Abstract
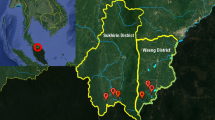
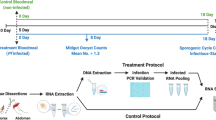
In Indonesia, 29 anopheline species are vectors of human malarias. Many of these morphologically indistinguishable vectors are in species complexes requiring molecular analysis to identify. Data on vectors in North Sumatra remain limited. This study investigated mosquitoes of the Maculatus Subgroup in Ujung Bandar Village, Langkat Regency, to identify them and their potential roles in malaria transmission. Mosquitoes collected via human landing catches (July 2022 – June 2023) were analysed using ITS2 PCR assays and a phylogenetic analysis. Screening for Plasmodium DNA was performed on mosquito heads and thoraces using RT-qPCR targeting the 18S rRNA, followed by species-specific nested PCR. All 234 specimens morphologically identified An. maculatus s.l. were confirmed as An. maculatus s.s., being genetically consistent with populations from Indonesia and mainland Southeast Asia. Four (1.7%; 95% CI: 0.5–4.3%) An. maculatus specimens were positive for Plasmodium genus DNA, with one specimen positive for the DNA of both Plasmodium knowlesi and Plasmodium inui. This represents the first detection of zoonotic malaria parasite DNA in wild-caught An. maculatus. These findings suggest that An. maculatus may be a potential vector of both zoonotic and human malarias in North Sumatra.
Similar content being viewed by others
Data availability
The datasets used during this study are available in the JCU Research Data repository ([https://doi.org/10.25903/raek-qs16](https:/doi.org/10.25903/raek-qs16), the DOI is under embargo). The DNA sequence data generated in this study are available in the GenBank repository. The An. maculatus ITS2 sequences from North Sumatra have been deposited under accession numbers PQ601060–PQ601066, and the cox1 sequences under accession numbers PQ596587–PQ596590. All datasets are publicly accessible via the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/).
References
World Health Organization. Global Vector Control Response 2017–2030 (World Health Organization, 2017).
Norris, D. E. Mosquito-borne diseases as a consequence of land use change. EcoHealth 1, 19–24, (2004). https://doi.org/10.1007/s10393-004-0008-7
World Health Organization. Manual on Practical Entomology in malaria, Part 1 and Part II (World Health Organization, 1975).
Burkot, T. R. et al. Identification of malaria-infected mosquitoes by a two-site enzyme-linked immunosorbent assay. Am. J. Trop. Med. Hyg. 33, 227–231. https://doi.org/10.4269/ajtmh.1984.33.227 (1984).
Snounou, G. et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 61, 315–320. https://doi.org/10.1016/0166-6851(93)90077-B (1993).
Foley, D. H., Rueda, L. M. & Wilkerson, R. C. Insight into global mosquito biogeography from country species records. J. Med. Entomol. 44, 554–567. https://doi.org/10.1093/jmedent/44.4.554 (2007).
O’Connor, C. T. & Sopa, T. A checklist of the mosquitoes of Indonesia (1981).
Elyazar, I. R. F. et al. Chapter three: the distribution and bionomics of Anopheles malaria vector mosquitoes in Indonesia. Adv. Parasitol. 83, 173–266. https://doi.org/10.1016/B978-0-12-407705-8.00003-3 (2013).
Kirnowardoyo, S. Status of Anopheles malaria vectors in Indonesia. Southeast. Asian J. Trop. Med. Public. Health. 16, 129–132 (1985).
Sinka, M. et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: occurrence data, distribution maps and bionomic precis. Parasit. Vectors. 4, 89. https://doi.org/10.1186/1756-3305-4-89 (2011).
Wibowo, A. A., Umniyati, S. R., Hutagalung, J. & Rahayu, T. Confirmation of Anopheles balabacensis as natural vector of malaria caused by Plasmodium knowlesi inhabits forested areas in Kecamatan Balik Bukit, Western Lampung Regency. E3S Web Conf. 151, 01028 (2020).
van de Straat, B. et al. Zoonotic malaria transmission and land use change in Southeast asia: what is known about the vectors. Malar. J. 21, 109. https://doi.org/10.1186/s12936-022-04129-2 (2022).
Ministry of Health Republic of Indonesia. Annual report 2022 Malaria. Ministry of Health Republic of Indonesia, Indonesia, (2022).
Nurwidayati, A., Purwanto, H. & Ambar Garjito, T. Roro Upiek Ngesti Wibawaning Astuti, R. The biodiversity of Anopheles and malaria vector control in indonesia: a review. BIO Web Conf. 101, 04004 (2024).
Garjito, T. A. et al. Genetic homogeneity of Anopheles maculatus in Indonesia and origin of a novel species present in central Java. Parasit. Vectors. 12 https://doi.org/10.1186/s13071-019-3598-1 (2019).
World Health Organization. Anopheline species complexes in South and South-East Asia. Vol.57 (WHO Regional Office for South-East Asia, 2007).
Ma, Y., Li, S. & Xu, J. Molecular identification and phylogeny of the maculatus group of Anopheles mosquitoes (Diptera: Culicidae) based on nuclear and mitochondrial DNA sequences. Acta Trop. 99, 272–280. https://doi.org/10.1016/j.actatropica.2006.09.005 (2006).
Rattanarithikul, R. & Green, C. A. Formal recognition of the species of the Anopheles maculatus group (Diptera: Culicidae) occurring in Thailand, including the descriptions of two new species and a preliminary key to females (1986).
Harbach RE. in Anopheles mosquitoes (ed Manguin Sylvie) Ch. 1 (IntechOpen, 2013).
Ali, R. S. M. et al. Molecular identification of mosquitoes of the Anopheles maculatus group of subgenus cellia (Diptera: Culicidae) in the Indonesian Archipelago. Acta Trop. 199 https://doi.org/10.1016/j.actatropica.2019.105124 (2019).
Harbach, R. E. The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bull. Entomol. Res. 94, 537–553. https://doi.org/10.1079/BER2004321 (2004).
Erlank, E., Koekemoer, L. L. & Coetzee, M. The importance of morphological identification of African anopheline mosquitoes (Diptera: Culicidae) for malaria control programmes. Malar. J. 17, 43. https://doi.org/10.1186/s12936-018-2189-5 (2018).
Ali, R. S. M. et al. Genetic and morphological evidence for a new species of the maculatus group of Anopheles subgenus cellia (Diptera: Culicidae) in Java, Indonesia. Parasit. Vectors. 12 https://doi.org/10.1186/s13071-019-3358-2 (2019).
World Health Organization. World Malaria Report 2024 (World Health Organization, 2025).
World Health Organization. World Malaria Report 2023 (World Health Organization, 2023).
WHO Malaria Policy Advisory Group. WHO Malaria Policy Advisory Group: Meeting report, 8–10 April 2025 (World Health Organization, 2025).
Dinas Kesehatan Provinsi Sumatera Utara. Kasus penyakit menurut kabupaten/kota dan jenis penyakit di provinsi Sumatera Utara,., <(2022). https://sumut.bps.go.id/id/statistics-table/3/YTA1Q1ptRmhUMEpXWTBsQmQyZzBjVzgwUzB4aVp6MDkjMw==/kasus-penyakit-menurut-kabupaten-kota-dan-jenis-penyakit-di-provinsi-sumatera-utara--2019.html?year=2022 (2023).
Aisyah, D. N. et al. The changing incidence of malaria in Indonesia: a 9-year analysis of surveillance data. Adv Public Health 2024, 2703477, https://doi.org/10.1155/adph/2703477 (2024).
Lubis, I. N. D. et al. Contribution of Plasmodium Knowlesi to multispecies human malaria infections in North Sumatera, Indonesia. J. Infect. Dis. 215, 1148–1155. https://doi.org/10.1093/infdis/jix091 (2017).
Fauziah, N. et al. Emerging malaria in indonesia: an overview of Plasmodium Knowlesi infections. Parasite Epidemiol. Control. 28, e00405. https://doi.org/10.1016/j.parepi.2024.e00405 (2025).
Singh, B. & Daneshvar, C. Human infections and detection of Plasmodium Knowlesi. Clin. Microbiol. Rev. 26, 165–184. https://doi.org/10.1128/cmr.00079-12 (2013).
Manguin, S., Garros, C., Dusfour, I., Harbach, R. E. & Coosemans, M. Bionomics, taxonomy, and distribution of the major malaria vector taxa of Anopheles subgenus cellia in Southeast asia: an updated review. Infect. Genet. Evol. 8, 489–503. https://doi.org/10.1016/j.meegid.2007.11.004 (2008).
Sallum, M., Peyton, E., Harrison, B. & Wilkerson, R. Revision of the leucosphyrus group of Anopheles (Cellia) (Diptera, Culicidae). Rev. Bras. Entomol. 49 https://doi.org/10.1590/S0085-56262005000500001 (2005).
Ang, J. X. D. et al. New vectors in Northern Sarawak, Malaysian Borneo, for the zoonotic malaria parasite, Plasmodium Knowlesi. Parasit. Vectors. 13, 472. https://doi.org/10.1186/s13071-020-04345-2 (2020).
Hawkes, F. M. et al. Vector compositions change across forested to deforested ecotones in emerging areas of zoonotic malaria transmission in Malaysia. Sci. Rep. 9, 13312. https://doi.org/10.1038/s41598-019-49842-2 (2019).
De Ang, J. X., Yaman, K., Kadir, K. A., Matusop, A. & Singh, B. New vectors that are early feeders for Plasmodium Knowlesi and other Simian malaria parasites in Sarawak, Malaysian Borneo. Sci. Rep. 11 https://doi.org/10.1038/s41598-021-86107-3 (2021).
Yanmanee, S. et al. Natural vectors of Plasmodium Knowlesi and other primate, avian and ungulate malaria parasites in Narathiwat Province, Southern Thailand. Sci. Rep. 13, 8875. https://doi.org/10.1038/s41598-023-36017-3 (2023).
Vidhya, P. T., Sunish, I. P., Maile, A. & Zahid, A. K. Anopheles sundaicus mosquitoes as vector for Plasmodium knowlesi, Andaman and nicobar Islands, India. Emerg. Infect. Dis. 25, 817–820. https://doi.org/10.3201/eid2504.181668 (2019).
Coatney, G. R., Collins, W. E., Warren, McW. & Contago, P. G. The Primate Malarias. 366 (U. S. Government Printing Office, 1971).
Jeslyn, W. P. et al. Molecular epidemiological investigation of Plasmodium Knowlesi in humans and macaques in Singapore. Vector Borne Zoonotic Dis. 11, 131–135. https://doi.org/10.1089/vbz.2010.0024 (2011).
van de Straat, B. Adapt or perish: defining malaria vector behaviours in a changing world PhD dissertation thesis, James Cook University, (2024).
Walton, C. et al. Genetic diversity and molecular identification of mosquito species in the Anopheles maculatus group using the ITS2 region of rDNA. Infect. Genet. Evol. 7, 93–102. https://doi.org/10.1016/j.meegid.2006.05.001 (2007).
Singh, S. et al. Anopheles (Cellia) maculatus group: its spatial distribution and molecular characterization of member species in north-east India. Acta Trop. 124, 62–70. https://doi.org/10.1016/j.actatropica.2012.06.011 (2012).
Beebe, N. W. DNA barcoding mosquitoes: advice for potential prospectors. Parasitology 145, 622–633. https://doi.org/10.1017/s0031182018000343 (2018).
Sinka, M. et al. A global map of dominant malaria vectors. Parasit. Vectors. 5 https://doi.org/10.1186/1756-3305-5-69 (2012).
Fang, Y., Shi, W. Q. & Zhang, Y. Molecular phylogeny of Anopheles hyrcanus group members based on ITS2 rDNA. Parasit. Vectors. 10, 417. https://doi.org/10.1186/s13071-017-2351-x (2017).
Castro, J. A., Picornell, A. & Ramon, M. Mitochondrial DNA: a tool for populational genetics studies. Int. Microbiol. 1, 327–332 (1998).
Moore, W. S. Inferring phylogenies from MtDNA variation: mitochondrial-gene Tress versus nuclear gene trees. Evolution 49, 718–726. https://doi.org/10.1111/j.1558-5646.1995.tb02308.x (1995).
Barcus, M. J. et al. Epidemic malaria in the menoreh hills of central Java. Am. J. Trop. Med. Hyg. 66, 287–292. https://doi.org/10.4269/ajtmh.2002.66.287 (2002).
Bangs, M. J. & Rusmiarto, S. Malaria Vector Incrimination in Indonesia Using CSP-ELISA from 1986 To 2007. (U.S. Naval Medical Research Unit No. 2 (Indonesia, 2007).
Poolphol, P. et al. Natural Plasmodium Vivax infections in Anopheles mosquitoes in a malaria endemic area of Northeastern Thailand. Parasitol. Res. 116, 3349–3359. https://doi.org/10.1007/s00436-017-5653-1 (2017).
Vythilingam, I. et al. Epidemiology of malaria in Attapeu Province, Lao PDR in relation to entomological parameters. Trans. R Soc. Trop. Med. Hyg. 99, 833–839. https://doi.org/10.1016/j.trstmh.2005.06.012 (2005).
Do Manh, C. et al. Vectors and malaria transmission in deforested, rural communities in north-central Vietnam. Malar. J. 9, 259. https://doi.org/10.1186/1475-2875-9-259 (2010).
Rahman, W. A., Che’Rus, A. & Ahmad, A. H. Malaria and Anopheles mosquitos in Malaysia. Southeast. Asian J. Trop. Med. Public. Health. 28, 599–605 (1997).
Alam, M. S. et al. Diversity of anopheline species and their Plasmodium infection status in rural Bandarban, Bangladesh. Parasit. Vectors. 5 https://doi.org/10.1186/1756-3305-5-150 (2012).
Alam, M. S. et al. Prevalence of anopheline species and their Plasmodium infection status in epidemic-prone border areas of Bangladesh. Malar. J. 9 https://doi.org/10.1186/1475-2875-9-15 (2010).
Doorenbos, W. B. Some experiences in the field of malaria. Med. J. Dutch East. Indies. I-III, 1229–1248 (1931).
Swellengrebel, N. H. & Rodenwaldt, E. The Anophelines of Dutch East Indies 3rd edn (Gustav Fischer, 1932).
Muenworn, V. et al. Biting activity and host preference of the malaria vectors Anopheles maculatus and Anopheles Sawadwongporni (Diptera: Culicidae) in Thailand. J. Vector Ecol. 34, 62–69. https://doi.org/10.1111/j.1948-7134.2009.00008.x (2009).
Tainchum, K., Kongmee, M., Manguin, S., Bangs, M. J. & Chareonviriyaphap, T. Anopheles species diversity and distribution of the malaria vectors of Thailand. Trends Parasitol. 31, 109–119. https://doi.org/10.1016/j.pt.2015.01.004 (2015).
Vythilingam, I. et al. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor, Malaysia (2009–2013): epidemiologic and entomologic analysis. Parasit. Vectors 7, https://doi.org/10.1186/1756-3305-7-436 (2014).
Jeyaprakasam, N. K. et al. High transmission efficiency of the simian malaria vectors and population expansion of their parasites Plasmodium cynomolgi and Plasmodium inui. PLoS Negl. Trop. Dis. 17, e0011438. https://doi.org/10.1371/journal.pntd.0011438 (2023).
Vythilingam, I. et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit. Vectors 1, https://doi.org/10.1186/1756-3305-1-26 (2008).
Collins, W. E., Contacos, P. G., Guinn, E. G. & Held, J. R. Transmission of Plasmodium fieldi by Anopheles maculatus, A. stephensi, and A. balabacensis balabacensis. J Parasitol 54, 376–376, (1968). https://doi.org/10.2307/3276956
Collins, W. E., Contacos, P. G., Guinn, E. G. & Held, J. R. Studies on the transmission of Simian malarias, I. Transmission of two strains of Plasmodium Inui by Anopheles maculatus and An. stephensi. J. Parasitol. 52, 664–668. https://doi.org/10.2307/3276425 (1966).
Collins, W. E., Contacos, P. G. & Guinn, E. G. Studies on the transmission of Simian malarias II. Transmission of the H strain of Plasmodium knowlesi by Anopheles balabacensis balabacensis. J. Parasitol. 53, 841–844 (1967).
Wharton, R. H., Eyles, D. E., Warren, M. & Cheong, W. H. Studies to determine the vectors of monkey malaria Malaya. Ann. Trop. Med. Parasitol. 58, 56–77. https://doi.org/10.1080/00034983.1964.11686215 (1964).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. https://doi.org/10.1373/clinchem.2008.112797 (2009).
McCall, M. N., McMurray, H. R., Land, H. & Almudevar, A. On non-detects in qPCR data. Bioinformatics 30, 2310–2316. https://doi.org/10.1093/bioinformatics/btu239 (2014).
Kralik, P. & Ricchi, M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front. Microbiol. 8 https://doi.org/10.3389/fmicb.2017.00108 (2017).
Sebayang, B. F. Changes in outdoor malaria and dengue vectors following human activities PhD dissertation thesis, James Cook University, (2025).
Imwong, M. et al. Spurious amplification of a Plasmodium Vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J. Clin. Microbiol. 47, 4173–4175. https://doi.org/10.1128/jcm.00811-09 (2009).
Harrison, G. F. et al. Plasmodium-specific molecular assays produce uninterpretable results and non-Plasmodium spp. Sequences in field-collected Anopheles vectors. Am. J. Trop. Med. Hyg. 89, 1117–1121. https://doi.org/10.4269/ajtmh.12-0581 (2013).
Lefterova Martina, I., Budvytiene, I., Sandlund, J., Färnert, A. & Banaei, N. Simple real-time PCR and amplicon sequencing method for identification of Plasmodium species in human whole blood. J. Clin. Microbiol. 53, 2251–2257. https://doi.org/10.1128/jcm.00542-15 (2015).
Ta, T. H. et al. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar. J. 13, 68. https://doi.org/10.1186/1475-2875-13-68 (2014).
Lucchi, N. W. et al. A new single-step PCR assay for the detection of the zoonotic malaria parasite Plasmodium knowlesi. PloS One. 7, e31848. https://doi.org/10.1371/journal.pone.0031848 (2012).
Isozumi, R. et al. Improved detection of malaria cases in Island settings of Vanuatu and Kenya by PCR that targets the Plasmodium mitochondrial cytochrome c oxidase III (cox3) gene. Parasitol. Int. 64, 304–308. https://doi.org/10.1016/j.parint.2014.09.006 (2015).
Kimura, M. et al. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol. Int. 46, 91–95. https://doi.org/10.1016/S1383-5769(97)00013-5 (1997).
Grigg, M. J. et al. Plasmodium Knowlesi detection methods for human infections-diagnosis and surveillance. Adv. Parasitol. 113, 77–130. https://doi.org/10.1016/bs.apar.2021.08.002 (2021).
World Health Organization. Global Technical Strategy for Malaria 2016–2030 (World Health Organization, 2015).
Harrison, L. E. et al. A multi-criteria framework for disease surveillance site selection: case study for Plasmodium Knowlesi malaria in Indonesia. R Soc. Open. Sci. 11, 230641. https://doi.org/10.1098/rsos.230641 (2024).
Tobin, R. J. et al. Updating estimates of Plasmodium Knowlesi malaria risk in response to changing land use patterns across Southeast Asia. PLoS Negl. Trop. Dis. 18, e0011570. https://doi.org/10.1371/journal.pntd.0011570 (2024).
Russell, T. L., Staunton, K. & Burkot, T. R. Standard operating procedure for performing human landing catch (HLC). protocols.io (2022).
World Health Organization. Malaria Entomology and Vector control. Guide for Participants (World Health Organization, 2013).
Silver, J. B. Mosquito Ecology: Field Sampling methods. Third Edition (Springer, 2008).
Beebe, N. W. & Saul, A. Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction - restriction fragment length polymorphism analysis. Am. J. Trop. Med. Hyg. 53, 478–481. https://doi.org/10.4269/ajtmh.1995.53.478 (1995).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. https://doi.org/10.1016/s0022-2836(05)80360-2 (1990).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal Omega. Mol. Syst. Biol. 7 https://doi.org/10.1038/msb.2011.75 (2011).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. https://doi.org/10.1007/bf01731581 (1980).
Saccone, C., De Giorgi, C., Gissi, C., Pesole, G. & Reyes, A. Evolutionary genomics in metazoa: the mitochondrial DNA as a model system. Gene 238, 195–209. https://doi.org/10.1016/S0378-1119(99)00270-X (1999).
Kamau, E. et al. Development of a highly sensitive genus-specific quantitative reverse transcriptase real-time PCR assay for detection and quantitation of Plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J. Clin. Microbiol. 49, 2946–2953. https://doi.org/10.1128/jcm.00276-11 (2011).
Braima, K. A. et al. Improved limit of detection for zoonotic Plasmodium Knowlesi and P. cynomolgi surveillance using reverse transcription for total nucleic acid preserved samples or dried blood spots. PLoS Negl. Trop. Dis. 19, e0012129. https://doi.org/10.1371/journal.pntd.0012129 (2025).
Lee, K. S. et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 7, e1002015 (2011).
Acknowledgements
We extend our gratitude to the community members of Dusun II and V in Ujung Bandar Village, Langkat Regency, North Sumatra, for their contributions during fieldwork and their hospitality. We thank Hidayatullah Nasution, Panusunan Hasibuan, and Ledy Afrida Sinaga for their contributions to the fieldwork and morphological identification of mosquitoes. We are grateful to Mhd Ihza Fahrezi and Kania Haura Chalisaturahmi (Universitas Sumatra Utara) for their assistance with laboratory assays. We thank Nigel Beebe (University of Queensland) and Neil Lobo (University of Notre Dame, USA) for their valuable discussions and input on the phylogenetic analyses.
Funding
This study was funded by the ZOOMAL project (‘Evaluating zoonotic malaria and agricultural land use in Indonesia’; #LS-2019-116), Australian Centre for International Agricultural Research, Australian Government. BFS was supported by a James Cook University Postgraduate Research Scholarship. MJG is supported by a NHMRC EL2 Investigator grant.
Author information
Authors and Affiliations
Contributions
BFS, TRB, TLR and MJG conceived and designed the study. TRB, TLR, TAG, INDL and MJG supervised and advised on fieldwork. BvdS, AK and BFS conducted field studies and mosquito collections. BFS and AMH conducted the laboratory analyses with input from INDL and MJG. BFS, TAG and JAFW conducted the phylogenetic analyses. BFS wrote the original draft of the manuscript. TRB and TLR provided supervision and extensive editing of the manuscript. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The studies were conducted in accordance with the guidelines and regulations approved by Universitas Sumatera Utara Faculty of Medicine (application number 723/KEP/USU/2021) on 14 July 2021 and James Cook University (application number H8583) on 29 September 2021.
Consent to participate
Human landing catch (HLC) procedures followed the ethical approvals of Universitas Sumatera Utara and James Cook University. All collectors were adults (≥ 18 years) who provided written informed consent after being informed of the associated risks and benefits. Volunteers received training in mosquito collection using an oral aspirator, and all measures outlined in the ethics approvals to minimise exposure to mosquito bites were strictly implemented.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sebayang, B.F., van de Straat, B., Kurniawan, A. et al. Evidence incriminating Anopheles maculatus as a potential vector of Plasmodium knowlesi and Plasmodium inui. Sci Rep (2026). https://doi.org/10.1038/s41598-026-35946-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-35946-z