Abstract
Parkinson’s disease (PD) urgently requires blood-based markers that flag pathology before disabling motor decline. This study measured absolute concentrations of 144 plasma metabolites in 20 neurologically healthy adults and in 40 PD patients clinically classified as intermediate (PD-I) or progressive (PD-II). A multinomial logistic regression model was built to examine how changes in metabolite concentrations relate to disease stage and to assess their exploratory discriminative performance in this cohort. Five metabolites: glutamine, butyric acid, indoleacetic acid, phosphatidylcholine aa C40:2, and acylcarnitine C12:1 emerged as the smallest biomarker set that consistently separated controls, PD-I, and PD-II. When three non-motor manifestations often present in the prodromal phase (drooling, REM behavior disorder and depression) were added, the combined profile clearly distinguished controls from early-stage patients and improved classification of intermediate versus progressive disease. The selected metabolites play roles in gut-derived signaling, mitochondrial \(\beta\)-oxidation, and membrane lipid homeostasis, while the clinical variables mirror the recognized early spread of \(\alpha\)-synuclein pathology, together offering a coherent snapshot of systemic changes across PD progression. Because the panel can be quantified from a single small plasma aliquot and a brief clinical interview, it represents a promising exploratory finding that requires validation in larger, independent cohorts before any consideration for clinical application or pre-symptomatic screening.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is the second most prevalent neurodegenerative disease worldwide after Alzheimer’s disease (AD), affecting approximately 1% of people over 60 years of age1. This chronic and progressive disorder is characterized primarily by the loss of dopaminergic neurons in the substantia nigra pars compacta, alongside the pathological accumulation of Lewy bodies2. Clinically, PD is best recognized by its hallmark motor symptoms, including bradykinesia (slow movement), rigidity, resting tremor, and postural instability. In addition, patients often experience a broad spectrum of non-motor symptoms that significantly impact their quality of life, including cognitive deficits, autonomic dysfunction, mood disorders (depression and anxiety), sleep behavior disorders, and sensory abnormalities3,4,5. Despite advances in symptomatic treatments that focus primarily on motor impairments, there are currently no curative or disease-modifying therapies. This underscores the pressing need to deepen our understanding of PD’s underlying biochemical and physiological mechanisms.
The Global Burden of Disease, Injury and Risk Factors Study (GBD) reported an increase in PD cases from 2.5 million in 1990 to 6.1 million in 2016, with a further rise to 8.5 million by 2019. Over this period, the age-standardized prevalence rate also climbed, while the World Health Organization reported an 81% increase in disability-adjusted life years (DALYs) and a more than twofold rise in PD-related deaths since 2000. These trends reflect both population aging and improved diagnosis, emphasizing the importance of tracking PD epidemiology to guide public health interventions and allocate healthcare resources effectively6. PD’s etiology is multifactorial, involving intricate interactions between genetic susceptibilities and environmental influences. Familial PD linked to genetic mutations accounts for around 10–15% of cases, while idiopathic PD, with no clear genetic cause, represents the majority of cases. The diverse and heterogeneous molecular pathways involved in PD complicate its early diagnosis and the development of personalized therapeutic approaches7.
Metabolomics has become a crucial tool for uncovering biochemical alterations associated with neurodegenerative diseases such as PD, capturing metabolic changes resulting from disease progression and gene-environment interactions8. Both targeted and untargeted metabolomics studies conducted on biofluids such as cerebrospinal fluid (CSF), blood plasma, and urine have revealed disruptions in key metabolic pathways linked to energy metabolism, neurotransmitter synthesis, oxidative stress, inflammation, and cellular homeostasis in PD patients9,10,11,12. Significantly altered metabolites identified include increased plasma levels of \(\beta\)-hydroxybutyrate, an energy metabolism-related ketone body with neuroprotective properties, and decrease levels of essential amino acids such as tyrosine, leucine, and valine. Altered plasma tyrosine levels in PD patients reflect disruptions in dopamine synthesis and neurotransmission13,14. Plasma lipid metabolites, particularly lysophosphatidylcholines (lysoPCs), ceramides, and triglycerides, have also emerged as vital PD markers. These lipid alterations correlate with increased inflammation, oxidative stress, mitochondrial dysfunction, and disruptions in neuronal membrane integrity, which are significantly associated with motor symptom severity and disease progression15,16,17,18,19. Further metabolic disruptions involve purinergic pathways, specifically plasma metabolites like hypoxanthine (increased), xanthine (decreased), and inosine (decreased), which are thought to play roles in regulating oxidative stress and mitochondrial dysfunction15,20. Plasma metabolites related to inflammation, such as 12-KETE and ferulic acid, have shown altered levels in advanced PD stages, indicating their potential role in exacerbating neurodegenerative and inflammatory processes15. Similar metabolic disturbances have also been observed in AD, suggesting overlapping as well as distinct metabolic profiles among various neurodegenerative disorders. Notably, significant changes in energy metabolism metabolites, including lactic acid and pyruvate, and alterations in amino acid metabolism, such as valine and leucine, are common to both PD and AD, indicating shared metabolic disruptions21,22,23.
Despite the promising findings from metabolomic research, a critical limitation persists, as most studies rely on untargeted analyses or relative quantifications rather than absolute quantitative methodologies. This limitation restricts the robustness, reproducibility, and clinical applicability of findings, hindering the identification and validation of reliable biomarkers. Consequently, there is an urgent need for studies employing absolute quantitative metabolomic analyses to provide more precise and reproducible biomarkers, facilitating clinical translation and improving patient outcomes. The objective of this study is to perform an absolute quantitative analysis of the plasma metabolome in healthy controls and patients with intermediate and advanced stages of PD, aiming to identify specific metabolic alterations associated with the disease and to explore their relationship with disease progression.
Methods
Participant selection
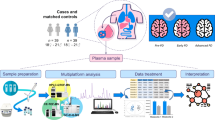
An overview of our experimental workflow from participant enrollment through clinical evaluation, metabolomic analysis and ultimately data analysis and biomarker discovery is shown in Fig. 1. In this cross-sectional comparative study, we included 40 patients with PD attending the Movement Disorder Clinic of the Instituto Nacional de Neurología y Neurocirugía Manuel Velasco Suárez in Mexico City. We only included patients who met the UK Parkinson’s Disease Society Brain Bank criteria for PD diagnosis24. At the time of inclusion, an expert neurologist clinically assessed patients using the Hoehn and Yahr score (H&Y) and the Unified Parkinson’s Disease Rating Scale (UPDRS) to evaluate the clinical stage of motor symptoms25,26. Beck’s Depression Inventory (BDI)27 was used to assess depressive symptoms in patients, and the Schwab and England Activities of Daily Living (SEADL) scale28 was used to evaluate participants’ independence in daily activities such as dressing, bathing, eating, and mobility. We used the H&Y score to divide PD patients into two groups: middle-stage PD (PD-I; n = 20) with a score of 1–3 and progressive-stage PD (PD-II; n = 20) with a score of 4–5.
The clinical interview documented each patient’s medical history, motor symptoms (tremor, rigidity, bradykinesia, postural instability), and non-motor symptoms (sleep disturbances, depression, anxiety, and drooling29,30) as well as pharmacological treatments. Among sleep-related manifestations, we specifically evaluated REM sleep behavior disorder (RBD), a highly specific non-motor feature of Parkinson’s disease. According to MDS clinical criteria31, RBD is characterized by the loss of normal REM atonia and the presence of vivid dreams accompanied by dream-enactment behaviors, reflecting dysregulation of pontine motor-inhibitory circuits. Medication status was documented for all patients; however, treatment regimens were heterogeneous in drug type, combinations, and dosage.
Twenty healthy controls, paired by age and sex with the patients, were also included in this study and each underwent the same neurological evaluation as the patient group. Exclusion criteria for both patients and controls were the absence of non-controlled chronic diseases, autoimmune diseases, infections, other neurological pathologies, and/or history of cancer.
Sample collection
For metabolomics analysis, we collected 6 mL of blood samples in a Vacutainer tube coated with K2-EDTA (BD). Samples were kept at 20\(^{\circ }\)C and processed to produce plasma within 1 hour of collection. The tubes were centrifuged at 2,000 x g for 10 minutes to generate the plasma samples. Aliquots of 500 \(\mu\)L of plasma were made and stored at −80 \(^{\circ }\)C until use.
Ethics
This study was approved by the Ethics Committee at the Instituto Nacional de Medicina Genómica (protocol number CEI 2024/09). The study adhered to all regulations for research on human subjects established by Mexican laws and the Declaration of Helsinki. All participants read and signed an informed consent.
Targeted metabolomic analysis
The targeted metabolomics workflow was based on previously established LC-MS/MS strategies for absolute quantification originally developed for other biological matrices and subsequently adapted for plasma analysis32,33,34,35.
A custom LC/DFI-MS/MS assay (TMIC PRIME ASSAY) was used for the targeted identification and quantification of up to 144 different endogenous metabolites from 50 \(\upmu\)L of plasma, including amino acids and amino acid derivatives, biogenic amines, acylcarnitines, glycerophospholipids, sphingomyelins, and organic acids. The method uses chemical derivatizations (for organic acids, amino acids, amino acid derivatives, and biogenic amines), analyte extraction, and LC separation (or DFI analysis for lipids), combined with selective mass-spectrometric detection using multiple reaction monitoring (MRM) pairs to identify and quantify metabolites. OptimaTM LC-MS grade water (Fisher Scientific, Ottawa, ON, CA) was used as a solvent for preparing stock solutions, calibration mixtures, QC mixtures, and a working isotope-labeled internal standard (ISTD) solution mixture. The working ISTD mixture included all stable-isotope–labeled analogues required for absolute quantification. These ISTDs included: D2-ornithine, 15N-histidine, D3-creatinine, D3-DOPA, D4-dopamine, 13C-tyrosine, 13C-D3-methionine, D3-proline, D4-serotonin, D4-putrescine, D3-sarcosine, 13C2-taurine, D4-tyramine, 15N-alanine, 13C6-arginine, 15N-asparagine, D3-aspartic acid, D7-citrulline, D3-glutamic acid, D5-glutamine, 13C2-glycine, 13C-leucine, 15N-phenylalanine, 13C-serine, D2-threonine, 15N2-tryptophan, D8-valine, D9-TMAO, 15N2-uric acid, D8-spermine, D8-spermidine, D6-diacetylspermine, D6-ADMA, D9-choline, D9-betaine, 2-D1-lactate, D4-\(\beta\)-hydroxybutyrate, 13C-\(\alpha\)-ketoglutarate, D4-citric acid, 13C-butyric acid, 13C-propionic acid, D4-succinic acid, 13C2-D2-fumaric acid, 13C-pyruvic acid, D2-hippuric acid, methyl-D3-malonic acid, and D2-indole-3-acetic acid. The concentration of the internal standard was defined within the linear detection range of the instrument for both the IS and the target analyte, aiming for the low-to-middle range of the standard curve (Supplementary Table 1).
For organic acid analyses, 150 \(\upmu\)L of ice-cold methanol and 10 \(\upmu\)L of the ISTD mixture were added to 50 \(\upmu\)L of plasma sample for overnight protein precipitation at −20 \(^{\circ }\)C, followed by centrifugation at 13,000\(\times\)g for 20 min. A total of 50 \(\upmu\)L of supernatant was loaded into the center of a 96-deep-well plate, followed by the addition of a 3-nitrophenylhydrazine (3-NPH) derivatization reagent prepared as 250 mM 3-NPH, 150 mM EDC in methanol, and 7.5% pyridine in 75% aqueous methanol. Samples were incubated for 2 h at room temperature with shaking. After incubation, butylated hydroxytoluene stabilizer (2 mg/mL) and water were added before the LC–MS injection.
For amino acids, biogenic amines, glycerophospholipids, acylcarnitines, and sphingomyelins, samples were thawed on ice, vortexed, and centrifuged at 13,000\(\times\)g; 10 \(\upmu\)L of each sample was then loaded onto the center of the filter on the upper 96-well plate and dried in a stream of nitrogen. Subsequently, a 5% phenyl-isothiocyanate (PITC) solution prepared in ethanol:water:pyridine (1:1:1, v/v/v) was added for derivatization and incubated for 20 min at room temperature, after which the filter spots were dried again using an evaporator. Metabolite extraction was then achieved by adding 300 \(\upmu\)L of extraction solvent. Extracts were obtained by centrifugation into the lower 96-deep-well plate, followed by a dilution step with MS running solvent (0.2% formic acid in water, 0.2% formic acid in acetonitrile).
Mass spectrometric analysis was performed on an ABSciex 4000 QTrap® tandem mass spectrometry instrument (Applied Biosystems/MDS Analytical Technologies, Foster City, CA) equipped with an Agilent 1290 series UHPLC system (Agilent Technologies, Palo Alto, CA). An Agilent reversed-phase Zorbax Eclipse XDB C18 column (3.0 mm \(\times\) 100 mm, 1.8 \(\mu\)m particle size, 80 Å pore size) with a Phenomenex (Torrance, CA) A SecurityGuard C18 guard column (4.0 mm \(\times\) 3.0 mm) was used for LC-MS/MS analysis. The controlling software for the sample analysis was Analyst 1.7.2 (Applied Biosystems/MDS Analytical Technologies, Foster City, CA; https://sciex.com/products/software/analyst-software).
The HPLC parameters used for the LC-MS/MS analysis of the PITC panel were as follows - solvent A: 0.2% (v/v) formic acid in water, and solvent B: 0.2% (v/v) formic acid in acetonitrile. The gradient profile for this UHPLC solvent run was as follows: t = 0 min, 0% B; t = 0.5 min, 0% B; t = 5.5 min, 95% B; t = 6.5 min, 95% B; t = 7.0 min, 0% B; and t = 9.5 min, 0% B. The column oven was set at 50 \(^{\circ }\)C. The flow rate was 500 \(\upmu\)L/min, and the sample injection volume was ten \(\upmu\)L.
The mass spectrometer was set to a positive electrospray ionization mode with a scheduled multiple reaction monitoring (MRM) scan. The IonSpray voltage was set at 5500 V and the temperature at 500 \(^{\circ }\)C. The curtain gas (CUR), ion source gas 1 (GS1), and ion source gas 2 (GS2) were set to 20 psi, 40 psi, and 50 psi, respectively. The collision gas (CAD) was set to a “medium” setting, corresponding to the instrument’s internal arbitrary units used by SCIEX systems. The entrance potential (EP) was set to 15 V. The declustering potential (DP), collision energy (CE), collision cell exit potential (CXP), MRM precursor ion (Q1), and fragment ion (Q3) were optimized individually for each analyte and isotope-labeled ISTD.
For DFI-MS/MS analysis, the UHPLC autosampler was connected directly to the MS ion source by red PEEK tubing. The DFI buffer mentioned above was used as the mobile phase, while the flow rate was programmed as follows: t = 0 min, 30 \(\upmu\)L/min; t = 1.6 min, 30 \(\upmu\)L/min; t = 2.4 min; 200 \(\upmu\)L/min; t = 2.8 min, 200 \(\upmu\)L/min; and t = 3.0 min, 30 \(\upmu\)L/min. The sample injection volume was 20 \(\upmu\)L. The IonSpray voltage was set at 5500 V and the temperature at 200 \(^{\circ }\)C. The CUR, GAS1, GAS2, and CAD were set at 20, 40, 50, and medium, respectively. The EP and CXP were set at 10 and 15 V for positive mode and −10 and −15 V, respectively, for negative mode.
Likewise, the DP, CE, Q1, and Q3 were optimized and set individually for each analyte and ISTD. For the analysis of organic acids by LC-MS/MS, the solvents used were (A) 0.01% (v/v) formic acid in water and (B) 0.01% (v/v) formic acid in acetonitrile. The gradient profile was as follows: t = 0 min, 25% B; t = 6.0 min, 65% B; t = 6.3 min, 90% B; t = 6.5 min, 100% B; t = 7.0 min, 100% B; t = 7.5 min, 25% B; t = 12.0 min, 25% B. The column oven was set to 40 \(^{\circ }\)C. The flow rate was 400 \(\upmu\)L/min, and the sample injection volume was 10 \(\upmu\)L. The mass spectrometer was set to a negative electrospray ionization mode with scheduled MRM scanning. The IonSpray voltage was set at −4500 V and the temperature at 400 \(^{\circ }\)C. The CUR, GAS1, GAS2, and CAD were set separately at 20, 30, 30 and medium. The EP was set at −10 V, and the DP, CE, CXP, Q1, and Q3 were optimized and set individually for all the analytes and isotope-labeled ISTDs.
Data analysis
All metabolite concentrations measured by the MS instrument were first log-transformed to stabilize variance and then autoscaled (mean centered and divided by the standard deviation) for Partial least squares discriminant analysis (PLS-DA). PLS-DA was performed in Metaboanalyst 6.036(https://www.metaboanalyst.ca/) to assess group separation between controls (CTL), intermediate-stage (PD-I) and progressive-stage (PD-II) Parkinson’s cohorts. 10-fold cross-validation (balanced stratified splits) was used to optimize the number of latent components and to evaluate classification accuracy. Variable importance in projection (VIP) scores were calculated from the final PLS-DA model, and metabolites with VIP scores greater than 1.5 were considered key discriminants.
Univariate analysis of each metabolite was carried out by one-way ANOVA on the log-transformed data, followed by Tukey’s honest significant difference (HSD) post hoc tests to identify pairwise differences among CTL, PD-I, and PD-II. Adjusted p-values (Holm-Bonferroni family-wise error rate (FWER)) <0.05 were deemed significant. To account for confounding by age and gender, each of the GA-selected metabolites was further tested with Analysis of Covariance (ANCOVA). Specifically, for each metabolite, we fitted the linear model \(metabolite \sim C(Label) + Age + C(Gender)\), using Type-II ANOVA to extract the group effect p-value. Metabolites with ANCOVA p <0.05 for the label term were retained as age and gender-adjusted features for downstream modeling.
For biomarker discovery, we implemented a penalized Genetic Algorithm (GA) in Python 3.8 (https://www.python.org) using the DEAP37 library (https://deap.readthedocs.io). Each individual in the GA population was a binary mask over the full metabolite feature set, encoding inclusion (1) or exclusion (0) of each variable. The GA parameters were: population size = 2,500; maximum generations = 500; elitism size = 2; tournament selection (size = 3); two-point crossover (probability = 0.5); bit-flip mutation (probability = 0.2); and a stagnation stop after 50 generations without fitness improvement. To enforce parsimony, individuals selecting zero or >8 features received a large negative fitness penalty. Fitness of each individual was defined as the mean Matthews Correlation Coefficient (MCC) from 10-fold cross-validation of a multinomial logistic-regression classifier (scikit-learn v1.638, LogisticRegression with solver = lbfgs, multi_class = multinomial, and max_iter = 10000; https://scikit-learn.org). The GA convergence history (best and mean MCC per generation) was monitored, and upon termination, the best individual defined an eight-metabolite signature. During each iteration of the genetic algorithm, candidate metabolite subsets encoded as binary masks were evaluated using cross-validated MCC. Individuals with higher fitness were preferentially retained, and new subsets were generated through tournament selection, two-point crossover, and bit-flip mutation. This iterative process produced successive populations enriched for high-performing solutions and continued until the predefined stopping criteria were met, yielding a compact subset of metabolites that maximized cross-validated performance under the model constraints. A final logistic-regression model was then trained using only the GA-selected features. Model performance was evaluated in combination with an exhaustive grid search within each 10-fold Cross Validation (CV) to tune the regularization strength of the GA-selected feature model. Using scikit-learn v1.6, we specified a multinomial logistic-regression estimator with solver = lbfgs, penalty = l2, and max_iter = 10000, and searched over \(C \in \{0.001, 0.01, 0.1, 0.5, 0.8, 1, 2, 3, 4, 10, 20, 30\}\) with the MCC scorer make_scorer(matthews_corrcoef). For each CV fold, the best hyperparameters were selected on the training split, then the model was refit and evaluated on the held-out split. Across the 10 CV folds, we computed and averaged. All figures were generated in Python using matplotlib v3.1039 (https://matplotlib.org) and seaborn v0.1340 (https://seaborn.pydata.org). Statistical tests were two-tailed, and \(p<0.05\) was considered significant unless otherwise stated.
To complement the GA-based feature selection, we implemented an MCC-optimized LASSO-regularized multinomial logistic regression to evaluate the stability of individual metabolites under an embedded penalization framework. All models were fitted using 10-fold stratified cross-validation and optimized for the Matthews correlation coefficient (MCC). To quantify feature stability, we performed 1,000 bootstrap resamples (sampling with replacement); in each resample, a LASSO model was fitted and metabolites with non-zero coefficients in any class were recorded. Selection frequencies were calculated as the proportion of bootstrap iterations in which each metabolite received a non-zero coefficient.
Multinomial and binary logistic regression
To distinguish among CTL, PD I, and PD II, we first fitted a multinomial logistic regression model to the data. Given a feature vector
where in our case \(p\) consists of the five log-transformed metabolite concentrations, the model computes for each class \(i\) a linear “score” (logit):
where \(\beta _{i0}\) is the intercept and \(\beta _{ij}\) is the coefficient for feature \(x_j\) in class \(i\).
Next, these three values \(\ell _1(\textbf{x}),\;\ell _2(\textbf{x}),\;\ell _3(\textbf{x})\) are converted into class probabilities \(\hat{p}_i(\textbf{x})\) via the softmax transformation:
so that \(\hat{p}_1(\textbf{x}) + \hat{p}_2(\textbf{x}) + \hat{p}_3(\textbf{x}) = 1\). Each sample is then assigned to the class with the highest \(\hat{p}_i(\textbf{x})\).
For the pairwise binary comparisons (CTL vs. PD I and PD I vs. PD II), we fitted separate binary logistic regression models with two feature sets (metabolites alone or metabolites plus clinical covariates and age). In each binary case, the model takes
and transforms this single logit into a probability via the logistic (sigmoid) function:
so that \(\hat{p}(\textbf{x})\in (0,1)\) represents the predicted probability of the positive class (PD I or PD II, respectively). A 10-fold stratified cross-validation procedure (random_state=42) was used to estimate classification performance. In each fold, the model was trained on 90% of the samples and tested on the held-out 10%. We recorded accuracy, precision, recall, \(F_1\) score, specificity, and Receiver Operating Characteristic Area Under the Curve (ROC AUC) on each held-out fold, and then reported the mean ± standard deviation across all 10 folds. After cross-validation, we refitted each binary model on the full dataset (for the chosen feature set) to obtain point estimates of \(\beta _0, \beta _1, \dots , \beta _p\). We then applied nonparametric bootstrap (1,000 replicates) by resampling \(N\) subjects with replacement, refitting the binary logistic model on each bootstrap sample, and recording the resulting coefficients to compute 95% confidence intervals for each parameter.
Results
Demographic and clinical characteristics
Tables 1 and 2 shows the demographic profile and prevalence of clinical and neurological symptoms among intermediate-stage (PD-I) and progressive-stage (PD-II) Parkinson’s disease patients. PD-II patients exhibited significantly higher rates of drooling, RBD, depression and hypomimia compared with PD-I patients. In contrast, tremor onset in the legs was more common in the PD-I group, and the mixed tremor–akinetic syndrome occurred more frequently in PD-II patients. No significant differences were observed for memory loss, falls, constipation, rigidity, or hypophonia. Additionally, PD-II patients were significantly older than both controls and PD-I participants.
Clinical severity scores across control and PD groups
Table 3 summarizes scores on the H&Y stage, UPDRS, and the SEADL scale for controls, PD-I, and PD-II. Both PD cohorts scored significantly worse than controls on all scales. Control participants had minimal impairment, intermediate-stage patients demonstrated moderate impairment, whereas progressive-stage patients exhibited severe deficits.
Plasma metabolome profiling across Parkinson disease stages
To investigate whether the clinical stage in PD is reflected in the circulating plasma metabolome, we first applied Partial Least Squares Discriminant Analysis (PLS-DA) to the log-transformed and auto-scaled dataset. The PLS-DA scores plot (Fig. 2) shows clear separation of controls (CTL) from both PD-I and PD-II, with partial overlap between the two disease stages. Variable Importance in Projection (VIP) scores, (Fig. 2), highlight the top discriminant metabolites (VIP > 1.5) and their stage-dependent abundance trends. The most relevant metabolites identified were: glycine, glutamine, PC aa C40:2, PC aa C40:1, sarcosine, asparagine, methylmalonic acid, putrescine, acylcarnitine C4, LysoPC a C18:1, propionic acid and hippuric acid. These compounds increase monotonically from CTL through PD-I to PD-II. Other relevant metabolites include: indoleacetic acid, glutamic acid, and carnitines C9, C3OH and C5, which decrease from CTL to PD stages. C5OH and LysoPC a C24:0 peak in PD-I before declining in PD-II.
Plasma metabolome discrimination and key drivers in Parkinson’s disease. a) PLS-DA scores plot of plasma metabolite profiles showing clear separation of control (CTL), intermediate-stage (PD-I), and progressive-stage (PD-II) groups along components 1 (6.5%) and 2 (9.5%). Ellipses denote 95% confidence intervals and arrows mark loading vectors for metabolites driving the highest separation between groups. b) VIP-score ranking of the top discriminant metabolites (VIP > 1.5), with adjacent heatmap tiles indicating relative mean abundance in CTL, PD-I, and PD-II (blue = low, red = high). Metabolite concentrations were log-transformed and autoscaled.
We then performed a one-way ANOVA on each metabolite (log-transformed concentrations) across the three groups, followed by Holm–Bonferroni post hoc tests to identify which pairwise contrasts drove the overall effects. A total of 22 metabolites showed significant group differences: Glycine, Glutamine, Glutamic acid, Total dimethylarginine, Lysine, Sarcosine, Butyric acid, Succinic acid, Fumaric acid, Methylmalonic acid, Indole acetic acid, PC aa C40:2, PC aa C40:1, C3:1, C4, C3OH, C6:1, C5OH, C5MDC, C9, C10:1, and C12:1. After adjusting for age and gender using ANCOVA, only 17 metabolites remained significant: butyric acid, C10:1, C12:1, C3OH, C5MDC, C5OH, C6:1, C9, glutamic acid, glutamine, glycine, indole acetic acid, lysine, PC aa C40:1, PC aa C40:2, sarcosine, and succinic acid. This indicates that five metabolites (total dimethylarginine, fumaric acid, methylmalonic acid, C3:1, and C4) were no longer significant after covariate adjustment (Table 4).
Among the remaining metabolites, plasma glycine was significantly elevated in PD-I and PD-II versus CTL. Plasma glutamine levels increased in both PD-I and PD-II relative to CTL, whereas plasma glutamic acid levels showed a tendency to decrease in both Parkinson stages compared to CTL. Plasma indoleacetic acid was lower in both PD-I and PD-II compared to CTL, and lysine was lower in PD-I than CTL. Sarcosine was elevated in both PD-I and PD-II versus CTL. Plasma butyric acid levels were reduced in PD-I but increased when comparing PD-I to PD-II. Succinic acid concentrations decreased in PD-I. The plasma levels of the acylcarnitine C3OH were lower in PD-I, while C5MDC was elevated in PD-I compared to CTL but decreased in PD-II relative to PD-I. Several other acylcarnitines exhibited alterations: C5OH was increased in both PD-I and PD-II compared to CTL, C6:1 was reduced in PD-I, and both C9 and C10:1 were decreased in PD-I, with C9 also decreasing in PD-II. C12:1 levels were lower in PD-I. Lastly, plasma phosphatidylcholines PC aa C40:2 and PC aa C40:1 were elevated in PD-I, with PC aa C40:2 also elevated in PD-II. Moreover, PC aa C40:1 levels increased further in PD-II compared to PD-I.
Biomarker panel discovery via penalized genetic algorithm
To summarize our stage-specific metabolomic findings into a compact exploratory biomarker panel, we first applied a penalized genetic algorithm (GA) to the full, log-transformed metabolite matrix. The GA was configured with a population size of 2,500, a maximum of 500 generations with a heavy penalty for solutions exceeding eight metabolites optimizing the Matthews correlation coefficient (MCC) (Fig. 3a). After convergence, in generation 120, the GA returned an eight-metabolite panel: glutamine, total dimethylarginine, butyric acid, methylmalonic acid, indole acetic acid, PC aa C40:2, acylcarnitine C4, and acylcarnitine C12:1.
Across independent GA runs under this configuration, the algorithm converged to similar high-performing solutions, with panels typically containing between five and eight metabolites and sharing a recurrent core subset. The penalization of models with more than eight variables restricted the search to compact combinations and yielded a small group of metabolites that were repeatedly selected in the best-performing panels.
Only five metabolites remained significant at p < 0.05 in the ANCOVA model adjusting for age and sex (Table 4): glutamine, butyric acid, indole acetic acid, PC aa C40:2, and C12:1. Figure 3b displays violin–dot plots of the sex and age-adjusted residuals for these metabolites. Figure 3c displays a t-SNE projection of all subjects using only these five ANCOVA-significant metabolites. Misclassified samples (by the final multiclass logistic-regression model built on those five features) are circled, and solid ellipses represent 95% confidence intervals for each diagnostic group. Most CTL samples form a tight cluster separate from PD-I and PD-II, with only a small number of misclassifications highlighted. Figure 3 d shows the one-versus-rest ROC curves for CTL, PD-I, and PD-II based on the logistic-regression model trained on the five ANCOVA-significant metabolites. Shaded areas indicate ±1 standard deviation (\(\delta\)) around each curve. The per-class AUCs (mean ± SD across folds) are 0.99 ± 0.04 (CTL), 0.86 ± 0.20 (PD-I), and 0.89 ± 0.12 (PD-II). The micro-averaged AUC is 0.90, and the macro-averaged AUC is 0.91.
The fitted logit functions are, for an observation x (vector of the five metabolite levels):
-
CTL: [3.6328 + (−1.3071) Glutamine + (0.5826) Butyric acid + (1.2816) Indole acetic acid + (−0.8892) PC aa C40:2 + (0.7906) C12:1]
-
PD-I: [−1.0859 + (0.0564) Glutamine + (−1.0952) Butyric acid + (−0.6249) Indole acetic acid + (0.2503) PC aa C40:2 + (−1.0600) C12:1]
-
PD-II: [−2.5469 + (1.2507) Glutamine + (0.5126) Butyric acid + (−0.6568) Indole acetic acid + (0.6389) PC aa C40:2 + (0.2694) C12:1]
Biomarker discovery strategy and metabolites selection. (a) Schematic of the penalized genetic-algorithm (GA) feature-selection pipeline, constrained to select at most eight features, used to optimize a multiclass logistic-regression classifier. (b) Violin–dot plots of the five metabolites that reached significance in ANCOVA (adjusted for age and gender; \(p<0.05\)). Each violin shows the distribution of the residuals (after adjusting for age and gender) for controls (CTL), intermediate-stage (PD-I), and progressive-stage (PD-II). Medians are indicated by horizontal bars; individual residuals are shown as dots; asterisks denote pairwise significance (*\(p<0.05\)) of post-hoc Holm–Bonferroni. (c) t-SNE projection of all subjects using only the five selected metabolites; misclassified samples are circled. Solid ellipses are 95% confidence intervals for each group. (d) One-vs-rest ROC curves for CTL, PD-I and PD-II, plus micro- and macro-averaged curves; AUC values are annotated in the legend.
To further evaluate the stability of the metabolite selection, we examined the results from the LASSO stability-selection analysis. The five metabolites with the highest selection frequencies across the 1,000 MCC-optimized LASSO bootstrap models (butyric acid, indole acetic acid, glycine, C4, and PC aa C40:2; Supplementary Table 2) showed overlap with the GA-derived five-metabolite set (glutamine, butyric acid, indole acetic acid, PC aa C40:2, and C12:1), differing only by glycine versus C12:1 and C4 versus glutamine. As shown in Supplementary Table 3, the GA-derived panel yielded higher mean accuracy, macro-averaged precision, recall, F1 score, MCC, and ROC AUC than the LASSO Top-5 panel, although the overall performance of both panels was comparable.
Binary logistic-regression models incorporating metabolites and clinical manifestations
Based on our five ANCOVA-significant metabolites, we next incorporated three clinical manifestations that differed between groups (drooling, RBD, and depression as found in Table 2, together with age (Table 1), into two separate binary logistic-regression models: one to distinguish controls (CTL) from intermediate-stage PD (PD-I), and another to distinguish PD-I from progressive-stage PD (PD-II). Each model used the same five metabolite predictors: glutamine, butyric acid, indole acetic acid, PC aa C40:2, and C12:1, plus the three binary clinical variables and age. The final logit equations were:
-
Logit for CTL vs PD-I (PD-I = 1):
$$\begin{aligned} {\begin{matrix} {{\,\textrm{logit}\,}}\bigl (P(\mathrm {PD\text {-}I}\mid x)\bigr ) & = 1.0379 + (0.8144)\,\text {Glutamine} - (0.8948)\,\text {Butyric acid} - (0.9876)\,\text {Indole acetic acid}\\ & \quad + (0.5385)\,\text {PC aa C40:2} - (0.9584)\,\text {C12:1} + (1.6084)\,\text {Drooling}\\ & \quad + (1.2118)\,\text {RBD} + (0.1990)\,\text {Depression} - (0.0805)\,\text {Age}. \end{matrix}} \end{aligned}$$ -
Logit for PD-I vs PD-II (PD-II = 1):
$$\begin{aligned} {\begin{matrix} {{\,\textrm{logit}\,}}\bigl (P(\mathrm {PD\text {-}II}\mid x)\bigr ) & = -28.7145 - (0.0117)\,\text {Glutamine} + (0.4840)\,\text {Butyric acid} - (0.1350)\,\text {Indole acetic acid}\\ & \quad + (0.1291)\,\text {PC aa C40:2} - (0.0624)\,\text {C12:1} + (0.8657)\,\text {Drooling}\\ & \quad + (0.6719)\,\text {RBD} + (0.3062)\,\text {Depression} + (0.4754)\,\text {Age}. \end{matrix}} \end{aligned}$$
Figure 4 illustrates the out-of-fold (OOF) probability distributions of each classifier alongside its corresponding ROC curve performance of the combined model (Metabolites + Clinical). The CTL vs PD-I bar chart (panel a) plots each subject’s normalized mean OOF probability of PD-I, with green bars for true CTL, blue for true PD-I, and red outlines indicating the four individuals misclassified in at least one of the ten cross-validation folds; the corresponding ROC curve shows a mean AUC of 0.97, confirming the excellent separation between controls and intermediate-stage patients. In panels (b)–(d), the PD-I vs PD-II bar chart (panel d) displays normalized mean OOF probabilities of PD-II (green = PD-I, blue = PD-II), highlighting four misclassified samples, and its ROC curve (panel d) achieves a mean AUC of 1.00, demonstrating an excellent discrimination of early versus progressive disease despite more subtle differences. Table 5 clearly shows that adding clinical variables to the five-metabolite panel further improves overall metrics, notably specificity.
Out-of-fold probability distributions and ROC curves for CTL vs PD-I and PD-I vs PD-II classifiers. Per-sample out-of-fold (OOF) probabilities (normalized to [0,1]) for each binary comparison. Bars are sorted by ascending mean OOF probability. In (a), green bars indicate true CTL and orange bars indicate true PD-I; in (b), green bars indicate true PD-I, and green bars indicate true PD-I. Any bar outlined in red denotes a subject misclassified in at least one of the 10 stratified cross-validation folds. (c), (d) Mean receiver-operating characteristic (ROC) curves obtained by averaging TPR vs. FPR across the 10 folds, with a shaded band representing ±1 SD of the fold-wise TPR at each FPR. Panel (c) corresponds to CTL vs PD-I, and panel (d) to PD-I vs PD-II. The mean area under the ROC curve (AUC) is reported in each legend.
Discussion
Quantitative metabolomics, which involves the precise measurement of absolute metabolite concentrations rather than relative abundances, remains a crucial and yet underutilized approach in PD research. PD metabolomics studies have historically been dominated by non-quantitative untargeted approaches. These studies reveal broad metabolic perturbations5,15,20,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58, but suffer from variability due to sample handling, instrument differences, matrix effects, ion suppression effects and normalization strategies. This variability complicates biomarker validation and impedes regulatory approval for clinical use. Quantitative metabolomics overcomes these hurdles by providing universal, robust, absolute concentrations that can be directly correlated with disease severity, treatment response, and progression59,60,61,62,63. In addition, absolute quantitation facilitates cross-study meta-analyses, the establishment of reference ranges, and longitudinal patient monitoring. These are critical elements for personalized medicine. Expanding quantitative metabolomics in PD will deepen mechanistic insights, enhance biomarker reliability, and accelerate the integration of metabolic signatures into clinical workflows, ultimately improving early diagnosis and enabling more precise therapeutic interventions.
This study identified a panel of three polar metabolites (glutamine, butyric acid, and indoleacetic acid) and two lipids (PC aa C40:2, and acylcarnitine C12:1) in plasma that were significantly different between PD patients and controls, after being adjusted for age and sex.
The increase in plasma glutamine levels in the intermediate and progressive PD groups corroborates prior evidence linking alterations in glutamine metabolism to excitotoxicity and mitochondrial dysfunction in PD pathophysiology64. In PD, elevated glutamine levels have been consistently reported in blood, urine, and cerebrospinal fluid (CSF) across eight studies62,65,66,67,68,69,70,71. This amino acid serves as a key precursor for neurotransmitters, supporting the glutamine–glutamate–GABA cycle that is critical for maintaining synaptic function.
The gut-brain axis emerges prominently in PD metabolic alterations, with butyric acid and indoleacetic acid (both microbiota-derived metabolites) showing significant deviations in PD patients. Butyric acid, a short-chain fatty acid, exhibits reduced plasma concentrations in early stages of PD, consistent with reports of gut microbial dysbiosis impairing neuroprotective and anti-inflammatory mechanisms72,73. Its subsequent partial restoration in advanced stages may reflect complex host-microbiome interactions during disease progression. Indoleacetic acid (IAA), a tryptophan metabolite with immunomodulatory roles, is consistently diminished in PD, supporting hypotheses that altered microbial metabolism contributes to systemic inflammation and intestinal barrier dysfunction implicated in PD pathology74. These findings reinforce the potential of gut-derived metabolites as biomarkers linking peripheral metabolic states to central neurodegeneration. In PD, dysregulation of the gut-brain axis is increasingly recognized as being important, with changes in colon permeability, inflammation, and motility preceding motor symptoms. Reduced levels of butyrate and IAA-producing bacteria in PD patients, which lead to diminished intestinalbarrier function, have been postulated to enable prion-like spread of \(\alpha\)-synuclein from the gut-to-brain via the enteric nervous system75.
Mitochondrial dysfunction is captured by alterations in acylcarnitine C12:1, reflecting disrupted fatty acid \(\beta\)-oxidation and energy homeostasis, a pattern previously documented in PD cohorts44,53,56. This lipid precursor not only highlights mitochondrial metabolic inefficiencies but also serves as an accessible peripheral marker that correlates with disease stage, offering insights into systemic manifestations of neuronal bioenergetic deficits. In line with this observation, Saiki et al76 found a decrease in long chain acylcarnitines (LCACs) in the early stage of Parkinson’s disease, suggesting primary changes in the skeletal muscles, unrelated to exercise and medication. Although not included in the final model, we found nine dysregulated acylcarnitine’s, and most of them (except C4 and C5OH) were decreased in PD-I.
Phosphatidylcholines play crucial roles in maintaining membrane integrity and regulating lipid homeostasis, making them essential components of neuronal membranes. They also participate actively in cell signaling and responses to oxidative stress. Recent Mendelian randomization analyses77,78 have linked higher serum levels of certain phosphatidylcholine species with a lower risk of PD. However, studies examining phosphatidylcholine levels in PD patients have produced conflicting results; while some report decreased levels, particularly in brain tissue and plasma, others have observed increased concentrations79, likely reflecting differences in phosphatidylcholine species, tissues analyzed, and metabolic contexts. Supporting the relevance of our identified phosphatidylcholine, a study using a sleep-deprived rotenone-induced rat model of PD identified PC aa C40:2 as significantly altered in the diseased state80. Intriguingly, this same lipid has also been implicated in AD; Mapstone et al. identified PC aa C40:2 as part of a lipid panel capable of predicting progression from normal cognition to mild cognitive impairment or AD in older adults with approximately 90% accuracy81. Similarly, PC aa C40:2 levels were significantly reduced in plasma samples from amyotrophic lateral sclerosis (ALS) patients relative to healthy controls82. Collectively, the recurring disruption of PC metabolism across these neurodegenerative disorders highlights a shared underlying theme of lipid dysregulation in neurodegeneration.
Combining these five metabolites into a biomarker panel provides a multidimensional view of metabolic perturbations in PD, encompassing gut microbiome-derived metabolites, mitochondrial and energy metabolism markers, and markers of membrane and signaling function. This integrative approach improves sensitivity and specificity by capturing diverse biochemical pathways affected in PD. Previous metabolomics studies have underscored the individual potential of several of these metabolites as biomarkers; however, their combined predictive power remains underexplored83. This five-metabolite panel exhibited an excellent performance, with an AUC macro of 0.91.
In addition to the final five-metabolite panel, several metabolites (notably glycine and its N-methyl derivative sarcosine) appeared repeatedly in intermediate analysis and showed nominal group differences, but were ultimately excluded once we enforced very small, clinically feasible panels. Their omission therefore reflects statistical and practical constraints (limited sample size, parsimony penalties, and redundancy among correlated predictors) rather than a conclusion that these metabolites lack biological relevance. In fact, converging metabolomics studies have consistently reported increased glycine levels in plasma, urine and CSF of PD patients, and have highlighted disruption of glycine–serine–threonine metabolism as a recurring biochemical signature in PD83. Glycine is an inhibitory neurotransmitter amino acid that can modulate dopaminergic and glutamatergic transmission and participates in glutathione synthesis, linking it to neurotransmitter imbalance and oxidative stress, both central to PD pathophysiology47. Sarcosine, as a methylated glycine derivative, integrates into one-carbon metabolism, and recent targeted analyses in PD cohorts have shown that sarcosine and betaine track homocysteine increases and cognitive status (including PD dementia), suggesting that perturbations in methyl-group transfer and one-carbon flux are relevant to disease progression84. Within this context, the transient selection of glycine and sarcosine in our early models likely reflects broader amino acid and one-carbon metabolic disturbances that are only partially captured by the final minimal panel. These metabolites should therefore be viewed as candidates for further investigation in larger, independent cohorts, particularly using targeted and pathway-based approaches that can more rigorously evaluate their contribution to PD-related metabolic signatures.
We complemented the metabolite panel with three clinical manifestations (drooling, RBD and depression) that were found significantly different between intermediate Parkinson and progressive Parkinson and controls (Table 2). According to Braak staging85, the progression of PD follows a predictable pattern of \(\alpha\)-synuclein pathology spreading from the peripheral and lower brain stem regions toward the cortex. These symptoms are categorized into autonomic, sensory, sleep-related, neuropsychiatric, and cognitive domains. In line with this framework, we selected characteristic non-motor symptoms that emerge temporally across Braak stages 1 and 2, reflecting their occurrence during early and prodromal stages of the disease. Autonomic dysfunction is a fundamental and often early feature of PD, impacting more than 90% of patients and involving various physiological systems86,87. These impairments commonly emerge before the onset of motor symptoms, reflecting the widespread neurodegeneration that extends beyond the basal ganglia. Despite its prevalence and clinical significance, autonomic dysfunction is frequently overlooked and inadequately addressed, typically recognized only when symptoms reach a severe and disruptive stage. Drooling affects between 37–84% of PD patients and at early stage. RBD are among the most prevalent and disabling non-motor symptoms of PD, affecting between 60% and 98% of patients. Neuropsychiatric symptoms are now widely acknowledged as integral features of PD, frequently appearing during the prodromal phase and significantly impacting both quality of life and the overall course of the disease88. The addition of these non-motor symptoms to the metabolite panel increased the performance of most of the metrics (Table 5).
The integration of targeted quantitative metabolomics with multivariate modeling provided a structured framework for identifying metabolic signals associated with PD stages. Across analyses, several metabolites showed consistent deviations in PD and converged onto pathways related to neurotransmitter cycling, gut–microbiome metabolism, mitochondrial function, and membrane lipid homeostasis. These biochemical domains are well aligned with established PD pathophysiology and reinforce the biological interpretability of the findings.
The final five-metabolite panel (glutamine, butyric acid, indoleacetic acid, PC aa C40:2, and acylcarnitine C12:1) captures complementary aspects of PD-related dysregulation, spanning amino acid metabolism, short-chain fatty acids, tryptophan–microbial metabolism, lipid remodeling, and mitochondrial \(\beta\)-oxidation. Although the present work was not designed to establish a clinically deployable classifier, the internal consistency of these metabolic shifts highlights their potential translational value and supports their further assessment in independent, larger, and longitudinal cohorts. A validated metabolic panel could eventually contribute to earlier detection, improved staging, and more precise monitoring of metabolic disturbances linked to neurodegeneration.
Study limitations
This study has several limitations that should be considered when interpreting the results. The relatively small sample size (n = 60) limits statistical power and increases the risk of overfitting, even under cross-validation. In addition, the PD-II group was significantly older than PD-I and controls, and age-related metabolic variation may partially contribute to the observed differences despite ANCOVA adjustment. Medication status was documented; however, treatment regimens varied widely across participants, including heterogeneity in dopaminergic therapies (e.g., levodopa, pramipexole) and other medications. This variability limits our ability to control for pharmacological effects on plasma metabolite levels and may introduce bias, thereby restricting the generalizability of the findings. Also, the GA-based models in this study should be viewed as exploratory tools for identifying candidate metabolite combinations. Future validation in larger cohorts using simpler, small-sample-appropriate frameworks will be essential to confirm the reproducibility and potential clinical utility of the identified metabolite panel.
Despite these limitations, the study also has several strengths. We used targeted, absolute quantitative LC–MS/MS with internal standards and stringent quality control, adjusted key comparisons for age and sex, and applied complementary feature-selection approaches (GA and LASSO), which consistently converged on a compact and biologically plausible metabolite subset. Although the results should be interpreted as exploratory and hypothesis-generating, the reproducible cross-validated performance, the agreement between independent algorithms, and the alignment of identified metabolites with known PD-related pathways support the biological coherence of the observed trends and justify future validation in larger, independent, and clinically well-characterized cohorts.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Su, D. et al. Projections for prevalence of parkinson’s disease and its driving factors in 195 countries and territories to 2050: modelling study of global burden of disease study 2021. BMJ 388, https://doi.org/10.1136/bmj-2024-080952 (2025). https://www.bmj.com/content/388/bmj-2024-080952.full.pdf.
Wakabayashi, K., Tanji, K., Mori, F. & Takahashi, H. The lewy body in parkinson’s disease: molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology 27, 494–506. https://doi.org/10.1111/j.1440-1789.2007.00803.x (2007).
Dahabiyeh, L. A., Nimer, R. M., Rashed, M., Wells, J. D. & Fiehn, O. Serum-based lipid panels for diagnosis of idiopathic parkinson’s disease. Metabolites 13, https://doi.org/10.3390/metabo13090990 (2023).
Ma, Z.-l. et al. Biomarkers of parkinson; disease: From basic research to clinical practice. Aging and disease 15, 1813–1830, https://doi.org/10.14336/ad.2023.1005 (2024).
Lokhov, P. G. et al. Application of clinical blood metabogram for diagnosis of early-stage parkinson’s disease: a pilot study. Front. Mol. Biosci. Volume 11 - 2024, https://doi.org/10.3389/fmolb.2024.1407974 (2024).
Luo, Y. et al. Global, regional, national epidemiology and trends of parkinson’s disease from 1990 to 2021: findings from the global burden of disease study 2021. Front. Aging Neurosci. Volume 16 - 2024, https://doi.org/10.3389/fnagi.2024.1498756 (2025).
Zafar, S. & Yaddanapudi, S. S. Parkinson disease. In StatPearls [Internet] (StatPearls Publishing, Treasure Island (FL), 2025). Updated 2023 Aug 7.
Lanznaster, D., Dingeo, G., Samey, R. A., Emond, P. & Blasco, H. Metabolomics as a crucial tool to develop new therapeutic strategies for neurodegenerative diseases. Metabolites 12, https://doi.org/10.3390/metabo12090864 (2022).
Ostrakhovitch, E. A., Ono, K. & Yamasaki, T. R. Metabolomics in parkinson’s disease and correlation with disease state. Metabolites 15, https://doi.org/10.3390/metabo15030208 (2025).
Luan, H. et al. Comprehensive urinary metabolomic profiling and identification of potential noninvasive marker for idiopathic parkinson’s disease. Sci. Reports 5, 13888. https://doi.org/10.1038/srep13888 (2015).
Willkommen, D. et al. Metabolomic investigations in cerebrospinal fluid of parkinson’s disease. PLoS One 13, e0208752. https://doi.org/10.1371/journal.pone.0208752 (2018).
Plewa, S. et al. The metabolomic approach reveals the alteration in human serum and cerebrospinal fluid composition in parkinson’s disease patients. Pharm. (Basel) 14, https://doi.org/10.3390/ph14090935 (2021).
Judd, J. M. et al. Inflammation and the pathological progression of alzheimer’s disease are associated with low circulating choline levels. Acta Neuropathol 146, 565–583. https://doi.org/10.1007/s00401-023-02616-7 (2023).
Abdik, E. & Çakir, T. Transcriptome-based biomarker prediction for parkinson’s disease using genome-scale metabolic modeling. Sci. Reports 14, 585. https://doi.org/10.1038/s41598-023-51034-y (2024).
Santos, W. T. et al. Metabolomics unveils disrupted pathways in parkinson’s disease: Toward biomarker-based diagnosis. ACS Chem. Neurosci. 15, 3168–3180. https://doi.org/10.1021/acschemneuro.4c00355 (2024).
Yin, K. F. et al. Causal association and mediating effect of blood biochemical metabolic traits and brain image-derived endophenotypes on alzheimer’s disease. Heliyon 10, e27422. https://doi.org/10.1016/j.heliyon.2024.e27422 (2024).
Vardarajan, B. et al. Lysophosphatidylcholines are associated with p-tau181 levels in early stages of alzheimer’s disease. Res Sq https://doi.org/10.21203/rs.3.rs-3346076/v1 (2024).
Pan, X. et al. Plasma metabolites distinguish dementia with lewy bodies from alzheimer’s disease: a cross-sectional metabolomic analysis. Front. Aging Neurosci. Volume 15 - 2023, https://doi.org/10.3389/fnagi.2023.1326780 (2024).
Dahabiyeh, L. A., Nimer, R. M., Wells, J. D., Abu-Rish, E. Y. & Fiehn, O. Diagnosing parkinson’s disease and monitoring its progression: Biomarkers from combined gc-tof ms and lc-ms/ms untargeted metabolomics. Heliyon 10, e30452. https://doi.org/10.1016/j.heliyon.2024.e30452 (2024).
de Lope, E. G. et al. Comprehensive blood metabolomics profiling of parkinson’s disease reveals coordinated alterations in xanthine metabolism. npj Park. Dis. 10, 68, https://doi.org/10.1038/s41531-024-00671-9 (2024).
Ambeskovic, M. et al. Metabolomic signatures of alzheimer’s disease indicate brain region-specific neurodegenerative progression. Int. J. Mol. Sci. 24, 14769 (2023).
Nielsen, J. E. et al. Serum metabolic signatures for alzheimer’s disease reveal alterations in amino acid composition: a validation study. Metabolomics 20, 12. https://doi.org/10.1007/s11306-023-02078-8 (2024).
Schweickart, A. et al. Serum and csf metabolomics analysis shows mediterranean ketogenic diet mitigates risk factors of alzheimer’s disease. NPJ Metab Health Dis 2, 15. https://doi.org/10.1038/s44324-024-00016-3 (2024).
Kimpara, T. & Takeda, A. Clinical diagnostic criteria for parkinson’s disease. Neurol. Ther. 37, 10–15, https://doi.org/10.15082/jsnt.37.1_10 (2020).
Hoehn, M. M. & Yahr, M. D. Parkinsonism: onset, progression and mortality. Neurology 17, 427–42. https://doi.org/10.1212/wnl.17.5.427 (1967).
Status and recommendations. Disease, M. D. S. T. F. o. R. S. f. P. The unified parkinson’s disease rating scale (updrs). Mov. Disord. 18, 738–750. https://doi.org/10.1002/mds.10473 (2003).
Beck, A. T., Ward, C. H., Mendelson, M., Mock, J. & Erbaugh, J. An inventory for measuring depression. Arch Gen Psychiatry 4, 561–71. https://doi.org/10.1001/archpsyc.1961.01710120031004 (1961).
Schwab, R. S. & England, A. C. Projection technique for evaluating surgery in parkinson’s disease. In Gillingham, F. J. & Donaldson, I. M. L. (eds.) Third Symposium on Parkinson’s Disease, 152–157 (E. & S. Livingstone, Edinburgh, 1969).
Proulx, M., de Courval, F. P., Wiseman, M. A. & Panisset, M. Salivary production in parkinson’s disease. Mov Disord 20, 204–7. https://doi.org/10.1002/mds.20189 (2005).
Srivanitchapoom, P., Pandey, S. & Hallett, M. Drooling in parkinson’s disease: a review. Park. Relat Disord 20, 1109–18. https://doi.org/10.1016/j.parkreldis.2014.08.013 (2014).
Chaudhuri, K. R. et al. The movement disorder society nonmotor rating scale: Initial validation study. Mov Disord 35, 116–133. https://doi.org/10.1002/mds.27862 (2020).
Zheng, J., Zhang, L., Johnson, M., Mandal, R. & Wishart, D. S. Comprehensive targeted metabolomic assay for urine analysis. Anal. Chem. 92, 10627–10634. https://doi.org/10.1021/acs.analchem.0c01682 (2020).
López-Hernández, Y. et al. The urinary metabolome of newborns with perinatal complications. Metabolites 14, 41. https://doi.org/10.3390/metabo14010041 (2024).
Zubkowski, A. et al. Quantitative comparison of whole blood, plasma and serum metabolomes across different blood collection methods. Metabolomics 21, 146. https://doi.org/10.1007/s11306-025-02345-w (2025).
López-Hernández, Y. et al. Targeted metabolomics identifies high performing diagnostic and prognostic biomarkers for covid-19. Sci. Reports 11, 14732. https://doi.org/10.1038/s41598-021-94171-y (2021).
Pang, Z. et al. Metaboanalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 52, W398–W406, https://doi.org/10.1093/nar/gkae253 (2024).
Fortin, F.-A., De Rainville, F.-M., Gardner, M.-A., Parizeau, M. & Gagné, C. DEAP: Evolutionary algorithms made easy. J. Mach. Learn. Res. 13, 2171–2175 (2012).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Hunter, J. D. Matplotlib: A 2d graphics environment. Comput. Sci. & Eng. 9, 90–95. https://doi.org/10.1109/MCSE.2007.55 (2007).
Waskom, M. L. seaborn: statistical data visualization. J. Open Source Softw. 6, 3021, https://doi.org/10.21105/joss.03021 (2021).
Bogdanov, M. et al. Metabolomic profiling to develop blood biomarkers for parkinson’s disease. Brain 131, 389–396. https://doi.org/10.1093/brain/awm304 (2008).
Hatano, T., Saiki, S., Okuzumi, A., Mohney, R. P. & Hattori, N. Identification of novel biomarkers for parkinson’s disease by metabolomic technologies. J. Neurol. Neurosurg. & Psychiatry 87, 295–301. https://doi.org/10.1136/jnnp-2014-309676 (2015).
LeWitt, P. A., Li, J., Lu, M., Guo, L. & Auinger, P. Metabolomic biomarkers as strong correlates of parkinson disease progression. Neurology 88, 862–869. https://doi.org/10.1212/WNL.0000000000003663 (2017).
Burté, F. et al. Metabolic profiling of parkinson’s disease and mild cognitive impairment. Mov. Disord. 32, 927–932. https://doi.org/10.1002/mds.26992 (2017).
Han, W., Sapkota, S., Camicioli, R., Dixon, R. A. & Li, L. Profiling novel metabolic biomarkers for parkinson’s disease using in-depth metabolomic analysis. Mov. Disord. 32, 1720–1728. https://doi.org/10.1002/mds.27173 (2017).
Zhao, H. et al. Potential biomarkers of parkinson’s disease revealed by plasma metabolic profiling. J. Chromatogr. B 1081–1082, 101–108. https://doi.org/10.1016/j.jchromb.2018.01.025 (2018).
Stoessel, D. et al. Promising metabolite profiles in the plasma and csf of early clinical parkinson’s disease. Front. Aging Neurosci. 10, https://doi.org/10.3389/fnagi.2018.00051 (2018).
Okuzumi, A. et al. Metabolomics-based identification of metabolic alterations in PARK2. Annals Clin. Transl. Neurol. 6, 525–536. https://doi.org/10.1002/acn3.724 (2019).
Picca, A. et al. Circulating amino acid signature in older people with parkinson’s disease: A metabolic complement to the EXosomes in parkinson disease ( EXPAND) study. Exp. Gerontol. 128, 110766. https://doi.org/10.1016/j.exger.2019.110766 (2019).
Molsberry, S. et al. Plasma metabolomic markers of insulin resistance and diabetes and rate of incident parkinson’s disease. J. Park. Dis 10, 1011–1021. https://doi.org/10.3233/JPD-191896 (2020).
Lokhov, P. G., Trifonova, O. P., Maslov, D. L., Lichtenberg, S. & Balashova, E. E. Diagnosis of parkinson’s disease by a metabolomics-based laboratory-developed test (ldt). Diagnostics 10, 332. https://doi.org/10.3390/diagnostics10050332 (2020).
Dong, M.-X., Wei, Y.-D. & Hu, L. Lipid metabolic dysregulation is involved in parkinson’s disease dementia. Metab. Brain Dis. 36, 463–470. https://doi.org/10.1007/s11011-020-00665-5 (2021).
Albillos, S. M. et al. Plasma acyl-carnitines, bilirubin, tyramine and tetrahydro-21-deoxycortisol in parkinson’s disease and essential tremor: A case-control biomarker study. Park. & Relat. Disord. 91, 167–172. https://doi.org/10.1016/j.parkreldis.2021.09.014 (2021).
Talavera Andújar, B. n. et al. Studying the parkinson’s disease metabolome and exposome in biological samples through different analytical and cheminformatics approaches: A pilot study. Anal. Bioanal. Chem. 414, 7399–7419, https://doi.org/10.1007/s00216-022-04207-z (2022).
Zhang, J. D., Xue, C., Kolachalama, V. B. & Donald, W. A. Interpretable machine learning on metabolomics data reveals biomarkers for parkinson’s disease. ACS Cent. Sci. 9, 1035–1045. https://doi.org/10.1021/acscentsci.2c01468 (2023).
Carrillo, F. et al. Multiomics approach discloses lipid and metabolite profiles associated with parkinson’s disease stages and applied therapies. Neurobiol. Dis. 202, 106698. https://doi.org/10.1016/j.nbd.2024.106698 (2024).
Tkachenko, K., González-Sáiz, J. M. & Pizarro, C. Untargeted lipidomics reveals potential biomarkers in plasma samples for the discrimination of patients affected by parkinson’s disease. Molecules 30, 850. https://doi.org/10.3390/molecules30040850 (2025).
Liu, S. et al. Unraveling the relation of parkinson’s disease and metabolites: A combined analysis of stool and plasma metabolites based on untargeted metabolomics technology. CNS Neurosci. & Ther.31, —, https://doi.org/10.1111/cns.70424 (2025).
Chang, K.-H. et al. Alterations of metabolic profile and kynurenine metabolism in the plasma of parkinson’s disease. Mol. Neurobiol. 55, 6319–6328. https://doi.org/10.1007/s12035-017-0845-3 (2018).
D’Andrea, G. et al. Different circulating trace amine profiles in De Novo and treated parkinson’s disease patients. Sci. Reports 9, —, https://doi.org/10.1038/s41598-019-42535-w (2019).
Klatt, S. et al. A six-metabolite panel as potential blood-based biomarkers for parkinson’s disease. npj Park. Dis. 7, 94, https://doi.org/10.1038/s41531-021-00239-x (2021).
Chang, K.-H. et al. Alterations of sphingolipid and phospholipid pathways and ornithine level in the plasma as biomarkers of parkinson’s disease. Cells 11, 395. https://doi.org/10.3390/cells11030395 (2022).
D’Ascenzo, N. et al. Metabolomics of blood reveals age-dependent pathways in parkinson’s disease. Cell & Biosci. 12, —, https://doi.org/10.1186/s13578-022-00831-5 (2022).
Wang, J., Wang, F., Mai, D. & Qu, S. Molecular mechanisms of glutamate toxicity in parkinson’s disease. Front. Neurosci. 14, 585584. https://doi.org/10.3389/fnins.2020.585584 (2020).
Schulte, E. C. et al. Alterations in lipid and inositol metabolisms in two dopaminergic disorders. PLOS ONE 11, e0147129. https://doi.org/10.1371/journal.pone.0147129 (2016).
Dong, M.-X., Hu, L., Wei, Y.-D. & Chen, G.-H. Metabolomics profiling reveals altered lipid metabolism and identifies a panel of lipid metabolites as biomarkers for parkinson’s disease related anxiety disorder. Neurosci. Lett. 745, 135626. https://doi.org/10.1016/j.neulet.2021.135626 (2021).
Yilmaz, A. et al. Metabolic profiling of csf from people suffering from sporadic and LRRK2 parkinson’s disease: A pilot study. Cells 9, 2394. https://doi.org/10.3390/cells9112394 (2020).
Kumari, S. et al. Identification of potential urine biomarkers in idiopathic parkinson’s disease using NMR. Clin. Chimica Acta 510, 442–449. https://doi.org/10.1016/j.cca.2020.08.005 (2020).
Wuolikainen, A. et al. Multi-platform mass spectrometry analysis of the csf and plasma metabolomes of rigorously matched amyotrophic lateral sclerosis, parkinson’s disease and control subjects. Mol. BioSystems 12, 1287–1298. https://doi.org/10.1039/c5mb00711a (2016).
Nagesh Babu, G. et al. Serum metabolomics study in a group of parkinson’s disease patients from northern india. Clin. Chimica Acta 480, 214–219, https://doi.org/10.1016/j.cca.2018.02.022 (2018).
Toczylowska, B., Zieminska, E., Michalowska, M., Chalimoniuk, M. & Fiszer, U. Changes in the metabolic profiles of the serum and putamen in parkinson’s disease patients - In Vitro and In Vivo NMR spectroscopy studies. Brain Res. 1748, 147118. https://doi.org/10.1016/j.brainres.2020.147118 (2020).
Unger, M. M. et al. Short chain fatty acids and gut microbiota differ between patients with parkinson’s disease and age-matched controls. Park. & Relat. Disord. 32, 66–72. https://doi.org/10.1016/j.parkreldis.2016.08.019 (2016).
Bedarf, J. R. et al. Functional implications of microbial and viral gut metagenome changes in early-stage L-dopa-naïve parkinson’s disease patients. Genome Medicine 9, 39. https://doi.org/10.1186/s13073-017-0428-y (2017).
Shao, Y. et al. Comprehensive metabolic profiling of parkinson’s disease by liquid chromatography-mass spectrometry. Mol. Neurodegener. 16, 4. https://doi.org/10.1186/s13024-021-00425-8 (2021).
Oliver, P. J., Civitelli, L. & Hu, M. T. The gut-brain axis in early parkinson’s disease: From prodrome to prevention. J. Neurol. 272, 1477–1492. https://doi.org/10.1007/s00415-025-13138-5 (2025).
Saiki, S. et al. Decreased long-chain acylcarnitines from insufficient β-oxidation as potential early diagnostic markers for parkinson’s disease. Sci. Reports 7, 1389. https://doi.org/10.1038/s41598-017-06767-y (2017).
Zhang, H., Cao, F., Yu, J., Liang, Y. & Wu, Y. Investigating plasma lipid profiles in association with parkinson’s disease risk. npj Park. Dis. 11, 7, https://doi.org/10.1038/s41531-025-00955-8 (2025).
Qin, Y. et al. Plasma lipidome, circulating inflammatory proteins, and parkinson’s disease: A mendelian randomisation study. Front. Aging Neurosci. 16, 1424056. https://doi.org/10.3389/fnagi.2024.1424056 (2024).
Miletić Vukajlović, J. et al. Increased plasma phosphatidylcholine/lysophosphatidylcholine ratios in patients with parkinson’s disease. Rapid Commun. Mass Spectrom. 34, e8595. https://doi.org/10.1002/rcm.8595 (2020).
Fagotti, J. et al. Chronic sleep restriction in the rotenone parkinson’s disease model in rats reveals peripheral early-phase biomarkers. Sci. Reports 9, 1898. https://doi.org/10.1038/s41598-018-37657-6 (2019).
Mapstone, M. et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat. Medicine 20, 415–418. https://doi.org/10.1038/nm.3466 (2014).
Chang, K.-H. et al. Altered metabolic profiles of the plasma of patients with amyotrophic lateral sclerosis. Biomedicines 9, 1944 (2021).
Luo, X., Liu, Y., Balck, A., Klein, C. & Fleming, R. M. T. Identification of metabolites reproducibly associated with parkinson’s disease via meta-analysis and computational modelling. npj Park. Dis. 10, 38, https://doi.org/10.1038/s41531-024-00732-z (2024).
Kalecký, K. & Bottiglieri, T. Targeted metabolomic analysis in parkinson’s disease brain frontal cortex and putamen with relation to cognitive impairment. npj Park. Dis. 9, 84, https://doi.org/10.1038/s41531-023-00531-y (2023).
Braak, H. et al. Staging of brain pathology related to sporadic parkinson’s disease. Neurobiol. Aging 24, 197–211. https://doi.org/10.1016/S0197-4580(02)00065-9 (2003).
Malek, N. et al. Autonomic dysfunction in early parkinson’s disease: Results from the united kingdom tracking parkinson’s study. Mov. Disord. Clin. Pract. 4, 509–516. https://doi.org/10.1002/mdc3.12454 (2016).
Pfeiffer, R. F. Autonomic dysfunction in parkinson’s disease. Neurotherapeutics 17, 1464–1479. https://doi.org/10.1007/s13311-020-00897-4 (2020).
Peña-Zelayeta, L. et al. Redefining non-motor symptoms in parkinson’s disease. J. Pers. Medicine 15, 172. https://doi.org/10.3390/jpm15050172 (2025).
Acknowledgements
JJOV thanks the financial support from the UNAM Postdoctoral Program DGAPA.
Funding
This research was funded by Secretaría de Ciencia, Humanidades, Tecnología e Innovación ((64382 and CF-2023-I-1226)); INMEGEN internal funds (Basal 2025); Secretaría de Salud (FPIS2024-INMEGEN-6940); Genome Alberta (a division of Genome Canada) (grant number TMIC MC4); The Canadian Institutes of Health Research (CIHR) (grant number FS 148461); and The Canada Foundation for Innovation (CFI) (grant number MSIF 35456).
Author information
Authors and Affiliations
Contributions
Conceptualization was undertaken by José Pedro Elizalde-Díaz, Yamilé López-Hernández, Juan José Oropeza Valdez, and Eduardo Martínez Martínez; Methodology by Juan José Oropeza Valdez, Yamilé López-Hernández, Osbaldo Resendis Antonio, Rupasri Mandal, and José Pedro Elizalde-Díaz; Investigation by Juan José Oropeza Valdez, José Pedro Elizalde-Díaz, Jaquelin Leyva Hernández, Laura Adalid-Peralta, and Mayela Rodríguez-Violante; Data Curation by Juan José Oropeza Valdez and José Pedro Elizalde-Díaz; Formal Analysis by Juan José Oropeza Valdez, Osbaldo Resendis Antonio, and Rupasri Mandal; Software and Validation by Rupasri Mandal; Visualization by Juan José Oropeza Valdez and Osbaldo Resendis Antonio; Resources/Clinical Investigation by Mayela Rodríguez-Violante and Laura Adalid-Peralta; Writing – Original Draft by Juan José Oropeza Valdez and Yamilé López-Hernández; Writing – Review & Editing by all authors; Supervision by Yamilé López-Hernández, Eduardo Martínez Martínez, and David S. Wishart; Project Administration by Yamilé López-Hernández, David S. Wishart, and Eduardo Martínez Martínez; and Funding Acquisition by David S. Wishart, Yamilé López-Hernández, and Eduardo Martínez Martínez.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Oropeza Valdez, J.J., Elizalde-Díaz, J.P., Antonio, O.R. et al. Association of a five-metabolite and early-symptom profile with Parkinson’s disease and its clinical progression. Sci Rep 16, 5885 (2026). https://doi.org/10.1038/s41598-026-36756-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-026-36756-z