Abstract
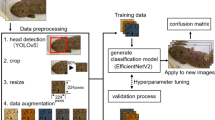
The yellow-spotted mountain newt (Neurergus derjugini), an endangered amphibian endemic to the Zagros Mountains, faces critical threats from habitat loss and climate change. Effective conservation requires reliable population monitoring, yet traditional marking methods are invasive and impractical. This study presents a non-invasive, image-based approach combining geometric computer vision and deep learning for individual identification and population estimation. We captured 549 adult N. derjugini in their natural habitat, photographing dorsal patterns under standardized conditions. A geometric pipeline (HSV thresholding, morphological operations) extracted yellow spot features (area, circularity, count), achieving 93% detection accuracy. Three convolutional neural networks (CNNs)—DenseNet121, EfficientNetB0, and InceptionV3—were fine-tuned for phenotypic classification, with DenseNet121 attaining the highest accuracy (99.11%) and AUC (0.98). Region-specific analysis showed optimal performance when combining head and trunk patterns (96.32% accuracy). A mark-recapture framework, applied to two sampling sessions (n = 332 and 217 individuals), identified 65 recaptures, yielding a Lincoln-Petersen population estimate of 1108 individuals. Our results demonstrate that deep learning outperforms traditional methods in robustness and scalability, particularly under variable field conditions. This study advances amphibian conservation by providing a rapid, ethical, and scalable tool for monitoring endangered species. Future directions include expanding datasets for temporal stability validation and deploying mobile applications for real-time field use. By integrating AI with ecological research, this work highlights the transformative potential of automated identification in biodiversity conservation.
Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study, including annotated images, extracted morphometric features, and training/validation/test sets, are available from the corresponding author upon reasonable request. The source code for image preprocessing, morphometric analysis, deep learning classification, and population estimation is included in the supplementary materials (Supplementary Files 1 and 2). Additionally, a compiled standalone executable (.exe) of the image analysis application, along with installation instructions, will be made publicly accessible via the project’s GitHub repository.
References
Karczmarski, L., Chan, S. C. Y., Chui, S. Y. S. & Cameron, E. Z. Individual identification and photographic techniques in mammalian ecological and behavioural research—Part 2: field studies and applications. Mamm. Biol. 102, 1047–1054 (2022).
Alberts, S. C. Social influences on survival and reproduction: Insights from a long-term study of wild baboons. J. Anim. Ecol. 88, 47–66 (2019).
Jolly, G. M. The estimation of animal abundance and related parameters. at (1974).
Williams, B. K., Nichols, J. D., Conroy, M. J. & Conroy, M. J. Analysis and Management of Animal Populations (Academic, 2002).
Takaya, K. Individual identification of Japanese giant salamanders (Andrias japonicus) and detection of their hybrids by image recognition using deep learning. Sci. Rep. 13, 16212 (2024).
Grant, E. H. C. et al. Quantitative evidence for the effects of multiple drivers on continental-scale amphibian declines. Sci. Rep. 6, 25625 (2016).
Stuart, S. N. et al. Status and trends of amphibian declines and extinctions worldwide. Science. 306, 1783–1786 (2004).
Schmidt, B. R. Count data, detection probabilities, and the demography, dynamics, distribution, and decline of amphibians. Comptes Rendus Biol. 326, 119–124 (2003).
Schmidt, B. R., Schaub, M. & Anholt, B. R. Why you should use capture-recapture methods when estimating survival and breeding probabilities: on bias, temporary emigration, overdispersion, and common toads. Amphibia-Reptilia 23, 375–388 (2002).
Heyer, R., Donnelly, M. A., Foster, M. & Mcdiarmid, R. Measuring and Monitoring Biological Diversity: Standard Methods for AmphibiansSmithsonian Institution,. (2014).
Bailey, L. L., Simons, T. R. & Pollock, K. H. Estimating detection probability parameters for plethodon salamanders using the robust capture-recapture design. J. Wildl. Manag. 68, 1–13 (2004).
Cove, M. V. & Spínola, R. M. Pairing noninvasive surveys with capture-recapture analysis to estimate demographic parameters for Dendrobates auratus (Anura: Dendrobatidae) from an altered habitat in Costa Rica. Phyllomedusa 12, 107–115 (2013).
Burley, N., Krantzberg, G. & Radman, P. Influence of colour-banding on the conspecific preferences of zebra finches. Anim. Behav. 30, 444–455 (1982).
Funk, W. C., Donnelly, M. A. & Lips, K. R. Alternative views of amphibian toe-clipping. Nature 433, 193 (2005).
Gauthier-Clerc, M. et al. Long-term effects of flipper bands on Penguins. Proc. R Soc. B Biol. Sci. 271, S423–S426 (2004).
Moorhouse, T. P. & Macdonald, D. W. Indirect negative impacts of radio-collaring: Sex ratio variation in water voles. J. Appl. Ecol. 42, 91–98 (2005).
Zemanova, M. A. Poor implementation of non-invasive sampling in wildlife genetics studies. Rethink Ecol. 4, 119–132 (2019).
Zemanova, M. A. Towards more compassionate wildlife research through the 3Rs principles: Moving from invasive to non-invasive methods. Wildlife Biol. 1–17 (2020). (2020).
Zemanova, M. A., Martín, R. L. & Leenaars, C. H. C. The impact of toe-clipping on animal welfare in amphibians: A systematic review. Glob Ecol. Conserv. 59, e03582 (2025).
Gibbons, W. J. & Andrews, K. M. PIT tagging: Simple technology at its best. Bioscience 54, 447–454 (2004).
Green, M. L., Ting, T. F. & Manjerovic, M. B. Mateus-Pinilla, N. Noninvasive alternatives for DNA collection from threatened rodents. Nat. Sci. 05, 18–26 (2013).
Banerjee, P. et al. Reinforcement of environmental DNA based methods (sensu stricto) in biodiversity monitoring and conservation: A review. Biology (Basel). 10, 1223 (2021).
Rees, H. C., Maddison, B. C., Middleditch, D. J., Patmore, J. R. M. & Gough, K. C. The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 51, 1450–1459 (2014).
Choo, Y. R. et al. Best practices for reporting individual identification using camera trap photographs. Glob Ecol. Conserv. 24, e01294 (2020).
Gardiner, R. Z., Doran, E., Strickland, K., Carpenter-Bundhoo, L. & Frère, C. A face in the crowd: A non-invasive and cost effective photo-identification methodology to understand the fine scale movement of Eastern water Dragons. PLoS One. 9, e96992 (2014).
Moro, D. & MacAulay, I. Computer-Aided pattern recognition of large reptiles as a noninvasive application to identify individuals. J. Appl. Anim. Welf. Sci. 17, 125–135 (2014).
Gamble, L., Ravela, S. & McGarigal, K. Multi-scale features for identifying individuals in large biological databases: an application of pattern recognition technology to the marbled salamander Ambystoma opacum. J. Appl. Ecol. 45, 170–180 (2008).
Bendik, N. F., Morrison, T. A., Gluesenkamp, A. G., Sanders, M. S. & O’Donnell, L. J. Computer-Assisted photo identification outperforms visible implant elastomers in an endangered Salamander, eurycea Tonkawae. PLoS One. 8, e59424 (2013).
Speed, C. W., Meekan, M. G. & Bradshaw, C. J. A. Spot the match–wildlife photo-identification using information theory. Front. Zool. 4, 2 (2007).
Wäldchen, J. & Mäder, P. Machine learning for image based species identification. Methods Ecol. Evol. 9, 2216–2225 (2018).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 61, 85–117 (2015).
Bichell, L. M. V., Krzyszczyk, E., Patterson, E. M. & Mann, J. The reliability of pigment pattern-based identification of wild bottlenose dolphins. Mar. Mammal Sci. 34, 113–124 (2018).
Osterrieder, S. K., Kent, C. S., Anderson, C. J. R., Parnum, I. M. & Robinson, R. W. Whisker spot patterns: A noninvasive method of individual identification of Australian sea lions (Neophoca cinerea). J. Mammal. 96, 988–997 (2015).
Ferreira, A. C. et al. Deep learning-based methods for individual recognition in small birds. Methods Ecol. Evol. 11, 1072–1085 (2020).
Patel, A. et al. Revealing the unknown: Real-time recognition of Galápagos snake species using deep learning. Animals 10, 806 (2020).
Pennycuick, C. J. & Rudnai, J. A method of identifying individual lions Panthera Leo with an analysis of the reliability of identification. J. Zool. 160, 497–508 (1970).
Anderson, C. J. R., Roth, J. D. & Waterman, J. M. Can whisker spot patterns be used to identify individual Polar bears? J. Zool. 273, 333–339 (2007).
Chui, S. Y. S. & Karczmarski, L. Everyone matters: identification with facial wrinkles allows more accurate inference of elephant social dynamics. Mamm. Biol. 102, 645–666 (2022).
Schofield, G., Katselidis, K. A., Dimopoulos, P. & Pantis, J. D. Investigating the viability of photo-identification as an objective tool to study endangered sea turtle populations. J. Exp. Mar. Bio Ecol. 360, 103–108 (2008).
Birenbaum, Z. et al. Facial recognition software for ecological studies of harbor seals. Ecol. Evol. 12, e8851 (2022).
Chen, P. et al. A study on giant panda recognition based on images of a large proportion of captive pandas. Ecol. Evol. 10, 3561–3573 (2020).
Clapham, M., Miller, E., Nguyen, M. & Darimont, C. T. Automated facial recognition for wildlife that lack unique markings: A deep learning approach for brown bears. Ecol. Evol. 10, 12883–12892 (2020).
Schofield, D. et al. Chimpanzee face recognition from videos in the wild using deep learning. Sci. Adv. 5, eaaw0736 (2019).
Takaya, K., Taguchi, Y. & Ise, T. Individual identification of endangered amphibians using deep learning and smartphone images: Case study of the Japanese giant salamander (Andrias japonicus). Sci. Rep. 13, 16212 (2023).
I. U. C. N. Neurergus derjugini. IUCN Red List. Threatened Species. 2023, eT88373273A50759480. https://doi.org/10.2305/IUCN.UK.2023-1.RLTS.T88373273A50759480.en (2023). Accessed on 03 January 2024.
Vaissi, S., Parto, P. & Sharifi, M. Ontogenetic changes in spot configuration (numbers, circularity, size and asymmetry) and lateral line in Neurergus microspilotus (Caudata: Salamandridae). Acta Zool. 99, 9–19 (2018).
Faul, C., Wagner, N. & Veith, M. Successful automated photographic identification of larvae of the European fire Salamander, Salamandra salamandra. Salamandra 58, 52–63 (2022).
Lunghi, E. et al. Photographic database of the European cave salamanders, genus Hydromantes. Sci. Data. 7, 171 (2020).
Romiti, F. et al. Photographic identification method (PIM) using natural body marks: A simple tool to make a long story short. Zool. Anz. 266, 136–147 (2017).
Schoen, A., Boenke, M. & Green, D. M. Tracking toads using photo identification and image-recognition software. Herpetol Rev 46, (2015).
Waye, H. L. Can a tiger change its spots? A test of the stability of spot patterns for identification of individual tiger salamanders (Ambystoma tigrinum). Herpetol Conserv. Biol. 8, 419–425 (2013).
Sharifi, M. & Afroosheh, M. Studying migratory activity and home range of adult Neurergus microspilotus (Nesterov, 1916) in the Kavat Stream, Western Iran, using photographic identification. Herpetozoa 27, 77–82 (2014).
Afroosheh, M., Akmali, V., Esmaeili-Rineh, S. & Sharifi, M. Distribution and abundance of the endangered yellow spotted mountain Newt Neurergus microspilotus (Caudata: Salamandridae) in Western Iran. Herpetol Conserv. Biol. 11, 52–60 (2016).
Royle, J. A. & Dorazio, R. M. Hierarchical Modeling and Inference in Ecology: The Analysis of Data from Populations, Metapopulations and Communities. (2008). https://doi.org/10.1016/B978-0-12-374097-7.X0001-4
Acknowledgements
We are deeply grateful to Ms. Shima Vaissi for her invaluable assistance during fieldwork. Her contributions were instrumental to the successful completion of this project.
Funding
This work was financially supported by the Department of Environment, Islamic Republic of Iran, and Razi University.
Author information
Authors and Affiliations
Contributions
S.V. conceived and designed the study. S.V., Z.R., P.F., P.D., M.S.M., and Z.T.K conducted fieldwork and collected data. P.F. performed data analysis in collaboration with S.V. S.V. wrote the manuscript. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All methods in this study were carried out in accordance with relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rahmdel, Z., Vaissi, S., Faramarzi, P. et al. Deep learning based individual identification and population estimation of the yellow spotted mountain newt (Neurergus derjugini). Sci Rep (2026). https://doi.org/10.1038/s41598-026-36092-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-36092-2