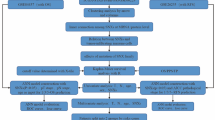
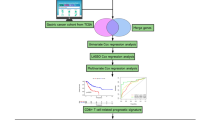
Abstract
Interferon-γ-inducible protein 30 (IFI30, also known as lysosomal thiol reductase, GILT) plays a key role in antigen processing by reducing disulfide bonds. However, its biological significance in gastric cancer (GC) has not been systematically elucidated. This study integrated pan-cancer multi-omics data, including transcriptomics (TCGA-STAD, GEO), genomics (whole-exome somatic mutations, copy number alterations), immune profiling, single-cell RNA sequencing, and transcription factor prediction to comprehensively characterize the dysregulation of IFI30 in GC. Downstream pathway involvement was inferred through weighted gene co-expression network analysis (WGCNA), gene set enrichment analysis (GSEA), and phosphoproteomic correlation mapping. The immune microenvironment was analyzed using CIBERSORTx, TIMER2.0, and re-annotated spatial transcriptomics. Multi-omics interrogation revealed that IFI30 is markedly up-regulated in gastric adenocarcinoma (STAD) relative to normal gastric mucosa. Across TCGA-GTEx and three validation cohorts, IFI30 mRNA and protein levels were significantly higher in tumours, with robust diagnostic performance (AUC = 0.92). Copy-number amplification—not point mutation—was the principal genomic driver of over-expression and was accompanied by heightened genome instability and co-occurrence of TP53 and PIK3CA alterations. Single-cell RNA-seq pinpointed IFI30 enrichment in dendritic cells, CD8⁺ T cells and macrophages, forming dense ligand-receptor networks that link innate and adaptive immunity. WGCNA and pathway analyses showed that IFI30-high tumours converge on antigen presentation, cytokine/chemokine, JAK–STAT and NF-κB signalling while activating epithelial-mesenchymal transition, cell-cycle and hypoxia programmes. IFI30 correlated strongly with multiple steps of the cancer–immunity cycle and with PD-L1, SPI1, FOXP3 and IRF1 expression. Pharmacogenomic profiling indicated resistance to MAPK- and cell-cycle inhibitors yet increased sensitivity to EGFR and PI3K/AKT blockade. IFI30-based signatures outperformed TIDE, TMB and PD-L1 in predicting immune-checkpoint-blockade response and were enriched in MSI-H tumours. In vitro, IFI30 protein was abundant in six gastric-cancer cell lines, and shRNA-mediated knock-down curtailed proliferation. Collectively, these findings establish IFI30 as a genomically driven, immunologically active and therapeutically actionable biomarker in gastric cancer. IFI30 is a copy-number–driven oncogenic and immunomodulatory gene that is markedly over-expressed in gastric adenocarcinoma. Its high expression integrates tumor-intrinsic programs (cell cycle, EMT, hypoxia) with tumor-extrinsic immune activation, predicts differential drug sensitivities, and outperforms established biomarkers in forecasting response to immune-checkpoint blockade—particularly in MSI-high disease. These findings nominate IFI30 as a promising diagnostic marker and therapeutic target.
Similar content being viewed by others
References
Yasuda, T. & Wang, Y. A. Gastric cancer immunosuppressive microenvironment heterogeneity: implications for therapy development[J]. Trends In Cancer 10(7), 627–642 (2024).
López, M. J. et al. Characteristics of gastric cancer around the world[J]. Critical Reviews In Oncology/hematology 181, 103841 (2023).
Smyth, E. C. et al. Gastric cancer[J]. Lancet (London, England) 396(10251), 635–648 (2020).
Zhang Z, Shao S, Luo H, et al. The functions of cuproptosis in gastric cancer: therapy, diagnosis, prognosis[J]. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 2024, 177: 117100.
Rugge, M. et al. Steps forward in understanding gastric cancer risk[J]. Gut 72(9), 1802–1803 (2023).
Song J, Zhu J, Jiang Y, et al. Advancements in immunotherapy for gastric cancer: Unveiling the potential of immune checkpoint inhibitors and emerging strategies[J]. Biochimica Et Biophysica Acta. Reviews On Cancer, 2025, 1880(2): 189277.
Abbas M, Faggian A, Sintali D N, et al. Current and future biomarkers in gastric cancer[J]. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 2018, 103: 1688-1700.
Sun, Y. et al. Advances and challenges in gastric cancer testing: the role of biomarkers[J]. Cancer Biology & Medicine 22(3), 212–230 (2025).
Singh, R., Jamieson, A. & Cresswell, P. GILT is a critical host factor for Listeria monocytogenes infection[J]. Nature 455(7217), 1244–1247 (2008).
Watts C. Immunology. Antigen presentation--losing its shine in the absence of GILT[J]. Science (New York, N.Y.), 2001, 294(5545): 1294-1295.
Rausch, M. P. & Hastings, K. T. GILT modulates CD4+ T-cell tolerance to the melanocyte differentiation antigen tyrosinase-related protein 1[J]. The Journal of Investigative Dermatology 132(1), 154–162 (2012).
Brottveit, M. et al. Mucosal cytokine response after short-term gluten challenge in celiac disease and non-celiac gluten sensitivity[J]. The American Journal of Gastroenterology 108(5), 842–850 (2013).
Becker, J. C. & Schrama, D. Control of central and peripheral tolerance to melanocyte differentiation antigens by GILT[J]. The Journal of Investigative Dermatology 132(1), 15–17 (2012).
Zhang, S. et al. Interferon Gamma Inducible Protein 30: from biological functions to potential therapeutic target in cancers[J]. Cellular Oncology (Dordrecht, Netherlands) 47(5), 1593–1605 (2024).
Fan, Y., Wang, X. & Li, Y. IFI30 expression predicts patient prognosis in breast cancer and dictates breast cancer cells proliferation via regulating autophagy[J]. International Journal of Medical Sciences 18(14), 3342–3352 (2021).
Chen, Y. et al. Interferon-γ inducible protein 30 promotes the epithelial-mesenchymal transition-like phenotype and chemoresistance by activating EGFR/AKT/GSK3β/β-catenin pathway in glioma[J]. CNS Neuroscience & Therapeutics 29(12), 4124–4138 (2023).
Kanehisa, M. et al. KEGG: biological systems database as a model of the real world[J]. Nucleic Acids Research 53(D1), D672–D677 (2025).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms[J]. Protein Science : a Publication of the Protein Society 28(11), 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Research 28(1), 27–30 (2000).
Guan, W.-L., He, Y. & Xu, R.-H. Gastric cancer treatment: recent progress and future perspectives[J]. Journal of Hematology & Oncology 16(1), 57 (2023).
Duan, Y. et al. Helicobacter pylori and gastric cancer: mechanisms and new perspectives[J]. Journal of Hematology & Oncology 18(1), 10 (2025).
Zeng, Y. & Jin, R. U. Molecular pathogenesis, targeted therapies, and future perspectives for gastric cancer[J]. Seminars In Cancer Biology 86(Pt 3), 566–582 (2022).
Khalili-Tanha, G. et al. Diagnostic, prognostic, and predictive biomarkers in gastric cancer: from conventional to novel biomarkers[J]. Translational Research : the Journal of Laboratory and Clinical Medicine 274, 35–48 (2024).
Yuan, W. et al. The role of MAPK pathway in gastric cancer: unveiling molecular crosstalk and therapeutic prospects[J]. Journal of Translational Medicine 22(1), 1142 (2024).
Leja, M. Potential of personalised approaches in gastric cancer prevention[J]. Gut 72(12), 2225–2226 (2023).
Hou W, Zhao Y, Zhu H. Predictive Biomarkers for Immunotherapy in Gastric Cancer: Current Status and Emerging Prospects[J]. International Journal of Molecular Sciences, 2023, 24(20).
Jiang, L. et al. Comprehensive single-cell pan-cancer atlas unveils IFI30+ macrophages as key modulators of intra-tumoral immune dynamics[J]. Frontiers In Immunology 16, 1523854 (2025).
Li, L. et al. IFI30 as a key regulator of PDL1 immunotherapy prognosis in breast cancer[J]. International Immunopharmacology 133, 112093 (2024).
Ye, C. et al. Prognostic value of gamma-interferon-inducible lysosomal thiol reductase expression in female patients diagnosed with breast cancer[J]. International Journal of Cancer 150(4), 705–717 (2022).
Funding
This work was supported by a grant from the Clinical Science Foundation project of Anhui Medical University:2023xkj169 and Scientific research project of colleges and universities in Anhui Province:2023AH050668.
Author information
Authors and Affiliations
Contributions
Minzhi Sun, Weiwei Yuan, and Ruizhi Zhaowang performed the experiments, collected data, and conducted the formal analysis.Qing Liu assisted with data acquisition and provided technical support.Xiao Yuan designed and supervised the study, guided data interpretation, and critically revised the manuscript for important intellectual content. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Due to the nature of this design, no additional ethical approval was needed. Consequently, the Local Ethics Committee waived the ethical approval. This study analyzed publicly available data from multiple independent cohorts.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Q., Yuan, W., Zhaowang, R. et al. Copy-number amplification drives IFI30 overexpression and coordinated immune activation, identifying a novel diagnostic and therapeutic target in gastric adenocarcinoma. Sci Rep (2026). https://doi.org/10.1038/s41598-026-37574-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-37574-z