Abstract
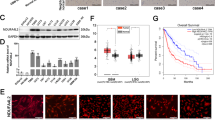
The current study explores the expression, functional significance, and underlying mechanisms of the mitochondrial protein NDUFS4 (NADH:ubiquinone oxidoreductase subunit S4) in glioma cells. TCGA shows that elevated NDUFS4 expression is consistently observed in glioma tissues, correlating with advanced tumor grade and diminished patient survival. Single-cell RNA sequencing further localizes this elevated expression primarily to glioma cells, where NDUFS4 co-expressed genes are integral to cellular respiration and mitochondrial ATP synthesis. These findings were corroborated in patient tissues and various primary and established glioma cell types, confirming consistent NDUFS4 overexpression. Genetic silencing (via shRNA) or CRISPR/Cas9-mediated knockout of NDUFS4 impaired mitochondrial function, evidenced by reduced oxygen consumption rate, inhibited mitochondrial complex I activity and ATP production and increased oxidative stress. NDUFS4 depletion also suppressed glioma cell proliferation, migration, and invasion, while promoting apoptosis. This inhibitory effect is specific to malignant cells, sparing non-cancerous astrocytes. Conversely, NDUFS4 overexpression enhanced mitochondrial activity and promoted aggressive malignant phenotypes in primary and immortalized glioma cells. Further multi-omics integration and experimental investigation established COX5B (cytochrome c oxidase subunit 5B) as an important downstream effector of NDUFS4. shRNA-induced silencing of COX5B replicated the outcomes of NDUFS4 depletion in primary glioma cells, and crucially, restoring COX5B in NDUFS4-silenced glioma cells abrogated the anti-glioma effects. In vivo studies demonstrated that NDUFS4 silencing effectively impeded intracranial growth of patient-derived glioma xenografts by compromising mitochondrial function, downregulating COX5B, inhibiting proliferation and inducing apoptosis. Collectively, these comprehensive data underscore NDUFS4’s essential role in glioma progression and position it as a promising therapeutic target for this aggressive malignancy.
Similar content being viewed by others
Data availability
Data are provided within the manuscript or supplementary information files.
References
Weller, M. et al. Glioma. Nat. Rev. Dis. Prim. 10, 33 (2024).
Weller, M. et al. Glioma. Nat. Rev. Dis. Prim. 1, 15017 (2015).
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Omuro, A. & DeAngelis, L. M. Glioblastoma and other malignant gliomas: a clinical review. JAMA 310, 1842–1850 (2013).
Reifenberger, G., Wirsching, H. G., Knobbe-Thomsen, C. B. & Weller, M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat. Rev. Clin. Oncol. 14, 434–452 (2017).
Lim, M., Xia, Y., Bettegowda, C. & Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 15, 422–442 (2018).
Hotchkiss, K. M. et al. A brave new framework for glioma drug development. Lancet Oncol. 25, e512–e519 (2024).
Yang, K. et al. Glioma targeted therapy: insight into future of molecular approaches. Mol. Cancer 21, 39 (2022).
Xu, S., Tang, L., Li, X., Fan, F. & Liu, Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 476, 1–12 (2020).
Chen, L., Zhang, H., Shang, C. & Hong, Y. The role and applied value of mitochondria in glioma-related research. CNS Neurosci. Ther. 30, e70121 (2024).
Ordys, B. B., Launay, S., Deighton, R. F., McCulloch, J. & Whittle, I. R. The role of mitochondria in glioma pathophysiology. Mol. Neurobiol. 42, 64–75 (2010).
Iuso, A. et al. Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J. Biol. Chem. 281, 10374–10380 (2006).
van de Wal, M. A. E. et al. Ndufs4 knockout mice with isolated complex I deficiency engage a futile adaptive brain response. Biochim. Biophys. Acta Proteins Proteom. 1873, 141055 (2025).
Yin, Z., Agip, A. A., Bridges, H. R. & Hirst, J. Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4(-/-) mouse. EMBO J. 43, 225–249 (2024).
van de Wal, M. A. E. et al. Ndufs4 knockout mouse models of Leigh syndrome: pathophysiology and intervention. Brain 145, 45–63 (2022).
Ortigoza-Escobar, J. D. et al. Ndufs4 related Leigh syndrome: a case report and review of the literature. Mitochondrion 28, 73–78 (2016).
Shil, S. K. et al. Ndufs4 ablation decreases synaptophysin expression in hippocampus. Sci. Rep. 11, 10969 (2021).
Sonsalla, G. et al. Direct neuronal reprogramming of NDUFS4 patient cells identifies the unfolded protein response as a novel general reprogramming hurdle. Neuron 112, 1117–1132 e1119 (2024).
Cheng, T. et al. NDUFS4 promotes tumor progression and predicts prognosis in gastric cancer. Carcinogenesis 43, 980–987 (2022).
Kloosterhof, N. K., Bralten, L. B., Dubbink, H. J., French, P. J. & van den Bent, M. J. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma?. Lancet Oncol. 12, 83–91 (2011).
Han, S. et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br. J. Cancer 122, 1580–1589 (2020).
Abdelfattah, N. et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 13, 767 (2022).
Wang, Y. et al. G protein inhibitory alpha subunit 2 is a molecular oncotarget of human glioma. Int. J. Biol. Sci. 19, 865–879 (2023).
Liu, F. et al. YME1L overexpression exerts pro-tumorigenic activity in glioma by promoting Galphai1 expression and Akt activation. Protein Cell 14, 223–229 (2023).
Guo, Y. Z. et al. TIMM44 is a potential therapeutic target of human glioma. Theranostics 12, 7586–7602 (2022).
Wang, Y. et al. Neuronal-driven glioma growth requires Galphai1 and Galphai3. Theranostics 11, 8535–8549 (2021).
Hu, T. & Xi, J. Identification of COX5B as a novel biomarker in high-grade glioma patients. Onco Targets Ther. 10, 5463–5470 (2017).
Gao, S. P. et al. High expression of COX5B is associated with poor prognosis in breast cancer. Future Oncol. 13, 1711–1719 (2017).
Gao, S. P. et al. Loss of COX5B inhibits proliferation and promotes senescence via mitochondrial dysfunction in breast cancer. Oncotarget 6, 43363–43374 (2015).
Bachman, N. J., Yang, T. L., Dasen, J. S., Ernst, R. E. & Lomax, M. I. Phylogenetic footprinting of the human cytochrome c oxidase subunit VB promoter. Arch. Biochem. Biophys. 333, 152–162 (1996).
Deighton, R. F. et al. Interactions among mitochondrial proteins altered in glioblastoma. J. Neurooncol. 118, 247–256 (2014).
Dickinson, A. et al. The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ. 20, 1644–1653 (2013).
Watson, D. C. et al. GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Nat. Cancer 4, 648–664 (2023).
Mise, K. et al. NDUFS4 regulates cristae remodeling in diabetic kidney disease. Nat. Commun. 15, 1965 (2024).
Cai, S. et al. Mitochondrial dysfunction in macrophages promotes inflammation and suppresses repair after myocardial infarction. J. Clin. Invest. 133, e159498 (2023).
Chu, Y. D., Lim, S. N., Yeh, C. T. & Lin, W. R. COX5B-mediated bioenergetic alterations modulate cell growth and anticancer drug susceptibility by orchestrating claudin-2 expression in colorectal cancers. Biomedicines 10, 60 (2021).
Wang, X., Miao, D., Ye, S., Xian, H. & Ge, W. Abnormal expression of COX5B gene and disorder of mitochondrial function in cryptorchid rats. J. Cell Mol. Med. 28, e70234 (2024).
Mansilla, N., Racca, S., Gras, D. E., Gonzalez, D. H. & Welchen, E. The complexity of mitochondrial complex IV: an update of cytochrome c oxidase biogenesis in plants. Int. J. Mol. Sci. 19, 662 (2018).
Hinkelbein, J. et al. Decreased tissue COX5B expression and mitochondrial dysfunction during sepsis-induced kidney injury in rats. Oxid. Med. Cell Longev. 2017, 8498510 (2017).
Bourens, M., Fontanesi, F., Soto, I. C., Liu, J. & Barrientos, A. Redox and reactive oxygen species regulation of mitochondrial cytochrome C oxidase biogenesis. Antioxid. Redox Signal 19, 1940–1952 (2013).
Sun, X. et al. Role of post-translational modifications of Sp1 in cancer: state of the art. Front. Cell Dev. Biol. 12, 1412461 (2024).
Chang, W. C. & Hung, J. J. Functional role of post-translational modifications of Sp1 in tumorigenesis. J. Biomed. Sci. 19, 94 (2012).
Zhao, Y. et al. Mitochondrial carrier homolog 2 is important for mitochondrial functionality and non-small cell lung cancer cell growth. Cell Death Dis. 16, 95 (2025).
Zha, J., Li, J., Yin, H., Shen, M. & Xia, Y. TIMM23 overexpression drives NSCLC cell growth and survival by enhancing mitochondrial function. Cell Death Dis. 16, 174 (2025).
Cheng, F., Huang, H., Yin, S., Liu, J. S. & Sun, P. Expression and functional implications of YME1L in nasopharyngeal carcinoma. Cell Death Dis. 15, 423 (2024).
Ding, M. et al. Mfn2-mediated mitochondrial fusion alleviates doxorubicin-induced cardiotoxicity with enhancing its anticancer activity through metabolic switch. Redox Biol. 52, 102311 (2022).
Yao, J. et al. The requirement of phosphoenolpyruvate carboxykinase 1 for angiogenesis in vitro and in vivo. Sci. Adv. 8, eabn6928 (2022).
Liu, Y. Y. et al. microRNA-200a downregulation in human glioma leads to Galphai1 over-expression, Akt activation, and cell proliferation. Oncogene 37, 2890–2902 (2018).
Qian, J. et al. COL8A1 overexpression promotes glioma cell growth by activating focal adhesion kinase signaling cascade. NPJ Precis. Oncol. 8, 273 (2024).
Acknowledgements
This work is supported by National Natural Science Foundation of China (82273055) and Scientific Research Project of Jiangsu Provincial Health Commission (Z2022044).
Author information
Authors and Affiliations
Contributions
J.W., J.L., Y.L., and L.J. participated in the conception and design of the study. J.W., J.L., L.X., and L.J. performed experiments. J.W., J.L., L.X., Y.L., and L.J. drafted the manuscript and contributed to its critical revision. All listed authors provided substantial intellectual input and granted final approval of the submitted version to the journal.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, J., Li, J., Xu, L. et al. Mitochondrial complex I subunit NDUFS4 overexpression drives glioma progression by regulating mitochondrial function and COX5B. npj Precis. Onc. (2026). https://doi.org/10.1038/s41698-026-01281-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-026-01281-9