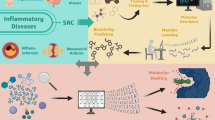
Abstract
The RNA cytosine-5 methyltransferase NSUN2 is an emerging therapeutic target in precision oncology, with aberrant overexpression driving tumor progression, metastasis, and therapy resistance across multiple malignancies. Despite its critical role in cancer biology, selective small-molecule inhibitors remain limited. We employed an AI-accelerated workflow to screen approximately 101 million compounds from the ZINC database using structure-based virtual screening. The AlphaFold2-predicted human NSUN2 structure was aligned with the experimentally determined M. jannaschii TRM4 homolog (PDB: 3A4T, 34.2% sequence identity, 1.82 Å RMSD). A CatBoost ensemble classifier trained on Morgan fingerprint descriptors with AutoDock Vina-derived labels achieved robust performance (training: recall 0.87, ROC-AUC 0.89; test: recall 0.71, ROC-AUC 0.85), with low test precision reflecting extreme class imbalance inherent to virtual screening. Multi-stage filtering identified 12,000 high-scoring compounds with binding affinities of −9.933 to −8.375 kcal/mol. ADMET profiling yielded 34 drug-like candidates with favorable pharmacokinetic and toxicological profiles. Molecular dynamics simulations over 50 nanoseconds validated binding stability of lead compounds ZINC-1000507789 and ZINC-1000507824. These structurally diverse non-covalent reversible inhibitors targeting the SAM cofactor binding pocket warrant experimental validation through biochemical assays and cellular studies to overcome therapeutic resistance in NSUN2-driven malignancies.
Similar content being viewed by others
Data availability
The results associated with this study are present in the paper or supplementary materials. All other materials used in the analyses are available upon reasonable request.
Code availability
The code has been uploaded in https://www.jianguoyun.com/p/DUENIX0Q887bChigiJ8GIAA.
References
Blanco, S. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014).
Frye, M., Harada, B. T., Behm, M. & He, C. RNA modifications modulate gene expression during development. Science. https://doi.org/10.1126/science.aau1646 (2018).
Xin, Yang et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. https://doi.org/10.1038/cr.2017.55 (2017).
Nikoletta, et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. https://doi.org/10.1371/journal.pbio.3000297 (2019).
Mei, L. et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 11, 270 (2020).
Su, J. et al. NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene 40, 5814–5828 (2021).
Hao, Chen et al. m(5)C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat. Commun. https://doi.org/10.1038/s41467-020-16722-7 (2020).
Yueqin, Wang et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol. Cancer https://doi.org/10.1186/s12943-023-01780-4 (2023).
Muzammil, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet https://doi.org/10.1016/j.ajhg.2012.03.023 (2012).
Abbasi-Moheb, L. et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet 90, 847–855 (2012).
Yoon, et al. RNA methyltransferase NSun2 deficiency promotes neurodegeneration through epitranscriptomic regulation of tau phosphorylation. Acta. Neuropathol. https://doi.org/10.1007/s00401-022-02511-7 (2022).
Blaze, J. et al. Neuronal Nsun2 deficiency produces tRNA epitranscriptomic alterations and proteomic shifts impacting synaptic signaling and behavior. Nat. Commun. https://doi.org/10.1038/s41467-021-24969-x (2021).
Chellamuthu, A., Sridhar, S. & Kumar, J. S. The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer. Cells 9, 1758 (2020).
Yongfeng, Tao et al. Chemical Proteomic Discovery of Isotype-Selective Covalent Inhibitors of the RNA Methyltransferase NSUN2. Angew. .Chem. Int. Ed. Engl. https://doi.org/10.1002/anie.202311924 (2023).
Carlsson, J. & Luttens, A. Structure-based virtual screening of vast chemical space as a starting point for drug discovery. Curr. Opin. Struct. Biol. 87, 102829 (2024).
Kitchen, D. B., Decornez, H., Furr, J. R. & Bajorath, J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 3, 935–949 (2004).
Xuan-Yu, Meng, Hong-Xing, Zhang, Mihaly, Mezei & Meng, Cui. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. https://doi.org/10.2174/157340911795677602 (2011).
Sterling, T. & Irwin, J. J. ZINC 15-Ligand Discovery for Everyone. J. Chem. Inf. Model 55, 2324–2337 (2015).
Ain Q. U., Aleksandrova A., Roessler F. D., Ballester P. J. Machine-learning scoring functions to improve structure-based binding affinity prediction and virtual screening. Wiley Interdiscip Rev. Comput. Mol. Sci. https://doi.org/10.1002/wcms.1225 (2016).
Zhe, Wang et al. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: the prediction accuracy of sampling power and scoring power. Phys Chem Chem Phys https://doi.org/10.1039/c6cp01555g (2016).
Gentile, F. et al. Deep Docking: A Deep Learning Platform for Augmentation of Structure Based Drug Discovery. ACS Cent. Sci. 6, 939–949 (2020).
Ton, A.T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol. Inform. 39, https://doi.org/10.1002/minf.202000028 (2020).
Xing, J. et al. NSun2 Promotes Cell Growth via Elevating Cyclin-Dependent Kinase 1 Translation. Mol. Cell Biol. 35, 4043–4052 (2015).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019).
Arman, et al. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature https://doi.org/10.1038/s41586-021-04220-9 (2021).
Guangfeng, Zhou et al. An artificial intelligence accelerated virtual screening platform for drug discovery. Nat Commun. https://doi.org/10.1038/s41467-024-52061-7 (2024).
Ballante, F., Marshall, G. R. An Automated Strategy for Binding-Pose Selection and Docking Assessment in Structure-Based Drug Design. J. Chem. Inf. Model https://doi.org/10.1021/acs.jcim.5b00603 (2015).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. https://doi.org/10.1002/jcc.21334 (2009).
Victor, et al. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. https://doi.org/10.1016/j.ejmech.2021.113705 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mihaly, Varadi et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab1061 (2021).
Janet, M., Thornton, Roman, A., Laskowski & Neera, Borkakoti. AlphaFold heralds a data-driven revolution in biology and medicine. Nat. Med. https://doi.org/10.1038/s41591-021-01533-0 (2021).
Heo, L. & Feig, M. Multi-state modeling of G-protein coupled receptors at experimental accuracy. Proteins 90, 1873–1885 (2022).
Walters, W. P., Murcko, A. A. & Murcko, M. A. Recognizing molecules with drug-like properties. Curr. Opin. Chem. Biol. 3, 384–387 (1999).
Chong, A. et al. Establishing the foundations for a data-centric AI approach for virtual drug screening through a systematic assessment of the properties of chemical data. (2024).
Luttens, A. et al. Rapid traversal of vast chemical space using machine learning-guided docking screens. Nat. Comput Sci. 5, 301–312 (2025).
Steven, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug. Discov. https://doi.org/10.1038/nrd3078 (2010).
Ahmet, et al. Recent applications of deep learning and machine intelligence on in silico drug discovery: methods, tools and databases. Brief Bioinform https://doi.org/10.1093/bib/bby061 (2018).
Yang, Y. et al. Efficient Exploration of Chemical Space with Docking and Deep Learning. J. Chem. Theory Comput 17, 7106–7119 (2021).
John, A. S., Roth, M. W. et al. ZINC20-A Free Ultralarge-Scale Chemical Database for Ligand Discovery. J. Chem. Inf. Model 60, 2155–2168 (2020).
Adrià, Cereto-Massagué et al. Molecular fingerprint similarity search in virtual screening. Methods https://doi.org/10.1016/j.ymeth.2014.08.005 (2014).
Ballester P. J., Mitchell J. B. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics https://doi.org/10.1093/bioinformatics/btq112 (2010).
Houston, D. R. & Walkinshaw, M. D. Consensus docking: improving the reliability of docking in a virtual screening context. J. Chem. Inf. Model https://doi.org/10.1021/ci300399w (2013).
Feher, M. Consensus scoring for protein-ligand interactions. Drug Discov. Today 11, 421–428 (2006).
Xiong, G. et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49, W5–W14 (2021).
Kola, I. & Landis, J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 3, 711–716 (2004).
Daina, A., Michielin, O. & Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717 (2017).
Michael, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. https://doi.org/10.1038/nrd4609 (2015).
Cheng, F., Li, W., Liu, G. & Tang, Y. In silico ADMET prediction: recent advances, current challenges and future trends. Curr. Top. Med Chem. 13, 1273–1289 (2013).
Schyman, P., Liu, R., Desai, V. & Wallqvist, A. vNN Web Server for ADMET Predictions. Front. Pharmacol. 8. https://doi.org/10.3389/fphar.2017.00889 (2017).
Ferreira L. L. G., Andricopulo A. D. ADMET modeling approaches in drug discovery. Drug Discov. Today https://doi.org/10.1016/j.drudis.2019.03.015 (2019).
Minjun, Chen, Jürgen, Borlak & Weida, Tong. High lipophilicity and high daily dose of oral medications are associated with significant risk for drug-induced liver injury. Hepatology https://doi.org/10.1002/hep.26208 (2012).
Redfern, W. S. et al. Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: evidence for a provisional safety margin in drug development. Cardiovasc Res 58, 32–45 (2003).
Tom, L. & Amy, P. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician. 3, 391–396 (2007).
Hospital, A., Goñi, J. R., Orozco, M. & Gelpí, J. L. Molecular dynamics simulations: advances and applications. Adv. Appl Bioinform Chem. 8, 37–47 (2015).
Scott, B. B., Thiberge, S. Y. & Guo, C. Molecular Dynamics Simulation for All. Neuron 100, 1045–1058 (2018).
Jacob, D. Durrant & J., A. McCammon. Molecular dynamics simulations and drug discovery. BMC Biol. https://doi.org/10.1186/1741-7007-9-71 (2011).
M., Karplus & J. A., A., McCammon. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9, 646–652 (2002).
Jeffrey, G. A. & Jeffrey, G. A. An introduction to hydrogen bonding. Vol. 12 (Oxford University Press New York, 1997).
Ron, et al. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. https://doi.org/10.1146/annurev-biophys-042910-155245 (2012).
Barducci, A., Bonomi, M. & Parrinello, M. Metadynamics. Wiley Interdiscip. Rev.: Computational Mol. Sci. 1, 826–843 (2011).
Sugita, Y. & Okamoto, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314, 141–151 (1999).
Frank, H., Niesen, Helena, Berglund & Masoud, Vedadi. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. https://doi.org/10.1038/nprot.2007.321 (2007).
Yang, L., Wang, B. & Gong, Z. Chemically modified non-coding RNAs in cancer. Expert Rev. Mol. Med 27, e19 (2025).
Li, P., Wang, W., Zhou, R., Ding, Y. & Li, X. The m(5) C methyltransferase NSUN2 promotes codon-dependent oncogenic translation by stabilising tRNA in anaplastic thyroid cancer. Clin. Transl. Med. 13. https://doi.org/10.1002/ctm2.1466 (2023).
Añazco-Guenkova, A. M., Miguel-López, B., Monteagudo-García, Ó, García-Vílchez, R. & Blanco, S. The impact of tRNA modifications on translation in cancer: identifying novel therapeutic avenues. NAR Cancer 6, 012 (2024).
Cully, M. Chemical inhibitors make their RNA epigenetic mark. Nat. Rev. Drug Discov. 18, 892–894 (2019).
Macarron, R. et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 10, 188–195 (2011).
Settles, B. Active learning literature survey. (2009).
Copeland, R. A. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists. Methods Biochem Anal. 46, 1–265 (2005).
Berdasco, M. & Esteller, M. Clinical epigenetics: seizing opportunities for translation. Nat. Rev. Genet. https://doi.org/10.1038/s41576-018-0074-2 (2018).
Topper, M. J., Vaz, M., Marrone, K. A., J. R., Brahmer, Julie, S. B, Baylin, Baylin. The emerging role of epigenetic therapeutics in immuno-oncology. Nat. Rev. Clin. Oncol. 17, 75–90 (2020).
The PyMOL Molecular Graphics System (2015).
Laboratory, R. Reduce - tool for adding and correcting hydrogens in PDB files, https://github.com/rlabduke/reduce/tree/master
Lab, F. Meeko: interface for AutoDock., https://github.com/forlilab/Meeko
Kuratani, M. et al. Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin. J. Mol. Biol. 401, 323–333 (2010).
Jianheng, Liu et al. Developmental mRNA m(5)C landscape and regulatory innovations of massive m(5)C modification of maternal mRNAs in animals. Nat. Commun. https://doi.org/10.1038/s41467-022-30210-0 (2022).
AlphaFold NSUN2 Structure, https://alphafold.ebi.ac.uk/.
RDKit: Open-source cheminformatics, https://github.com/rdkit/rdkit (2025).
Shuzhe, Wang, Jagna, Witek, Gregory, A., Landrum & Sereina, Riniker. Improving Conformer Generation for Small Rings and Macrocycles Based on Distance Geometry and Experimental Torsional-Angle Preferences. J. Chem. Inf. Model https://doi.org/10.1021/acs.jcim.0c00025 (2020).
Eastman, P. et al. OpenMM 8: Molecular Dynamics Simulation with Machine Learning Potentials. J. Phys. Chem. B 128, 109–116 (2024).
DuIvyTools, https://github.com/CharlesHahn/DuIvyTools.
Gowers, R. et al. MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations (2016).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput Chem. 32, 2319–2327 (2011).
Acknowledgements
This study was funded by the China Postdoctoral Science Foundation (Certificate Number: 2025M772149).
Author information
Authors and Affiliations
Contributions
Shuangqi Yu, Qiao Peng organized the data analyzed. Shengrong Long wrote the paper. WeiWei and Xiang Li proofread the article. Xiang Li and Shengrong Long reviewed the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, S., Peng, Q., Wei, W. et al. AI-driven virtual screening platform identifies novel NSUN2 inhibitor candidates for targeted cancer therapy: a computational drug discovery approach. npj Precis. Onc. (2026). https://doi.org/10.1038/s41698-026-01296-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-026-01296-2