Abstract
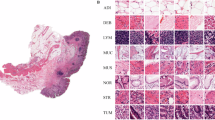
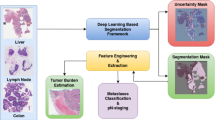
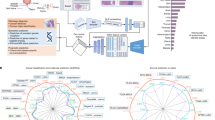
Deep learning is expected to aid pathologists in tasks such as tumour segmentation. We developed a general tumour segmentation model for histopathological images and examined its performance in different cancer types. The model was developed using over 20,000 whole-slide images from over 4000 patients with colorectal, endometrial, lung, or prostate carcinoma. Performance was validated in pre-planned analyses on external cohorts with over 3000 patients across six cancer types. Exploratory analyses included over 1500 additional patients from The Cancer Genome Atlas. Average Dice coefficient was over 80% in all validation cohorts with en bloc resection specimens and in The Cancer Genome Atlas cohorts. No performance loss was observed when comparing the general model with single-cancer models specialised in cancer types from the development set. In conclusion, extensive and rigorous evaluations demonstrate that generic tumour segmentation by a single model is possible across cancer types, patient populations, sample preparations and slide scanners.
Similar content being viewed by others
Code availability
The source code is made available at https://github.com/icgi/automatic-tumour-segmentation-in-WSIs.
Data availability
Materials from TCGA can be downloaded from the TCGA Research Network (https://www.cancer.gov/tcga). Individual patient-level data from the other materials can be made available to other researchers upon reasonable request by contacting the corresponding author, subject to approval by the relevant people or review board at the institutions that provided the original data.
References
Hanna, M. G. et al. Integrating digital pathology into clinical practice. Mod. Pathol. 35, 152–164 (2022).
Rajpurkar, P., Chen, E., Banerjee, O. & Topol, E. J. AI in health and medicine. Nat. Med. 28, 31–38 (2022).
Soerjomataram, I. & Bray, F. Planning for tomorrow: global cancer incidence and the role of prevention 2020–2070. Nat. Rev. Clin. Oncol. 18, 663–672 (2021).
World Health Organization and others. World Health Statistics 2023: Monitoring Health for the SDGs, Sustainable Development Goals (World Health Organization, 2023).
Niazi, M. K. K., Parwani, A. V. & Gurcan, M. N. Digital pathology and artificial intelligence. Lancet Oncol. 20, e253–e261 (2019).
Skrede, O.-J. et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 395, 350–360 (2020).
van Rijthoven, M., Balkenhol, M., Siliņa, K., Van Der Laak, J. & Ciompi, F. HookNet: multi-resolution convolutional neural networks for semantic segmentation in histopathology whole-slide images. Med. Image Anal. 68, 101890 (2021).
Ciga, O. & Martel, A. L. Learning to segment images with classification labels. Med. Image Anal. 68, 101912 (2021).
Schmitz, R. et al. Multi-scale fully convolutional neural networks for histopathology image segmentation: from nuclear aberrations to the global tissue architecture. Med. Image Anal. 70, 101996 (2021).
Khened, M., Kori, A., Rajkumar, H., Krishnamurthi, G. & Srinivasan, B. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci. Rep. 11, 1–14 (2021).
Ho, D. J. et al. Deep Interactive Learning-based ovarian cancer segmentation of H&E-stained whole slide images to study morphological patterns of BRCA mutation. J. Pathol. Inform. 14, 100160 (2023).
Frank, S. J. Accurate diagnostic tissue segmentation and concurrent disease subtyping with small datasets. J. Pathol. Inform. 14, 100174 (2023).
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at https://doi.org/10.48550/arXiv.2108.07258 (2021).
Chen, R. J. et al. Towards a general-purpose foundation model for computational pathology. Nat. Med. 30, 850–862 (2024).
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 181–188 (2024).
Campanella, G., Vanderbilt, C. & Fuchs, T. Computational pathology at health system scale–self-supervised foundation models from billions of images. in Proc. AAAI 2024 Spring Symposium on Clinical Foundation Models (AAAI, 2024).
Nechaev, D., Pchelnikov, A. & Ivanova, E. Hibou: a family of foundational vision transformers for pathology. Preprint at https://doi.org/10.48550/arXiv.2406.05074 (2024).
Filiot, A., Jacob, P., Mac Kain, A. & Saillard, C. Phikon-v2, a large and public feature extractor for biomarker prediction. Preprint at https://doi.org/10.48550/arXiv.2409.09173 (2024).
Zimmermann, E. et al. Virchow2: scaling self-supervised mixed magnification models in pathology. Preprint at https://doi.org/10.48550/arXiv.2408.00738 (2024).
Alber, M. et al. Atlas: a novel pathology foundation model by Mayo Clinic, Charité, and Aignostics. Preprint at https://doi.org/10.48550/arXiv.2501.05409 (2025).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 71, 209–249 (2021).
Ma, J. et al. Segment anything in medical images. Nat. Commun. 15, 654 (2024).
Kirillov, A. et al. Segment anything. In Proc. IEEE/CVF International Conference on Computer Vision, 4015–4026 (IEEE, 2023).
Risbridger, G. P., Davis, I. D., Birrell, S. N. & Tilley, W. D. Breast and prostate cancer: more similar than different. Nat. Rev. Cancer 10, 205–212 (2010).
Chen, Z. et al. Vision transformer adapter for dense predictions. In Proc. The Eleventh International Conference on Learning Representations (ICLR, 2023).
Cheng, B., Misra, I., Schwing, A. G., Kirillov, A. & Girdhar, R. Masked-attention mask transformer for universal image segmentation. In Proc. IEEE/CVF conference on computer vision and pattern recognition, 1290–1299 (IEEE, 2022).
Griebel, T., Archit, A. & Pape, C. Segment anything for histopathology. In Proc. Medical Imaging with Deep Learning (MIDL, 2025).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proc. International Conference on Medical image computing and computer-assisted intervention 234–241 (Springer, 2015).
Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Kleppe, A. et al. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 21, 199–211 (2021).
Dhiman, P. et al. The TRIPOD-P reporting guideline for improving the integrity and transparency of predictive analytics in healthcare through study protocols. Nat. Mach. Intell. 5, 816–817 (2023).
Bondi, J. et al. Expression and gene amplification of primary (A, B1, D1, D3, and E) and secondary (C and H) cyclins in colon adenocarcinomas and correlation with patient outcome. J. Clin. Pathol. 58, 509–514 (2005).
Merok, M. et al. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann. Oncol. 24, 1274–1282 (2013).
Kerr, D. J. et al. Rofecoxib and cardiovascular adverse events in adjuvant treatment of colorectal cancer. N. Engl. J. Med. 357, 360–369 (2007).
Dyrbekk, A. P. H. et al. Evaluation of ROS1 expression and rearrangements in a large cohort of early-stage lung cancer. Diagn. Pathol. 18, 70 (2023).
Wæhre, H. et al. Fifteen-year mortality after radical prostatectomy: which factors are available for patient counselling? Scand. J. Urol. 48, 123–130 (2014).
Kerr, R. S. et al. Adjuvant capecitabine plus bevacizumab versus capecitabine alone in patients with colorectal cancer (QUASAR 2): an open-label, randomised phase 3 trial. Lancet Oncol. 17, 1543–1557 (2016).
Hald, S. M. et al. LAG-3 in non–small-cell lung cancer: expression in primary tumors and metastatic lymph nodes is associated with improved survival. Clin. Lung Cancer 19, 249–259 (2018).
Cyll, K. et al. PTEN and DNA ploidy status by machine learning in prostate cancer. Cancers 13, 4291 (2021).
Skaland, I. et al. Validating the prognostic value of proliferation measured by Phosphohistone H3 (PPH3) in invasive lymph node-negative breast cancer patients less than 71 years of age. Breast Cancer Res. Treat. 114, 39–45 (2009).
Egeland, N. G. et al. Validation study of MARCKSL1 as a prognostic factor in lymph node-negative breast cancer patients. PLoS ONE 14, e0212527 (2019).
Lillesand, M. et al. Mitotic activity index and CD25+ lymphocytes predict risk of stage progression in non-muscle invasive bladder cancer. PLoS ONE 15, e0233676 (2020).
The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014).
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Abeshouse, A. et al. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015).
Liu, J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416 (2018).
Touvron, H., Vedaldi, A., Douze, M. & Jégou, H. Fixing the train-test resolution discrepancy. In Advances in Neural Information Processing Systems Vol. 32 (NeurIPS, 2019).
Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8, 679–698 (1986).
Buslaev, A. et al. Albumentations: fast and flexible image augmentations. Information 11. https://www.mdpi.com/2078-2489/11/2/125 (2020).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. In (eds Wallach, H. et al.) Advances in Neural Information Processing Systems 32, 8024–8035. http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-highperformance-deep-learning-library.pdf (2019).
Brock, A., De, S., Smith, S. L. & Simonyan, K. High-performance large-scale image recognition without normalization. In Proc. International Conference on Machine Learning 1059–1071 (ICML, 2021).
Brock, A., De, S. & Smith, S. L. Characterizing signal propagation to close the performance gap in unnormalized ResNets. In International Conference on Learning Representations (ICLR, 2021).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning 448–456 (ICML, 2015).
Wightman, R. PyTorch Image Models. https://github.com/rwightman/pytorch-image-models (2019).
Hu, J., Shen, L. & Sun, G. Squeeze-and-excitation networks. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 7132–7141 (IEEE, 2018).
Wang, Q. et al. ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks. In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 11531–11539 (IEEE, 2020).
Hendrycks, D. & Gimpel, K. Gaussian error linear units (GELUs). Preprint at https://doi.org/10.48550/arXiv.1606.08415 (2016).
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proc. European Conference on Computer Vision 801–818 (Springer, 2018).
Yakubovskiy, P. Segmentation Models Pytorch. https://github.com/qubvel/segmentation_models.pytorch. (2020).
Wu, Y. & He, K. Group normalization. In Proc. European Conference on Computer Vision 3–19 (Springer, 2018).
He, K., Zhang, X., Ren, S. & Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proc. IEEE International Conference on Computer Vision 1026–1034 (IEEE, 2015).
Sutskever, I., Martens, J., Dahl, G. & Hinton, G. On the importance of initialization and momentum in deep learning. In International Conference on Machine Learning, 1139–1147 (2013).
Loshchilov, I. & Hutter, F. Sgdr: Stochastic gradient descent with warm restarts. In Proc. International Confernce on Learning Representations (ICLR, 2017).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
Maier-Hein, L. et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 9, 5217 (2018).
Maier-Hein, L. et al. Metrics reloaded: recommendations for image analysis validation. Nat. Methods 21, 195–212 (2024).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Laake, P., Lydersen, S. & Veierød, M. B. Medical statistics in clinical and epidemiological research (Gyldendal akademisk, 2012).
Acknowledgements
The authors thank our dear friend and colleague Håvard Emil Danielsen, who passed away in the autumn of 2023; he was among the initiators of this study, facilitated access to materials, and contributed to early versions of the manuscript draft. We thank the laboratory and technical personnel at the Institute for Cancer Genetics and Informatics for essential sample preparation and assistance; Marian Seiergren and Paul Callaghan for assisting with figures. Akershus University hospital, Oslo University Hospital (Aker Hospital, Rikshospitalet, and the Norwegian Radium Hospital), Amsterdam Medical Center, Innsbruck Medical University, University Hospital of North Norway, Nordland Hospital Trust, Vestfold Hospital Trust, and Stavanger University Hospital for access to materials, the personnel at said institutions for sample preparation, and all contributing patients; the participating centres in the QUASAR 2 trial and the VICTOR trial, and all participating patients. This study was funded by The Research Council of Norway through its IKTPLUSS Lighthouse programme (grant number 259204). The Research Council of Norway had no role in study design, data collection, data analysis, data interpretation, writing the report, or the decision to submit the paper for publication.
Author information
Authors and Affiliations
Contributions
O.-J.S. and F.A. initiated the project. A.N., K.L., G.B.K., J.K., A.G.Z., O.T.B., L.-T.R.B., E.H.R., E.S.H., B.B., E.R., M.L., V.K., E.J. and D.J.K. provided access to samples and clinical and pathological data. M.P. annotated all the WSIs used in the study. L.V. annotated the validation cohort VBr2 from breast carcinoma. O.-J.S., M.P., M.X.I., T.S.H., and A.K. decided on the inclusions and exclusions of samples. O.-J.S. developed and implemented the segmentation method, conducted the statistical analyses, and wrote the first draft of the manuscript. O.-J.S., T.S.H., K.L., F.A. and A.K. revised the manuscript draft. All authors reviewed, contributed to, and approved the manuscript. All authors had full access to all the data in the study. O.-J.S. had the final responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing interests
O.-J.S., M.P., M.X.I., T.S.H., D.J.K., K.L., F.A. and A.K. report having shares in DoMore Diagnostics. K.L. reports being a board member in DoMore Diagnostics. O.-J.S., T.S.H. and K.L. report filing a patent application titled ‘Histological image analysis’ with International Patent Number PCT/EP2018/080828. O.-J.S. T.S.H., K.L. and A.K. report filing a patent application titled ‘Histological image analysis’ with International Patent Application Number PCT/EP2020/076090. The other authors have no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Skrede, OJ., Pradhan, M., Isaksen, M.X. et al. Generalisation of automatic tumour segmentation in histopathological whole-slide images across multiple cancer types. npj Precis. Onc. (2026). https://doi.org/10.1038/s41698-026-01311-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-026-01311-6