Abstract
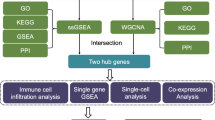
Spinal cord ischemia-reperfusion injury (SCII) often causes neurological damage and devastating sensory and motor dysfunction. Identifying key genes and signaling pathways in SCII progression may provide novel therapeutic targets. Two gene expression datasets (GSE138966 and GSE167274) were obtained from the Gene Expression Omnibus database. Differentially expressed genes were identified using R software, followed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Hub genes were screened via Venn analysis, and a protein-protein interaction (PPI) network was constructed using Cytoscape software. Key hub genes were validated by qRT-PCR in a rat SCII model. A total of 99 hub genes were identified, including 60 up-regulated and 39 down-regulated genes. KEGG analysis revealed significant enrichment in MAPK, cAMP, and Rap1 signaling pathways. PPI network analysis highlighted Ccl2, Mmp9, Itgb1, Timp1, Myd88, and Lgals3 as central nodes. qRT-PCR validation showed persistent up-regulation of Tnc, Thbs2, and S100a10 at 1 h, 24 h, and 48 h post-SCII; early up-regulation of Msn, Lcp1, Lcn2, and Akap12 at 1 h; and delayed up-regulation of Itga5 at 48 h (P < 0.05). This study identifies novel, key SCII-related genes that have been largely overlooked and, for the first time, defines their time-dependent expression patterns via in vivo experimental validation. Our findings provide crucial mechanistic insights and nominate promising therapeutic targets for SCII.
Similar content being viewed by others
Data availability
The datasets selected in our research can be found and downloaded for free online. GEO Database accession number: GSE138966, GSE167274. All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Sueda, T. & Takahashi, S. Spinal cord injury as a complication of thoracic endovascular aneurysm repair. Surg. Today. 48, 473–477. https://doi.org/10.1007/s00595-017-1588-5 (2018).
Kong, X. & Gao, J. Macrophage polarization: a key event in the secondary phase of acute spinal cord injury. J. Cell. Mol. Med. 21, 941–954. https://doi.org/10.1111/jcmm.13034 (2017).
Stenudd, M., Sabelstrom, H. & Frisen, J. Role of endogenous neural stem cells in spinal cord injury and repair. JAMA Neurol. 72, 235–237. https://doi.org/10.1001/jamaneurol.2014.2927 (2015).
Henmi, S. et al. Comparison of early patency rate and long-term outcomes of various techniques for reconstruction of segmental arteries during thoracoabdominal aortic aneurysm repair. Eur. J. Cardiothorac. Surg. https://doi.org/10.1093/ejcts/ezz015 (2019).
Jin, W., Botchway, B. O. A. & Liu, X. Curcumin can activate the Nrf2/HO-1 signaling pathway and scavenge free radicals in spinal cord injury treatment. Neurorehabil Neural Repair. 35, 576–584. https://doi.org/10.1177/15459683211011232 (2021).
Liu, W. Z., Ma, Z. J., Li, J. R. & Kang, X. W. Mesenchymal stem cell-derived exosomes: therapeutic opportunities and challenges for spinal cord injury. Stem Cell. Res. Ther. 12, 102. https://doi.org/10.1186/s13287-021-02153-8 (2021).
Orlov, Y. L., Anashkina, A. A., Klimontov, V. V. & Baranova, A. V. Medical Genetics, genomics and bioinformatics aid in Understanding molecular mechanisms of human diseases. Int. J. Mol. Sci. 22, 256. https://doi.org/10.3390/ijms22189962 (2021).
Wang, D. et al. Bioinformatics-Based analysis of the lncRNA-miRNA-mRNA network and TF regulatory network to explore the regulation mechanism in spinal cord Ischemia/Reperfusion injury. Front. Genet. 12, 650180. https://doi.org/10.3389/fgene.2021.650180 (2021).
Zhou, Z., Han, B., Jin, H., Chen, A. & Zhu, L. Changes in long non-coding RNA transcriptomic profiles after ischemia-reperfusion injury in rat spinal cord. PeerJ 8, e8293. https://doi.org/10.7717/peerj.8293 (2020).
Pavic, G. et al. Microglia contributes to remyelination in cerebral but not spinal cord ischemia. Glia 69, 2739–2751. https://doi.org/10.1002/glia.24068 (2021).
Ritchie, M. E. et al. K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. https://doi.org/10.1093/nar/gkv007 (2015).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573. https://doi.org/10.1093/bioinformatics/btq170 (2010).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Hu, X., Ni, S., Zhao, K., Qian, J. & Duan, Y. Bioinformatics-Led discovery of osteoarthritis biomarkers and inflammatory infiltrates. Front. Immunol. 13, 871008. https://doi.org/10.3389/fimmu.2022.871008 (2022).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. https://doi.org/10.1093/nar/gky1131 (2019).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Yin, F. et al. Transplantation of mesenchymal stem cells exerts anti-apoptotic effects in adult rats after spinal cord ischemia-reperfusion injury. Brain Res. 1561, 1–10. https://doi.org/10.1016/j.brainres.2014.02.047 (2014).
Basso, D. M., Beattie, M. S. & Bresnahan, J. C. Descending systems contributing to locomotor recovery after mild or moderate spinal cord injury in rats: experimental evidence and a review of literature. Restor. Neurol. Neurosci. 20, 189–218 (2002).
Yang, J. et al. An iron delivery pathway mediated by a Lipocalin. Mol. Cell. 10, 1045–1056. https://doi.org/10.1016/s1097-2765(02)00710-4 (2002).
Kuramoto, K., Liang, H., Hong, J. H. & He, C. Exercise-activated hepatic autophagy via the FN1-α5β1 integrin pathway drives metabolic benefits of exercise. Cell Metab. 35, 620–632 e625. https://doi.org/10.1016/j.cmet.2023.01.011 (2023).
Gelman, I. H. Emerging roles for SSeCKS/Gravin/AKAP12 in the control of cell Proliferation, cancer Malignancy, and barriergenesis. Genes Cancer. 1, 1147–1156. https://doi.org/10.1177/1947601910392984 (2010).
Morellini, F. & Schachner, M. Enhanced novelty-induced activity, reduced anxiety, delayed resynchronization to daylight reversal and weaker muscle strength in tenascin-C-deficient mice. Eur. J. Neurosci. 23, 1255–1268. https://doi.org/10.1111/j.1460-9568.2006.04657.x (2006).
Wabnitz, G. H. et al. Costimulation induced phosphorylation of L-plastin facilitates surface transport of the T cell activation molecules CD69 and CD25. Eur. J. Immunol. 37, 649–662. https://doi.org/10.1002/eji.200636320 (2007).
Madureira, P. A., O’Connell, P. A., Surette, A. P., Miller, V. A. & Waisman, D. M. The biochemistry and regulation of S100A10: a multifunctional plasminogen receptor involved in oncogenesis. J. Biomed. Biotechnol. 2012, 353687. https://doi.org/10.1155/2012/353687 (2012).
Huang, L., Wong, T. Y., Lin, R. C. & Furthmayr, H. Replacement of threonine 558, a critical site of phosphorylation of Moesin in vivo, with aspartate activates F-actin binding of Moesin. Regulation by conformational change. J. Biol. Chem. 274, 12803–12810. https://doi.org/10.1074/jbc.274.18.12803 (1999).
Serrador, J. M. et al. CD43 interacts with Moesin and Ezrin and regulates its redistribution to the uropods of T lymphocytes at the cell-cell contacts. Blood 91, 4632–4644 (1998).
Simantov, R., Febbraio, M. & Silverstein, R. L. The antiangiogenic effect of thrombospondin-2 is mediated by CD36 and modulated by histidine-rich glycoprotein. Matrix Biol. 24, 27–34. https://doi.org/10.1016/j.matbio.2004.11.005 (2005).
Alizadeh, A., Dyck, S. M. & Karimi-Abdolrezaee, S. Traumatic spinal cord injury: an overview of Pathophysiology, models and acute injury mechanisms. Front. Neurol. 10, 282. https://doi.org/10.3389/fneur.2019.00282 (2019).
Hutson, T. H. & Di Giovanni, S. The translational landscape in spinal cord injury: focus on neuroplasticity and regeneration. Nat. Rev. Neurol. 15, 732–745. https://doi.org/10.1038/s41582-019-0280-3 (2019).
Jiang, W. et al. CCL2 is a key regulator and therapeutic target for periodontitis. J. Clin. Periodontol. 50, 1644–1657. https://doi.org/10.1111/jcpe.13872 (2023).
Du, S. et al. X. A nerve injury-specific long noncoding RNA promotes neuropathic pain by increasing Ccl2 expression. J. Clin. Invest. 132, 689. https://doi.org/10.1172/jci153563 (2022).
Lee, J. Y., Kim, H. S., Choi, H. Y., Oh, T. H. & Yune, T. Y. Fluoxetine inhibits matrix metalloprotease activation and prevents disruption of blood-spinal cord barrier after spinal cord injury. Brain 135, 2375–2389. https://doi.org/10.1093/brain/aws171 (2012).
Rempe, R. G., Hartz, A. M. S. & Bauer, B. Matrix metalloproteinases in the brain and blood-brain barrier: versatile breakers and makers. J. Cereb. Blood Flow. Metab. 36, 1481–1507. https://doi.org/10.1177/0271678x16655551 (2016).
Wang, Q. et al. Myd88 knockdown with RNA interference induces in vitro immune hyporesponsiveness in dendritic cells from rhesus monkeys. Immunogenetics 74, 303–312. https://doi.org/10.1007/s00251-022-01260-x (2022).
Bayer, A. L. & Alcaide, P. MyD88: at the heart of inflammatory signaling and cardiovascular disease. J. Mol. Cell. Cardiol. 161, 75–85. https://doi.org/10.1016/j.yjmcc.2021.08.001 (2021).
Rong, J. et al. Long non-coding RNA KCNQ1OT1/microRNA-204-5p/LGALS3 axis regulates myocardial ischemia/reperfusion injury in mice. Cell. Signal. 66, 109441. https://doi.org/10.1016/j.cellsig.2019.109441 (2020).
Yan, L. et al. Hspb1 and Lgals3 in spinal neurons are closely associated with autophagy following excitotoxicity based on machine learning algorithms. PLoS One. 19, e0303235. https://doi.org/10.1371/journal.pone.0303235 (2024).
Tavares, L. P. et al. Blame the signaling: role of cAMP for the resolution of inflammation. Pharmacol. Res. 159, 105030. https://doi.org/10.1016/j.phrs.2020.105030 (2020).
Xu, Z. et al. ERK1/2 and p38 mitogen-activated protein kinase mediate iNOS-induced spinal neuron degeneration after acute traumatic spinal cord injury. Life Sci. 79, 1895–1905. https://doi.org/10.1016/j.lfs.2006.06.023 (2006).
Sun, X. et al. Activation of the Epac/Rap1 signaling pathway alleviates blood-brain barrier disruption and brain damage following cerebral ischemia/reperfusion injury. Int. Immunopharmacol. 117, 110014. https://doi.org/10.1016/j.intimp.2023.110014 (2023).
Midwood, K. S., Chiquet, M., Tucker, R. P. & Orend, G. Tenascin-C at a glance. J. Cell. Sci. 129, 4321–4327. https://doi.org/10.1242/jcs.190546 (2016).
Midwood, K. et al. Tenascin-C is an endogenous activator of Toll-like receptor 4 that is essential for maintaining inflammation in arthritic joint disease. Nat. Med. 15, 774–780. https://doi.org/10.1038/nm.1987 (2009).
Chang, Z. et al. THBS2 promotes gastric cancer progression and stemness via the Notch signaling pathway. Am. J. Cancer Res. 14, 3433–3450. https://doi.org/10.62347/uxwk4038 (2024).
Adams, J. C. & Lawler, J. The thrombospondins. Cold Spring Harb Perspect. Biol. 3, a009712. https://doi.org/10.1101/cshperspect.a009712 (2011).
Milosevic, A. et al. Cell- and region-specific expression of depression-related protein p11 (S100a10) in the brain. J. Comp. Neurol. 525, 955–975. https://doi.org/10.1002/cne.24113 (2017).
Okura, G. C., Bharadwaj, A. G. & Waisman, D. M. Recent advances in molecular and cellular functions of S100A10. Biomolecules 13, 569. https://doi.org/10.3390/biom13101450 (2023).
Satooka, H., Nagakubo, D., Sato, T. & Hirata, T. The ERM protein Moesin regulates CD8(+) regulatory T cell homeostasis and Self-Tolerance. J. Immunol. 199, 3418–3426. https://doi.org/10.4049/jimmunol.1700074 (2017).
Wang, Y. et al. LCP1 knockdown in monocyte-derived macrophages: mitigating ischemic brain injury and shaping immune cell signaling and metabolism. Theranostics 14, 159–175. https://doi.org/10.7150/thno.88678 (2024).
Liu, R. et al. NOX activation in reactive astrocytes regulates astrocytic LCN2 expression and neurodegeneration. Cell. Death Dis. 13, 371. https://doi.org/10.1038/s41419-022-04831-8 (2022).
Wang, G. et al. Lipocalin-2 released in response to cerebral ischaemia mediates reperfusion injury in mice. J. Cell. Mol. Med. 19, 1637–1645. https://doi.org/10.1111/jcmm.12538 (2015).
Cha, J. H. et al. W. AKAP12 mediates barrier functions of fibrotic scars during CNS repair. PLoS One. 9, e94695. https://doi.org/10.1371/journal.pone.0094695 (2014).
Chen, W. et al. Migrasome-related ITGA5 for predicting prognosis, immune infiltration and drug sensitivity of hepatocellular carcinoma. Apoptosis https://doi.org/10.1007/s10495-025-02103-2 (2025).
Acknowledgements
This study was supported by Zhenjiang Science and Technology Plan Project-Social Development (Grant No. SH2023061), the Research Fund of the First People’s Hospital of Zhenjiang (Grant No. Y2020012) and the Third Phase of “Jinshan Doctors” - Young Talents in Medical Field of Zhenjiang City.
Author information
Authors and Affiliations
Contributions
G.M. contributed to the study design, analysis, and interpretation of data and drafted the manuscript. L.H. and M.J. contributed to the study design and interpretation of the data, revised the manuscript and approved the final version. S.C. , W.L. and Y.J. critically revised the manuscript and approved the final manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, M., Liu, H., Sun, C. et al. Identification of key hub genes in spinal cord ischemia-reperfusion injury via integrated bioinformatics analysis and in vivo validation. Sci Rep (2026). https://doi.org/10.1038/s41598-026-39101-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-39101-6