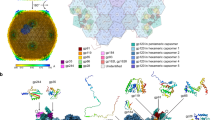
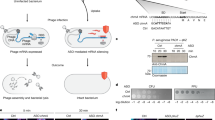
Abstract
Pseudomonas jumbophage phiKZ is prominent because of the proteinaceous nuclear structure that is formed during the infection cycle, protecting the replicating phage DNA from host nucleases. Progress has been made in deciphering the molecular basis of nucleus formation and other unique aspects of phiKZ biology, but the understanding of how it causes host lysis remains minimal. Here we present bioinformatic, physiological and molecular evidence for a “lysis cassette”, the expression of which is necessary and sufficient for the temporally regulated disruption of the host envelope. This cluster contains genes for the usual components of an MGL (multigene lysis) system, including the holin, endolysin, i-spanin and o-spanin. In addition, a fifth gene in the cluster, encoding a cytoplasmic protein, was found to accelerate the timing of holin-mediated lethality when expressed in trans. Evidence is provided that suggests this lysis regulator protein interacts with the cytoplasmic domain of the phiKZ class III holin. In support of this notion, alpha-fold analysis generated a high-confidence structure of a conserved holin-lysis regulator heterodimer complex. Infections at high multiplicity resulted in slower bulk culture lysis profiles than at low multiplicity, suggesting that phiKZ might have a lysis-inhibition system.
Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Protein sequences are provided in Figure 1, also available at NC_004629.1. Primer sequences are provided in Supplemental Table 1. Protein sequences used to create multi-sequence alignment are provided in Supplemental data 2 and 3.
References
Krylov, V. N. & Zhazykov, I. Z. [Pseudomonas bacteriophage phiKZ–possible model for studying the genetic control of morphogenesis]. Genetika 14, 678–685 (1978).
Mesyanzhinov, V. V. et al. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J. Mol. Biol. 317, 1–19 (2002).
Mendoza, S. D. et al. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature 577, 244–248 (2020).
Cahill, J. & Young, R. Phage lysis: multiple genes for multiple barriers. in 33–70. https://doi.org/10.1016/bs.aivir.2018.09.003 (2019).
Young, R. Phage lysis: three steps, three choices, one outcome. J. Microbiol. 52, 243–258 (2014).
Cahill, J. & Young, R. Release of Phages From Prokaryotic Cells. in Encyclopedia of Virology 501–518 https://doi.org/10.1016/B978-0-12-814515-9.00074-6 (Elsevier, 2021).
Gründling, A., Manson, M. D. & Young, R. Holins kill without warning. Proc. Natl. Acad. Sci. U S A. 98, 9348–9352 (2001).
Cahill, J. et al. Spatial and temporal control of lysis by the lambda holin. mBio 15, (2024).
Berry, J., Savva, C., Holzenburg, A. & Young, R. The lambda spanin components Rz and Rz1 undergo tertiary and quaternary rearrangements upon complex formation. Protein Sci. 19, 1967–1977 (2010).
Cahill, J. et al. Genetic analysis of the lambda spanins Rz and Rz1: identification of functional domains. G3 Genes|Genomes|Genetics. 7, 741–753 (2017).
Berry, J. D., Rajaure, M. & Young, R. Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol. Microbiol. 88, 35–47 (2013).
Miroshnikov, K. A., Faizullina, N. M., Sykilinda, N. N. & Mesyanzhinov, V. V. Properties of the endolytic transglycosylase encoded by gene 144 of Pseudomonas aeruginosa bacteriophage PhiKZ. Biochem. (Mosc). 71, 300–305 (2006).
Fokine, A., Miroshnikov, K. A., Shneider, M. M., Mesyanzhinov, V. V. & Rossmann, M. G. Structure of the bacteriophage phiKZ lytic transglycosylase gp144. J. Biol. Chem. 283, 7242–7250 (2008).
Krylov, V. et al. Phage phiKZ—The First of Giants. Viruses 13, 149 (2021).
Paradis-Bleau, C. et al. Peptidoglycan lytic activity of the Pseudomonas aeruginosa phage phiKZ gp144 lytic transglycosylase. FEMS Microbiol. Lett. 266, 201–209 (2007).
Chertkov, O. V. et al. Dual active site in the endolytic transglycosylase gp144 of bacteriophage PhiKZ. Acta Naturae. 9, 81–87 (2017).
Briers, Y. et al. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages φKZ and EL. Mol. Microbiol. 65, 1334–1344 (2007).
Kongari, R. et al. Phage spanins: diversity, topological dynamics and gene convergence. BMC Bioinform. 19, 326 (2018).
Juncker, A. S. et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662 (2003).
Braun, V. & Wu, H. C. Chapter 14 Lipoproteins, structure, function, biosynthesis and model for protein export. in 319–341 https://doi.org/10.1016/S0167-7306(08)60417-2 (1994).
Park, T., Struck, D. K., Deaton, J. F. & Young, R. Topological dynamics of holins in programmed bacterial lysis. Proceedings of the National Academy of Sciences 103, 19713–19718 (2006).
Grayhack, E. J. & Roberts, J. W. The phage λ Q gene product: activity of a transcription antiterminator in vitro. Cell 30, 637–648 (1982).
Cahill, J. et al. Suppressor analysis of the fusogenic lambda Spanins. J. Virol. 91, https://doi.org/10.1128/jvi.00413-17 (2017).
Chernyshov, S. V., Masulis, I. S. & Mikoulinskaia, G. V. From DNA to lytic proteins: transcription and translation of the bacteriophage T5 holin/endolysin Operon. World J. Microbiol. Biotechnol. 40, 256 (2024).
Summer, E. J. et al. Rz/Rz1 Lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 373, 1098–1112 (2007).
Moussa, S. H., Lawler, J. L. & Young, R. Genetic dissection of T4 Lysis. J. Bacteriol. 196, 2201–2209 (2014).
Tran, T. A. T., Struck, D. K. & Young, R. Periplasmic domains define Holin-Antiholin interactions in T4 Lysis Inhibition. J. Bacteriol. 187, 6631–6640 (2005).
Ramanculov, E. & Young, R. Genetic analysis of the T4 holin: timing and topology. Gene 265, 25–36 (2001).
Ramanculov, E. & Young, R. An ancient player unmasked: T4 rI encodes a t -specific antiholin. Mol. Microbiol. 41, 575–583 (2001).
Young, R. Bacteriophage holins: deadly diversity. J. Mol. Microbiol. Biotechnol. 4, 21–36 (2002).
Krieger, I. V. et al. The structural basis of T4 phage lysis control: DNA as the signal for lysis inhibition. J. Mol. Biol. 432, 4623–4636 (2020).
Schwarzkopf, J. M. F., Mehner-Breitfeld, D. & Brüser, T. A dimeric holin/antiholin complex controls lysis by phage T4. Front. Microbiol. 15, 1419106 (2024).
Wang, I. N., Smith, D. L. & Young, R. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54, 799–825 (2000).
Bavda, V. R. & Jain, V. Deciphering the role of holin in mycobacteriophage D29 physiology. Front Microbiol 11, 883 (2020).
Raab, R. et al. Mutational analysis of bacteriophage lambda lysis gene S. J. Bacteriol. 167, 1035–1042 (1986).
Reader, R. W. & Siminovitch, L. Lysis defective mutants of bacteriophage lambda: on the role of the S function in Lysis. Virology 43, 623–637 (1971).
Briers, Y., Peeters, L. M., Volckaert, G. & Lavigne, R. The Lysis cassette of bacteriophage фKMV encodes a signal-arrest-release endolysin and a Pinholin. Bacteriophage 1, 25–30 (2011).
Holt, A. et al. Phage-encoded cationic antimicrobial peptide required for Lysis. J. Bacteriol. 204, JB0021421 (2021).
Kongari, R., Snowden, J., Berry, J. D. & Young, R. Localization and regulation of the T1 unimolecular Spanin. J. Virol. 92, e00380–18 (2018).
Savva, C. G. et al. Stable micron-scale holes are a general feature of canonical holins. Mol. Microbiol. 91, 57–65 (2014).
To, K. H., Dewey, J., Weaver, J., Park, T. & Young, R. Functional analysis of a class I holin, P2 Y. J. Bacteriol. 195, 1346–1355 (2013).
Pang, T., Park, T. & Young, R. Mapping the pinhole formation pathway of S21. Mol. Microbiol. 78, 710–719 (2010).
Summer, E. J. et al. Genomic and biological analysis of phage Xfas53 and related prophages of Xylella fastidiosa. J. Bacteriol. 192, 179–190 (2010).
Bläsi, U. & Young, R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage Lysis. Mol. Microbiol. 21, 675–682 (1996).
Barenboim, M., Chang, C., Hajj, D., Young, R. & F. & Characterization of the dual start motif of a class II Holin gene. Mol. Microbiol. 32, 715–727 (1999).
Chan, A. et al. Bacteriophage genome-wide transposon mutagenesis. Preprint at. https://doi.org/10.1101/2025.11.23.690004 (2025).
Schmidt, C., Velleman, M. & Arber, W. Three functions of bacteriophage P1 involved in cell Lysis. J. Bacteriol. 178, 1099–1104 (1996).
Niazi, S. K. Critical assessment of AI-based protein structure prediction: fundamental challenges and future directions in drug discovery. Comput. Struct. Biotechnol. Rep. 2, 100064 (2025).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold3. Nature 630, 493–500 (2024).
Agarwal, V. & McShan, A. C. The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins. Nat. Chem. Biol. 20, 950–959 (2024).
Pleteneva, E. A. et al. Pseudolysogeny of Pseudomonas aeruginosa bacteria infected with φKZ-like bacteriophages. Russ J. Genet. 46, 20–25 (2010).
Abedon, S. T. Lysis of lysis-inhibited bacteriophage T4-infected cells. J. Bacteriol. 174, 8073–8080 (1992).
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177, 4121–4130 (1995).
Klotz, A., Kaczmarczyk, A. & Jenal, U. A. Synthetic Cumate-Inducible promoter for graded and homogenous gene expression in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 89, e0021123 (2023).
Carver, T., Harris, S. R., Berriman, M., Parkhill, J. & McQuillan, J. A. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469 (2012).
Hallgren, J. et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Preprint at. https://doi.org/10.1101/2022.04.08.487609 (2022).
Tsirigos, K. D., Peters, C., Shu, N., Käll, L. & Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407 (2015).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019).
Teufel, F. et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025 (2022).
Simm, D., Hatje, K. & Kollmar, M. Waggawagga: comparative visualization of coiled-coil predictions and detection of stable single α-helices (SAH domains). Bioinformatics 31, 767–769 (2015).
Zhang, N. & Young, R. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage λ. Mol. Gen. Genet. 262, 659–667 (1999).
Blattner, F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997).
Acknowledgements
The authors would like to thank the funding supported by the National Institute of General Medical Sciences, grant no. R35GM136396 to R.Y. and by the Center for Phage Technology, which is jointly supported by Texas A&M AgriLife Research. We also thank Joseph Bondy-Denomy for kindly sharing phiKZ phage lysates, and the lab of Dr. Urs Jenal at the University of Basel in Switzerland for providing the cumate inducible vector pQFT.
Funding
This work was supported by funding from the National Institute of General Medical Sciences, grant no. R35GM136396 to R.Y. and by the Center for Phage Technology, which is jointly supported by Texas A&M AgriLife Research.
Author information
Authors and Affiliations
Contributions
Conceptualization, P.M., J.W., R.Y.; Data Curation, P.M., J.W.; Formal Analysis, P.M., G.G., K.S., J.W.; Experimental works, P.M., J.W., G.G., K.S.; Investigation, P.M., J.W.; Methodology, P.M., J.W., R.Y.; Resources, P.M., R.Y.; Supervision, R.Y.; Writing – Original Draft, P.M., R.Y.; Writing – Review & Editing, P.M., R.Y.; Validation, P.M., R.Y.; Visualization, P.M.; Project Administration, P.M., R.Y.; Funding Acquisition, R.Y.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Manohar, P., Wan, J., Ganser, G. et al. The Lysis cassette of jumbophage PhiKZ. Sci Rep (2026). https://doi.org/10.1038/s41598-026-36188-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-36188-9