facilitating methods using AuNPs, BSA-coated AuNPs, and standard BSA were further compared. AuNPs provided partial restoration (TPH = 545,605.7 RFU), whereas BSA-coated AuNPs provided improved restoration (TPH = 682,938.3 RFU) with harmonious heterozygous peaks (mean PHR = 0.90). Standard BSA had the highest restoration (TPH = 786,122.7 RFU; mean PHR = 0.92), restored alleles between dye channels. Statistical analysis revealed significant enhancement by BSA (p < 0.0001) and moderate enhancement by BSA-coated AuNPs (p = 0.0186). The BSA yielded optimal results, but it also exhibited larger sample differences. On the other hand, BSA-coated AuNPs offered more consistent facilitation at a very low concentration compared to BSA. This study thus explains the mode of PCR inhibition by melanin. It further demonstrates that BSA remains the most effective facilitator, but it also has its own limitations. In contrast, BSA-coated AuNPs offer a reliable nanotechnology-based method to overcome this limitation in forensic PCR applications.
Similar content being viewed by others
Data availability
I declare that the authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper. The molecular dynamics simulation movies used in this manuscript have been uploaded to the public data repository Zenodo to ensure transparency and accessibility, as per FAIR data principles. The Zenodo DOI is: https://doi.org/10.5281/zenodo.13933345. These movies are not part of any previously published or pending manuscripts. They serve as supporting data for the molecular interaction insights discussed in this submission and are being referenced here for the first time in a peer-reviewed publication. The data sharing on Zenodo is intended solely to comply with data availability standards and does not constitute prior or dual publication.
References
Mullis, K. B. The unusual origin of the polymerase chain reaction. Sci. Am. 262(4), 56–65 (1990).
Galluzzi, L. et al. Real-time PCR applications for diagnosis of leishmaniasis. Parasit. Vectors 11, 1–13 (2018).
Matsuda, K. PCR-based detection methods for single-nucleotide polymorphism or mutation: Real-time PCR and its substantial contribution toward technological refinement. Adv. Clin. Chem. 80, 45–72 (2017).
Serrano-Cumplido, A. et al. Application of the PCR number of cycle threshold value (Ct) in COVID-19. SEMERGEN 47(5), 337–341 (2021).
Shahi, S. et al. Polymerase chain reaction (PCR)-based methods: Promising molecular tools in dentistry. Int. J. Biol. Macromol. 117, 983–992 (2018).
Mirmajlessi, S. M. et al. Real-time PCR applied to study on plant pathogens: Potential applications in diagnosis-a review. Plant Prot. Sci. 51(4), 177–190 (2015).
Petralia, S. & Conoci, S. PCR technologies for point of care testing: Progress and perspectives. ACS Sen. 2(7), 876–891 (2017).
McDonald, C., Taylor, D. & Linacre, A. PCR in forensic science: A critical review. Genes 15(4), 438 (2024).
Weusten, J. & Herbergs, J. A stochastic model of the processes in PCR based amplification of STR DNA in forensic applications. Forensic Sci. Int. Genet. 6(1), 17–25 (2012).
Zhu, H. et al. PCR past, present and future. Biotechniques 69(4), 317–325 (2020).
Singh, J. et al. A critical review on PCR, its types and applications. Int. J. Adv. Res. Biol. Sci. 1(7), 65–80 (2014).
Vajpayee, K. et al. PCR inhibitors and facilitators-their role in forensic DNA analysis. Forensic Sci. Int. 349, 111773 (2023).
Herrling, T., Jung, K. & Fuchs, J. The role of melanin as protector against free radicals in skin and its role as free radical indicator in hair. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 69(5), 1429–1435 (2008).
Guo, L. et al. Recent advances and progress on melanin: From source to application. Int. J. Mol. Sci. 24(5), 4360 (2023).
Bashkatov, A. N. et al. Estimate of the melanin content in human hairs by the inverse Monte-Carlo method using a system for digital image analysis. Quantum Electron. 36(12), 1111 (2006).
Santos Nogueira, A. C. & Joekes, I. Hair color changes and protein damage caused by ultraviolet radiation. J. Photochem. Photobiol. B Biol. 74(2), 109–117 (2004).
Cordero, R. J. & Casadevall, A. Melanin. Curr. Biol. 30(4), R142–R143 (2020).
Eckhart, L. et al. Melanin binds reversibly to thermostable DNA polymerase and inhibits its activity. Biochem. Biophys. Res. Commun. 271(3), 726–730 (2000).
Suenaga, E. & Nakamura, H. Evaluation of three methods for effective extraction of DNA from human hair. J. Chromatogr. B 820(1), 137–141 (2005).
Yoshii, T. et al. Water-soluble eumelanin as a PCR-inhibitor and a simple method for its removal. Nihon Hoigaku Zasshi 47(4), 323–329 (1993).
Thompson, R. E., Duncan, G. & McCord, B. R. An investigation of PCR inhibition using Plexor(®) -based quantitative PCR and short tandem repeat amplification. J. Forensic Sci 59(6), 1517–1529 (2014).
Silva Almeida Vicente, A. L. et al. Comparison of protocols for removal of melanin from genomic DNA to optimize PCR amplification of DNA purified from highly pigmented lesions. Histol. Histopathol. 34(9), 1089–1096 (2019).
Yoshii, T. et al. PCR inhibitor: Water-soluble melanin, which inhibits DNA polymerases and DNases. In Advances in Forensic Haemogenetics: 15th Congress of the International Society for Forensic Haemogenetics (Internationale Gesellschaft für forensische Hämogenetik eV), Venezia, 13–15 October 1993. Springer (1994).
Alaeddini, R. Forensic implications of PCR inhibition—A review. Forensic Sci. Int. Genet. 6(3), 297–305 (2012).
Sarna, T., Swartz, H. M. & Zadlo, A. Interaction of melanin with metal ions modulates their cytotoxic potential. Appl. Magn. Reson. 53(1), 105–121 (2022).
Solano, F. Melanins: Skin pigments and much more—types, structural models, biological functions, and formation routes. New J. Sci. 2014(1), 498276 (2014).
Vajpayee, K., Sagar, D. & Dash, H. R. Forensic DNA typing: Inception, methodology, and technical advancements. In Forensic DNA typing: Principles, applications and advancements 3–26 (Springer, 2020).
Wang, H. et al. A re-evaluation of dilution for eliminating PCR inhibition in soil DNA samples. Soil Biol. Biochem. 106, 109–118 (2017).
Kreader, C. A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62(3), 1102–1106 (1996).
Farell, E. M. & Alexandre, G. Bovine serum albumin further enhances the effects of organic solvents on increased yield of polymerase chain reaction of GC-rich templates. BMC. Res. Notes 5(1), 1–8 (2012).
Schriewer, A., Wehlmann, A. & Wuertz, S. Improving qPCR efficiency in environmental samples by selective removal of humic acids with DAX-8. J. Microbiol. Methods 85(1), 16–21 (2011).
Hamza, I. A. & Leifels, M. Assessment of PCR inhibitor removal methods to monitor viruses in environmental water samples: DAX-8 outperforms competitors. Water Air Soil Pollut. 235(1), 20 (2023).
Zhang, Y. et al. Bovine thrombin enhances the efficiency and specificity of polymerase chain reaction. Biotechniques 57(6), 289–294 (2014).
Strien, J., Sanft, J. & Mall, G. Enhancement of PCR amplification of moderate GC-containing and highly GC-rich DNA sequences. Mol. Biotechnol. 54(3), 1048–1054 (2013).
Vajpayee, K., Paida, V. & Shukla, R. K. Nanoparticle-assisted PCR: Fundamentals, mechanisms, and forensic implications. Int. J. Legal Med. 139(3), 945–964 (2025).
Tiwari, P. M. et al. Functionalized gold nanoparticles and their biomedical applications. Nanomaterials 1(1), 31–63 (2011).
Li, Y. et al. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 7(5), 1116–1123 (1998).
Eom, S. H., Wang, J. & Steitz, T. A. Structure of Taq polymerase with DNA at the polymerase active site. Nature 382(6588), 278–281 (1996).
Shi, X. et al. Spectroscopic investigation of the interactions between gold nanoparticles and bovine serum albumin. Chin. Sci. Bull. 57(10), 1109–1115 (2012).
Rossen, L. et al. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17(1), 37–45 (1992).
Schrader, C. et al. PCR inhibitors–occurrence, properties and removal. J. Appl. Microbiol. 113(5), 1014–1026 (2012).
Sidstedt, M. et al. The impact of common PCR inhibitors on forensic MPS analysis. Forensic Sci. Int. Genet. 40, 182–191 (2019).
McCord, B., Pionzio, A. & Thompson, R. Analysis of the effect of a variety of PCR inhibitors on the amplification of DNA using real time PCR, melt curves and STR analysis. The U.S. Department of Justice, Washington DC, Document No. 249148 (2014).
Roskoski, R. Modulation of enzyme activity. In xPharm: The comprehensive pharmacology reference (eds Enna, S. J. & Bylund, D. B.) 1–11 (Elsevier, 2007).
Funes-Huacca, M. E. et al. A comparison of the effects of PCR inhibition in quantitative PCR and forensic STR analysis. Electrophoresis 32(9), 1084–1089 (2011).
Sidstedt, M. et al. Humic substances cause fluorescence inhibition in real-time polymerase chain reaction. Anal. Biochem. 487, 30–37 (2015).
Dash, H. R. et al. Useful autosomal STR marker sets for forensic and paternity applications in the Central Indian population. Ann. Hum. Biol. 48(1), 37–48 (2021).
Kumar, A. et al. Genomic diversity at 22 STR loci (extended CODIS STR) in the population of Rajasthan, India. Gene Rep. 23, 101150 (2021).
Glock, B. et al. Additional variability at the D12S391 STR locus in an Austrian population sample: Sequencing data and allele distribution. Forensic Sci. Int. 90(3), 197–203 (1997).
Biedermann, A. & Kotsoglou, K. N. Forensic science and the principle of excluded middle: “Inconclusive” decisions and the structure of error rate studies. Forensic Sci. Int. Synergy 3, 100147 (2021).
Giambernardi, T. A., Rodeck, U. & Klebe, R. J. Bovine serum albumin reverses inhibition of RT-PCR by melanin. Biotechniques 25(4), 564–566 (1998).
Hu, Q. et al. A comparison of four methods for PCR inhibitor removal. Forensic Sci. Int. Genet. 16, 94–97 (2015).
NedumpullyGovindan, P., Monticelli, L. & Salonen, E. Mechanism of taq DNA polymerase inhibition by fullerene derivatives: Insight from computer simulations. J. Phys. Chem. B 116(35), 10676–10683 (2012).
Lu, C. et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 17(7), 4291–4300 (2021).
Janek, J. & Kolafa, J. Novel barostat implementation for molecular dynamics. J. Chem. Phys. 160(18), 184111 (2024).
Kimling, J. et al. Turkevich method for gold nanoparticle synthesis revisited. J. Phys. Chem. B 110(32), 15700–15707 (2006).
Shrivastava, P., Jain, T. & Kumawat, R. K. Direct PCR amplification from saliva sample using non-direct multiplex STR kits for forensic DNA typing. Sci. Rep. 11(1), 7112 (2021).
Hedman, J., Ansell, R. & Nordgaard, A. A ranking index for quality assessment of forensic DNA profiles forensic DNA profiles. BMC Res. Notes 3, 290 (2010).
Acknowledgements
The authors gratefully acknowledge the Council of Scientific and Industrial Research (CSIR), Government of India, for awarding the Senior Research Fellowship (SRF) to Kamayani Vajpayee and Shriyansh Srivastava. The authors also thank Ahmedabad University, Indian Institute of Technology Gandhinagar, and the Central University of Gujarat, Gandhinagar, for providing laboratory space, infrastructure, and research resources essential for the successful completion of this study. We also thank Professor Souvik Sengupta, Associate Professor, School of Arts and Sciences, Ahmedabad University for his critics and suggestions that has improved the work presented.
Author information
Authors and Affiliations
Contributions
K.V. conceptualized the study, curated data, performed formal analysis, conducted investigations, developed methodology, contributed to validation and visualisation, and prepared figure 1, 3, and 4, wrote the original draft. S.S. (Shriyansh Srivastava) contributed to data curation, formal analysis, investigation, methodology, and writing of the original draft. S.S. (Shivkant Sharma) was involved in data curation. S.G. contributed to data curation, formal analysis, prepared figure 2, methodology, and writing of the original draft. A.S. participated in conceptualisation, data curation, formal analysis, investigation, methodology, validation, visualisation, and original draft writing. V.P. contributed to data curation. H.R.D. contributed to methodology, validation, and writing—review and editing. A.P. contributed to formal analysis, validation, and writing—review and editing. R.K.S. was involved in conceptualisation, data curation, formal analysis, investigation, methodology, validation, visualisation, and writing—review and editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vajpayee, K., Srivastava, S., Sharma, S. et al. Mechanistic insights into melanin-induced PCR inhibition and its NanoPCR-based mitigation. Sci Rep (2026). https://doi.org/10.1038/s41598-026-35010-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-35010-w