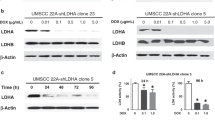
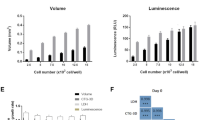
Abstract
The tumor cells frequently rely on glycolysis to produce adenosine 5′-triphosphate (ATP), even when sufficient oxygen is available to allow oxidative phosphorylation (the Warburg effect). In these malignancies, the breakdown of glucose to pyruvate, instead of reaching the mitochondria, is transformed to lactate by an enzyme called lactate dehydrogenase (LDH) and then expelled by the cells, further fuelling the tumour microenvironment (TME). LDH facilitates the translation of pyruvate to lactate, hence replenishing the required NAD + equivalents for the ongoing glycolysis process. Having a pivotal role in cancer cells’ prognosis and survival, and affecting the TME. To date, no inhibitors have yet been approved against the LDH. However, numerous clinical trials are ongoing, and results are yet to be awaited. Considering the existing gap, we present herein a high-throughput virtual screening (HTVS) approach to identify new compounds that effectively inhibit LDH activity. We generated the pharmacophore model based on 28 LDH enzyme inhibitors from previous literature. The model was used to screen 500,000 ligands in addition to their molecular docking and drug-likeness filtering. The analysis led to the identification of 5 hits, which were further subjected to the MD simulations. Further considering the outcome of molecular dynamics results, we selected ligands 15 and 422 to corroborate their anticancer potential via inhibiting the LDH enzyme. The biological validation revealed that both ligands, 15 and 422, possess IC50 values of 147.34 and 206.35 nM, respectively, against LDH. The anticancer potential analysis of DU-145 and PC-3 also established their anticancer properties, and both compounds were found to marginally elevate oxidative stress, change mitochondrial membrane potential, and induce apoptosis in DU-145 cells.
Similar content being viewed by others
Data availability
In silico raw files associated with molecular docking and dynamics will be provided upon request. Key structures associated with pharmacophore modelling and the glide score of top hits are presented in the supporting information file. All the high-definition individual images associated with MD simulations are also supplied.
References
Kiri, S. & Ryba, T. Cancer, metastasis, and the epigenome. Mol. Cancer. 23 (1), 154 (2024).
Wang, X. et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature 634 (8035), 970–978 (2024).
Baldo, B. A. & Pham, N. H. Non-targeted drugs for cancer therapy, Drug Allergy: Clinical Aspects, Diagnosis, Mechanisms, Structure-Activity Relationships, Springer2020, pp. 645–682.
Sawyers, C. Targeted cancer therapy. Nature 432 (7015), 294–297 (2004).
Casini, A. & Pöthig, A. Metals in cancer research: beyond platinum metallodrugs. ACS Cent. Sci. 10 (2), 242–250 (2024).
Garcia-Oliveira, P. et al. Status and challenges of Plant-Anticancer compounds in cancer treatment. Pharmaceuticals (Basel Switzerland) 14(2) (2021).
Lainé, A. L. & Passirani, C. Novel metal-based anticancer drugs: a new challenge in drug delivery. Curr. Opin. Pharmacol. 12 (4), 420–426 (2012).
Sarmento-Ribeiro, A. B., Scorilas, A., Goncalves, A. C., Efferth, T. & Trougakos, I. P. The emergence of drug resistance to targeted cancer therapies: clinical evidence. Drug Resist. Updates. 47, 100646 (2019).
Haider, T., Pandey, V., Banjare, N., Gupta, P. N. & Soni, V. Drug resistance in cancer: mechanisms and tackling strategies. Pharmacol. Rep. 72 (5), 1125–1151 (2020).
Kartal-Yandim, M., Adan-Gokbulut, A. & Baran, Y. Molecular mechanisms of drug resistance and its reversal in cancer. Crit. Rev. Biotechnol. 36 (4), 716–726 (2016).
Danesi, R. et al. Druggable targets meet oncogenic drivers: opportunities and limitations of target-based classification of tumors and the role of Molecular Tumor Boards, ESMO open 6(2) 100040. (2021).
Glorieux, C., Liu, S., Trachootham, D. & Huang, P. Targeting ROS in cancer: rationale and strategies. Nat. Rev. Drug Discovery. 23 (8), 583–606 (2024).
Feng, Y. et al. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 7 (12), 6124–6136 (2018).
Miao, P., Sheng, S., Sun, X., Liu, J. & Huang, G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. 65 (11), 904–910 (2013).
Hatami, H., Sajedi, A., Mir, S. M. & Memar, M. Y. Importance of lactate dehydrogenase (LDH) and monocarboxylate transporters (MCTs) in cancer cells. Health Sci. Rep. 6 (1), e996 (2023).
Claps, G. et al. The multiple roles of LDH in cancer. Nat. Reviews Clin. Oncol. 19 (12), 749–762 (2022).
Gallo, M. et al. Lactic dehydrogenase and cancer: an overview. Front. Biosci. (Landmark Ed). 20 (8), 1234–1249 (2015).
Le, A. et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. 107 (5), 2037–2042 (2010).
Granchi, C. et al. Assessing the differential action on cancer cells of LDH-A inhibitors based on the N-hydroxyindole-2-carboxylate (NHI) and malonic (Mal) scaffolds. Org. Biomol. Chem. 11 (38), 6588–6596 (2013).
Rai, G. et al. Discovery and optimization of potent, cell-active pyrazole-based inhibitors of lactate dehydrogenase (LDH). J. Med. Chem. 60 (22), 9184–9204 (2017).
Addadi, R. et al. Design, Synthesis, Characterization, in vitro biological Evaluation, and in Silico investigation of 4-Substituted arylidene pyrazolones through Docking, molecular dynamics Simulations, and DFT studies. ChemistrySelect 10 (28), e02315 (2025).
Momin, Y. & Beloshe, V. Pharmacophore modeling in drug design, Advances in Pharmacology, Elsevier2025, pp. 313–324.
Thakur, S. et al. Design and development of chromene-3-carboxylate derivatives as antidiabetic agents: exploring the antidiabetic potential via dual Inhibition of angiotensin II type 1 receptor and Neprilysin enzyme. Eur. J. Med. Chem. 117705. (2025).
Kapancık, S. et al. Chemical composition, cytotoxicity, and molecular Docking analyses of Thuja orientalis extracts. J. Mol. Struct. 1318, 139279 (2024).
Hazarika, S. et al. Investigation of antibacterial potential of natsiatum herpeticum Buch.-Ham. Ex Arn. Using in silico-in vitro approach. South. Afr. J. Bot. 164, 167–179 (2024).
Sanches, V. H. S. et al. Rational approach to new chemical entities with antiproliferative activity on Ab1 tyrosine kinase encoded by the BCR-ABL gene: an hierarchical biochemoinformatics analysis. Pharmaceuticals 17 (11), 1491 (2024).
Pradhan, T., Gupta, O. & Chawla, G. Identification of novel thiazolidine-4-one based hits as potential PPARγ modulators through in Silico workflow and validation through in vitro studies. J. Mol. Struct. 1339, 142391 (2025).
Abdullaha, M. et al. Methoxy-naphthyl-Linked N-Benzyl pyridinium styryls as dual cholinesterase inhibitors: Design, Synthesis, biological Evaluation, and Structure-Activity relationship. ACS Omega. 8 (20), 17591–17608 (2023).
Sharma, M., Thakur, S., Jadhav, H. R. & Bharate, S. B. Identification of azelastine and carvedilol as cholinesterase inhibitors via Structure-Based virtual screening of FDA-approved drugs. ChemistrySelect 8(28) (2023).
Srivastava, H. K. & Sastry, G. N. Molecular dynamics investigation on a series of HIV protease inhibitors: assessing the performance of MM-PBSA and MM-GBSA approaches. J. Chem. Inf. Model. 52 (11), 3088–3098 (2012).
Gopinath, P. & Kathiravan, M. K. Docking studies and molecular dynamics simulation of Triazole benzene sulfonamide derivatives with human carbonic anhydrase IX Inhibition activity. RSC Adv. 11 (60), 38079–38093 (2021).
Alimirah, F., Chen, J., Basrawala, Z., Xin, H. & Choubey, D. DU-145 and PC-3 human prostate cancer cell lines express androgen receptor: implications for the androgen receptor functions and regulation. FEBS Lett. 580 (9), 2294–2300 (2006).
Qiu, K. et al. Targeting USP10–FAK pathway sensitizes BCR-ABL + leukemia cells to tyrosine kinase inhibitors. Cell. Invest. 1 (2), 100017 (2025).
Li, W., Zhou, J. & Xu, Y. Study of the in vitro cytotoxicity testing of medical devices. Biomedical Rep. 3 (5), 617–620 (2015).
Mazlumoğlu, B. Ş. In vitro cytotoxicity test methods: MTT and NRU. Int. J. PharmATA. 3 (2), 50–53 (2023).
Allen, M., Millett, P., Dawes, E. & Rushton, N. Lactate dehydrogenase activity as a rapid and sensitive test for the quantification of cell numbers in vitro. Clin. Mater. 16 (4), 189–194 (1994).
Ocak, M. Changes in Polarity and Regeneration-Related gene expression in in vitro bone marrow mesenchymal stem cells in a rheumatoid arthritis injury model and Pharmacological modulation. Ahi Evran Med. J. 9 (1), 3–11 (2025).
Kumar, P., Nagarajan, A. & Uchil, P. D. Analysis of cell viability by the lactate dehydrogenase assay, Cold Spring Harbor Protocols 2018(6) pdb-prot095497. (2018).
Larsen, T. Determination of lactate dehydrogenase (LDH) activity in milk by a fluorometric assay. J. Dairy Res. 72 (2), 209–216 (2005).
Cai, W. et al. TRIM24 drives colorectal cancer progression via CAVIN2 degradation and ERK/RhoA pathway activation. Cell. Invest. 1 (3), 100033 (2025).
Eruslanov, E. & Kusmartsev, S. Identification of ROS using oxidized DCFDA and flow-cytometry, Advanced protocols in oxidative stress II, Springer2009, pp. 57–72.
Zhang, Q. Y. et al. Antioxidant dipeptide, cyclo (Phe-Phe), protects against bone dysplasia by suppression of phospholipid peroxidation. Cell. Invest. 1 (2), 100003 (2025).
Liou, G. Y., Kim, W. & Hobbs, T. M. Increased levels of oxidative stress in human prostate intraepithelial neoplasia and prostate cancer: evidence from 4-Hydroxyneonal detection and its Implications, antioxidants (Basel. Switzerland) 14(9) (2025).
Acknowledgements
The authors thank their respective institutions and the Hunan Provincial Health Commission for their support of the 2023 National Clinical Key Specialty Major Scientific Research Project (Z202318) and the National Key R&D Program of China (2024YFC3406800).
Funding
The work was supported by funding from the 2023 National Clinical Key Speciality Major Scientific Research Project (Z202318) and the National Key R&D Program of China (2024YFC3406800).
Author information
Authors and Affiliations
Contributions
Y.H. and Y.Z. conceived and designed the study. S.B. and U.P.Y. conducted the experiments and data collection. M.A.B. and A.V. performed the data analysis and interpretation. T.G.S. and N.B. contributed to the literature review and drafting of the manuscript. Y.H. and Y.Z. wrote the main manuscript text and prepared the figures. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, Y., Benni, S., Yadav, U.P. et al. Deploying the high-throughput virtual screening (HTVS) approach for the identification of new lactate dehydrogenase (LDH) inhibitors with anticancer assets. Sci Rep (2026). https://doi.org/10.1038/s41598-026-36385-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-36385-6