Abstract
Early Autism Spectrum Disorder (ASD) identification is crucial but resource-intensive. This study evaluated a novel two-stage multimodal AI framework for scalable ASD screening using data from 1242 children (18–48 months). A mobile application collected parent-child interaction audio and screening tool data (MCHAT, SCQ-L, SRS). Stage 1 differentiated typically developing from high-risk/ASD children, integrating MCHAT/SCQ-L text with audio features (AUROC 0.942). Stage 2 distinguished high-risk from ASD children by combining task success data with SRS text (AUROC 0.914, Accuracy 0.852). The model’s predicted risk categories strongly agreed with gold-standard ADOS-2 assessments (79.59% accuracy) and correlated significantly (Pearson r = 0.830, p < 0.001). Leveraging mobile data and deep learning, this framework demonstrates potential for accurate, scalable early ASD screening and risk stratification, supporting timely interventions.
Similar content being viewed by others
Introduction
Autism Spectrum Disorder (ASD) presents a growing global health challenge, characterized by a complex interplay of social-communication challenges, repetitive behaviors, and sensory processing differences. These characteristics significantly impact on the quality of life and long-term outcomes for individuals with ASD1. The heterogeneous nature of the disorder, with various presentations and varying severities, underscores the need for innovative diagnostic approaches, including those capable of identifying distinct behavioral phenotypes within the spectrum2. Crucially, early identification and intervention have been consistently shown to optimize developmental trajectories, reduce symptom severity, and improve overall well-being3,4. However, widely accepted diagnostic tools such as the Autism Diagnostic Observation Schedule (ADOS-2)5 and the Autism Diagnostic Interview-Revised (ADI-R) are resource-intensive, demanding substantial clinical expertise and extensive time6. This high resource requirement can delay or limit access to timely assessments, particularly in under-resourced areas, hindering the potential benefits of early intervention. Addressing this challenge, digital health innovations leveraging artificial intelligence (AI) offer a promising avenue for scalable, objective, and automated diagnostic support7,8,9,10.
It is crucial, however, to clarify that these AI tools are intended to aid in detection and risk stratification, not to replace comprehensive clinical diagnosis. Many existing AI applications rely on a single data modality. For instance, screening questionnaires like the M-CHAT-R/F11 and SCQ12, while widely used, depend on subjective parent reports which may not fully capture the complexity or subtle signs of ASD11,12. Moreover, traditional use of tools like the M-CHAT-R/F or SCQ often focuses primarily on the overall score, potentially overlooking the valuable semantic information contained within individual items. Drawing inspiration from work developing structured ASD phenotype descriptions using natural language processing (NLP)13, this study adopts a novel approach. We employ NLP techniques not just on the scores, but on the text of the screening questionnaires themselves, aiming to extract meaningful descriptions and identify specific behavioral traits associated with ASD-related terms used in the questions.
Other AI approaches analyzing facial expressions14 or neuroimaging data15,16 show potential but may require specialized equipment or yield results comparable to human experts, limiting widespread practical application for initial screening14. Integrating AI-based aids into primary care shows promise for enhancing efficiency17, but there remains a need for tools that leverage easily accessible, yet rich, data sources.
This study introduces and evaluates a novel multimodal AI framework designed to enhance the early screening and identification of ASD risk in young children. Crucially, this framework uniquely integrates two key data sources: (1) voice data extracted directly from videos capturing naturalistic parent-child interactions, and (2) semantically analyzed data derived from the text of standardized ASD screening questionnaires (e.g., M-CHAT-R/F, SCQ, SRS), processed using NLP. The clinical significance of this multimodal approach lies in its potential to create a more robust and reliable early detection signal. It combines objective, quantifiable vocal biomarkers related to language development and social communication, often altered in ASD18,19 captured during everyday interactions, with nuanced behavioral trait information extracted from validated screening instruments11,12. This synergy aims to improve the accuracy and reliability of early screening efforts compared to unimodal approaches, as discussed in related literature20,21,22,23.
Furthermore, our approach aligns with recent advancements in multimodal fusion techniques, such as those seen in robust multimodal emotion recognition with transformers (e.g., Zhu et al.11), cross-modal fusion for utterance-level analysis24 and contextual interaction-based multimodal emotion analysis with enhanced semantic information (e.g., Hazarika et al.25). Similarly, the use of contrastive learning for removing negative information in multimodal analysis (e.g., Wang et al.26) provides a foundation for developing more robust and reliable models.
From a clinical perspective, providing a more accurate risk assessment earlier in a child’s development holds substantial utility. Such a tool can assist clinicians in identifying children who warrant prioritized referral for comprehensive diagnostic evaluation. This facilitates more efficient use of limited diagnostic resources, potentially reducing lengthy wait times and enabling quicker access to essential early intervention services⁸. By leveraging readily available data like interaction videos and screening questionnaires (analyzed for content), the framework aims for practical applicability in diverse clinical settings.
Despite progress in AI for ASD7,9, and specific analyses of voice18,19,27 or multimodal data8,21,22,23,28, a research gap persists in developing and validating scalable AI tools that specifically integrate naturalistic voice recordings from interaction videos with semantically processed screening tool data for the explicit purpose of early ASD risk identification and stratification. This gap is particularly relevant in the context of ensuring trustworthy AI systems, as explored in fields like image super-resolution (e.g., Korkmaz et al.29), and leveraging advanced signal processing techniques, such as client-server based recognition systems for emotional and behavioral states (e.g., Zhu et al.30) or WiFi-based non-contact human presence detection (e.g., Zhang et al.31), and dynamic spectral graph anomaly detection (e.g., Zheng et al.32). These advancements highlight the potential for more sophisticated data acquisition and analysis in clinical settings.
To address this gap, this study introduces a novel two-stage multimodal AI framework that combines text data (e.g., MCHAT, SCQ, SRS), audio data from parent-child interactions, and hospital clinical information to enhance ASD screening and risk stratification. This framework builds upon prior work and addresses the limitations of unimodal approaches highlighted by Rahman et al. and others20. Specifically, the framework leverages advanced deep learning models like RoBERTa33, known for its strong performance in NLP tasks, and Whisper34, a state-of-the-art speech recognition model, to capture comprehensive information from text and audio data. Recent advances in speech processing, such as pre-trained speech embeddings, have further demonstrated the potential of leveraging audio data to understand spoken language development in children with ASD18. While other audio analysis methods, such as Audio Spectrogram Transformers (ASTs)35, have shown promise in various applications, the present framework utilizes Whisper for its superior speech recognition capabilities, particularly in handling the complexities of child speech and its demonstrated effectiveness in capturing subtle linguistic and prosodic features. The framework incorporates the following key innovations:
This framework introduces several key innovations. First, it features multimodal integration by merging semantically processed text-based survey data (via RoBERTa) with audio-derived features from parent-child interactions (processed by Whisper). This allows for a more comprehensive capture of social communication, language, and behavioral indicators than single-modality or score-based approaches. Second, our approach focuses on risk stratification, mapping prediction probabilities to clinical benchmarks such as ADOS-2 scores for actionable risk categorization into “Low Risk,” “Moderate Risk,” and “High Risk” groups6, thereby providing more reliable probability estimates in line with the recommendations of Nixon et al. on the importance of model calibration in deep learning36. Finally, we designed the framework with the potential for scalability and clinical utility, keeping in mind the potential for integration into a Clinical Decision Support System (CDSS). Such integration could support clinical decision-making in screening pathways, improve accessibility, and potentially alleviate clinical workload3,37,38. The CDSS integration is inspired by successful implementations in other domains, as reviewed by Abdar et al.39, highlighting the potential of AI to assist in clinical decision-making39. It also offers the potential for personalized treatment planning, a significant advancement in neurorehabilitation as suggested by Chang et al.40.
By integrating these specific multimodal data sources (semantically analyzed questionnaires and voice from videos), employing advanced deep learning tailored to each modality, explicitly addressing model calibration for reliable risk stratification, and considering practical clinical integration, this research aims to provide a scalable, automated tool. The goal is to significantly enhance the accuracy and efficiency of early ASD screening, thereby supporting clinicians in identifying at-risk children sooner. Demonstrating the feasibility and effectiveness of this AI-driven approach aims to pave the way for more accessible and timely support for ASD assessment pathways, ultimately contributing to improved developmental outcomes and quality of life for individuals with ASD and their families by facilitating earlier access to intervention.
Results
This section presents the results of the two-stage AI framework for ASD risk stratification, focusing on model performance, calibration, risk stratification, and correlation with clinical measures.
Stage 1 model performance: differentiating typically developing from at-risk children
The Stage 1 model, a multi-modal neural network based on the “RoBERTa-large” pre-trained model, was trained to distinguish between children with typical development (TD) and those in the combined High-Risk/ASD group. Performance was evaluated using 5-fold cross-validation, yielding an average AUROC of 0.942, accuracy of 0.86, precision of 0.85, recall of 0.85, and F1-score of 0.85 (Table 1). The consistently high AUC scores across all folds, illustrated in the ROC curves (Fig. 1), demonstrate the model’s strong and robust ability to discriminate between the groups.
These curves display the model’s performance in discriminating between typically developing children and those at risk for ASD across five cross-validation folds. Each Receiver Operating Characteristic (ROC) curve represents the performance of the model on a different held-out test set. The Area Under the Curve (AUC) for each fold is displayed in the legend, and the consistently high AUC scores demonstrate the model’s robust discriminative ability.
Auxiliary task: language delay prediction
The Stage 1 model also performed an auxiliary task of predicting language delay. The model achieved good performance on this task, with an average AUROC of 0.91, accuracy of 0.82, precision of 0.80, recall of 0.81, and F1-score of 0.80 across five folds (Table 2, Fig. 2). This further demonstrates its ability to extract meaningful clinical information from the input data.
These plots show the performance of the Stage 1 model on the secondary task of identifying language delay. The Receiver Operating Characteristic (ROC) curves are shown for five cross-validation folds, with each curve representing the model’s performance on a different held-out test set and the Area Under the Curve (AUC) for each fold displayed in the legend.
Stage 2 model performance: differentiating high-risk from ASD
The Stage 2 model, a fine-tuned RoBERTa-large model, was trained to differentiate between individuals at high risk (HR) for ASD and those diagnosed with ASD. This model integrated behavioral task success/failure data with textual data derived from the SRS.
The model was trained and evaluated using five different random seeds (Folds 100, 42, 2021, 7, and 12345) to assess the robustness of its performance. The results on held-out test sets for each fold are presented in Table 3 and Fig. 3.
The plots demonstrate the model’s robust ability to classify individuals as either High-Risk or diagnosed ASD across five separate evaluation runs. Each Receiver Operating Characteristic (ROC) curve shows the model’s performance when trained with a different random seed, with the Area Under the Curve (AUC) for each run displayed in the legend to confirm consistent, high performance.
The Stage 2 model demonstrated strong performance in differentiating between individuals diagnosed with ASD and those identified as high-risk (HR) for ASD. Across five-fold cross-validation, the model achieved an average AUC of 0.91 (range: 0.90–0.93) (Table 3). It also exhibited balanced overall performance, with average accuracy, precision, recall, and F1-score values of 0.85, 0.90, 0.90, and 0.90, respectively. Accuracy across folds ranged from 0.82 to 0.88, while AUC scores remained consistently high, ranging from 0.90 to 0.93, underscoring the model’s excellent discriminative ability. The ROC curves in Fig. 3 visually corroborate this strong performance, with all curves lying well above the chance line.
The model consistently excelled at correctly identifying individuals diagnosed with ASD, indicated by a generally high recall for class 1 (ASD) across all folds, ranging from 0.84 to 0.97. Precision for class 1 was also consistently high, ranging from 0.88 to 0.92. Performance on the High-Risk group (class 0) exhibited more variability. Notably, Fold 2021 demonstrated the highest precision (0.90) but the lowest recall (0.64) for this group. This suggests a potential trade-off between correctly identifying all high-risk individuals (high recall) and minimizing false positives (high precision) within this specific group. These variations may be attributed to the inherent heterogeneity of the High-Risk group or potentially reflect some sensitivity to the specific random seed used during training. However, the consistently high average AUC of 0.91 indicates that the model is generally robust and effective in distinguishing between individuals diagnosed with ASD and those at HR.
Calibration
To assess the reliability of the model’s predicted probabilities, calibration plots were generated. A perfectly calibrated model would produce a calibration plot with a diagonal line, indicating that the predicted probability matches the observed fraction of positive cases.
As shown in Fig. 4, the original model exhibits some degree of miscalibration, particularly in the higher probability range. To address this, isotonic regression was applied to calibrate the model’s predictions. Figure 4 (right panel) shows the calibration plot after applying isotonic regression. The calibrated model demonstrates improved calibration, with the curve aligning more closely to the ideal diagonal line.
These reliability curves compare the model’s probabilistic predictions to actual outcomes, showing significant improvement after calibration. The left panel shows the original, uncalibrated model, while the right panel shows the model’s improved calibration after applying isotonic regression, where its predictions align more closely with the ideal diagonal line of perfect calibration.
The Expected Calibration Error (ECE) for the original model was [0.14]. After isotonic regression calibration, the ECE was reduced to [<0.0001], indicating improved calibration performance.
These results suggest that calibration can enhance the reliability of the model’s predictions, making them more interpretable and clinically useful. The calibrated probabilities can be more confidently used to inform risk stratification and clinical decision-making.
Correlation between model predictions and ADOS scores
To assess the validity of the Stage 2 model’s predictions, we evaluated the correlation between the model’s output (mean calibrated probabilities across the five folds) and the clinically administered ADOS-2 total(T) score. The analysis revealed a strong and statistically significant positive correlation, with a Pearson correlation coefficient of r = 0.830 (p < 0.001) and a Spearman’s rank correlation of ρ = 0.889 (p < 0.001).
These results indicate a strong and statistically significant positive correlation (p < 0.001, using an alpha level of 0.05 throughout the study) between the model’s predictions and the severity of ASD symptoms as measured by ADOS given by the Fig. 5.
These visualizations confirm a strong, statistically significant positive correlation between the model’s output and clinical measures of ASD symptom severity. The scatter plot (left) and pair plot (right) both depict the relationship between the Stage 2 model’s mean calibrated probabilities and the clinically administered ADOS-2 TOTAL(T) scores, with the pair plot also showing the marginal distributions of each variable.
Risk stratification and threshold optimization
Building upon the strong correlation between model predictions and ADOS scores, we performed risk stratification to categorize participants into risk groups based on the model’s output and their ADOS-2 TOTAL(T) scores. Building upon the strong correlation with ADOS scores, we performed risk stratification to categorize participants into risk groups based on the model’s output and their ADOS-2 TOTAL(T) scores by optimizing thresholds for the model’s mean calibrated probability. The process aimed to best align the model’s output with clinical risk categories defined by ADOS-2 TOTAL(T) scores: Low Risk (score < 7), Moderate Risk (score ≥ 7 and ≤13), and HR (score > 13). The resulting optimized thresholds for the model’s probability were <0.40 for Low Risk, between 0.40 and 0.85 for Moderate Risk, and >0.85 for HR. As shown in Fig. 6, applying these thresholds to the model’s predictions yielded a maximum agreement (Accuracy) of 0.80 with the ADOS-based categories.
This graph illustrates the data-driven process used to identify the optimal probability thresholds for categorizing individuals into risk groups. The plot shows the agreement scores (Accuracy) between model-predicted risk categories and ADOS-based risk categories across a grid search of different threshold combinations, with the peak of the curve indicating the optimized thresholds (0.40, 0.85) that yielded the highest agreement.
This process of enhancing agreement from an uncalibrated state to an optimized one is detailed in Supplementary Fig. 16 and Supplementary Fig. 17, which show the progression from the initial agreement heatmap to the final calibrated cross-tabulation. The cross-tabulation heatmaps in Fig. 7 visualize the agreement between the model-predicted risk categories (using the optimized thresholds) and the ADOS-based risk categories.
The heatmaps provide a visual cross-tabulation of the model’s classification accuracy against the ground-truth risk categories defined by ADOS-2 scores. Using the optimized thresholds of 0.40 and 0.85, the left panel shows the classification results for individuals in the ADOS High Risk group, while the right panel shows the results for the ADOS Low and Moderate Risk groups.
Overall, the model demonstrates good agreement with ADOS-based risk categories, particularly for the High-Risk group (23 out of 37 correctly identified). This suggests that the model’s predictions align well with established clinical assessments, particularly for identifying individuals with more severe ASD symptoms. The discrepancies observed in the Moderate Risk group may reflect the inherent challenges in categorizing individuals with borderline symptom severity and highlight the potential value of the model in providing supplementary information to aid clinical judgment in such cases.
Comparison with existing literature
The Stage 1 model, designed to differentiate between typically developing (TD) children and those at HR for or diagnosed with ASD, achieved an impressive average AUROC of 0.942 across five folds. This performance surpasses many previous efforts that relied solely on traditional machine learning methods applied to parent-reported questionnaires, as highlighted in the review by Rahman et al.20. The high AUROC scores, consistently above 0.92 in each fold (Fig. 1, Table 1), indicate the model’s robustness and ability to generalize well to unseen data. Furthermore, the model’s strong performance on the auxiliary task of language delay prediction (average AUROC of 0.908, Fig. 2, Table 2) underscores its ability to extract meaningful and clinically relevant information from the multimodal input data. The successful integration of audio data, processed using the Whisper model, demonstrates the value of incorporating naturalistic observations of parent-child interactions, which capture subtle complexity in social communication and language development often missed by traditional assessment methods19,27.
The Stage 2 model, a fine-tuned RoBERTa-large model, further refined the classification by distinguishing between individuals at HR for ASD and those with a confirmed diagnosis. This model achieved an average AUROC of 0.914 and an average accuracy of 0.852 across five different training runs (Table 3, Fig. 3). These results are particularly noteworthy given the challenging nature of this classification task. The consistently high AUC scores across all folds (0.90 to 0.93) demonstrate the model’s robust discriminative ability and its potential for clinical utility. The strong positive correlation between the model’s predicted probabilities and ADOS-2 TOTAL(T) scores (Pearson r = 0.830, p < 0.001; Spearman ρ = 0.889, p < 0.001) (Fig. 5) further validates the clinical relevance of the model’s output, suggesting that it captures meaningful information about ASD symptom severity. These findings build upon prior work that has explored the use of AI in ASD diagnosis, such as those leveraging neuroimaging data or facial behavior analysis14,15,16, by demonstrating the power of combining textual and behavioral data within a deep learning framework.
Overall model performance
Our two-stage multimodal AI framework demonstrated robust performance in ASD risk stratification. An initial 3-class classification model (TD vs. High-Risk vs. ASD) was also explored, with performance metrics detailed in Supplementary Table 1 and corresponding confusion matrices and ROC curves in Supplementary Fig. 1. However, the two-stage binary classification approach was ultimately adopted for its superior performance and clinical interpretability.
Stage 1 Classification (TD vs. [High+ASD]): As detailed in Supplementary Table 2, the multimodal “Audio + Text with language delay” model (our Stage 1 model) achieved an AUROC of 0.9551 (95% CI: 0.8247–0.9812). Unimodal baselines showed lower performance: “Audio only” achieved an AUROC of 0.7691 (95% CI: 0.6758–0.8521), while “Text only” achieved 0.9418 (95% CI: 0.9020–0.9726). This highlights the significant contribution of questionnaire text and the value of multimodal fusion. The model “Audio + Text w/o language delay” also maintained a high AUROC of 0.9496 (95% CI: 0.9143–0.9777).
Stage 2 Classification (High-Risk vs. ASD): Performance metrics for Stage 2 models are presented in Supplementary Table 3. Our proposed Stage 2 model (“SRS + 5 Tasks S/F”) achieved an AUROC of 0.9317 (95% CI: 0.8585–0.9854). This multimodal approach significantly outperformed unimodal baselines: “Base SVM” (AUROC 0.8263), “SRS Only” (AUROC 0.8831), and “5 Tasks S/F Only” (AUROC 0.7607), confirming the synergistic effect of integrating both SRS and structured interaction task outcomes.
Error analysis and risk stratification
The error analysis utilized two datasets: confusion matrices in Supplementary Figs. 2–3 were computed on our original held-out test set, while visualizations in Supplementary Figs. 18–20 were generated using an independent, newly collected external cohort to assess Stage 1 true-vs-predicted labels (Supplementary Fig. 18), Stage 1 calibrated risk levels (Supplementary Fig. 19), and Stage 2 calibrated risk levels (Supplementary Fig. 20).
The first stage of the model (TD vs. non-TD) demonstrated high sensitivity on the original held-out set, achieving 100% sensitivity for non-TD detection. All true ASD (n = 11) and High-risk (n = 11) cases were correctly flagged as “Other,” while a 23% false-positive rate occurred among TD children (Supplementary Fig. 2). This performance held on the independent external cohort, where non-TD sensitivity remained at 100%. Crucially, these false positives almost exclusively mapped to the Low Risk category in the subsequent Stage 2 analysis (Supplementary Fig. 18). The risk calibration heatmap for the external cohort further showed that genuine non-TD cases occupied higher risk bins, while correctly classified TD children remained in the Low Risk bin (Supplementary Fig. 19).
In the second stage of the model (High-risk vs. ASD), detailed results for the original test set are presented in Supplementary Fig. 3. On the external cohort, the risk calibration demonstrated strong sensitivity for ASD, with 10 of 11 ASD cases (91%) classified as either HR or Medium Risk. In contrast, TD children remained almost entirely in the Low Risk category (12/13), while the High-risk group spanned all risk bins (Supplementary Fig. 20). The overall binary prediction performance for Stage 2 on this cohort is summarized in the confusion matrix in Supplementary Fig. 21.
In sum, by combining high-sensitivity binary classification with calibrated risk stratification—and validating on two independent cohorts—our two-stage framework offers a robust, clinically actionable pathway for early ASD screening with minimized unnecessary follow-ups and sustained accuracy.
Interpretability analysis
Micro-level interpretability was performed for the Stage 2 model using SHAP values and CLS attention mechanisms, providing sample-specific insights. This analysis focused particularly on the SRS + 5 Tasks Success/Failure model due to its high reliability (AUROC 0.9317).
Our SHAP analysis quantified the impact of specific features on model predictions. For predicting High-Risk (Class 0), task successes (e.g., “Success of Mimicked actions2” [+0.030], “Success of Reacted to snack” [+0.009]) consistently showed the largest positive contributions, while phrases indicating strong social skills like “usually looks up and pays attention when spoken to” (-0.006) decreased this likelihood (Supplementary Figs. 4, 5, 10, 11). Conversely, for predicting ASD (Class 1), these same task successes had negative SHAP values (e.g., “Success of Mimicked actions2” [-0.030]), actively decreasing the likelihood of an ASD diagnosis. Instead, phrases reflecting social-communication strengths such as “usually looks up and pays attention when spoken to” (+0.007) increased the ASD prediction (Supplementary Figs. 6, 7, 12, 13).
To further investigate the model’s focus, we analyzed the [CLS] token’s attention patterns. This confirmed that the model places significant emphasis on both explicit task outcomes and critical behavioral descriptors. High attention scores were consistently observed for phrases related to direct task performance like “Success of Mimicked actions2” (≈0.010) and “Success of Played catch” (≈0.0035) (Supplementary Figs. 8, 9). The model also paid high attention to core ASD-related features such as “child always different” (≈0.005), “speech tone” (≈0.0045), “not attending to social approaches from adults” (≈0.003), and adaptive behaviors like “self-care behavior” (≈0.013) (Supplementary Figs. 14, 15).
Discussion
This study introduces a novel two-stage multimodal AI framework for ASD screening and risk stratification, representing a significant advancement in early detection and intervention efforts. By integrating text-based parent-reported surveys, audio data from parent-child interactions, and clinical assessment data including ADOS module scores (utilized particularly for differentiating High-Risk and ASD groups in Stage 2), our framework addresses critical limitations of prior research that often relied on unimodal data sources³. To our knowledge, this is the first large-scale study (n = 1242) to employ such a comprehensive, AI-driven approach to systematically investigate the correlation between predictive phenotypes and ASD diagnostic outcomes. The results underscore the feasibility and effectiveness of leveraging advanced deep learning models, specifically RoBERTa33 and Whisper34, to achieve accurate ASD screening and risk stratification.
A key contribution of this research lies in developing a comprehensive ASD assessment framework that integrates the AI model’s predictions with ADOS-2 total(T) scores, which measure the severity of autism symptoms rather than ASD risk itself. This integration enables a clinically meaningful and fine-grained evaluation of symptom severity, aligning AI-driven assessments with established clinical scales. The framework demonstrated high agreement (79.59% accuracy) with ADOS-2-defined symptom severity categories, suggesting its potential to support clinical decision-making by providing structured insights into symptom presentation. Notably, the model exhibited high sensitivity in identifying individuals with elevated ADOS-2 total(T) scores (correctly identifying 23 out of 37 individuals in the highest severity category), underscoring its potential utility in prioritizing individuals for comprehensive diagnostic evaluations and addressing delays in ASD diagnosis.6,37.
The severity stratification framework provides multi-level clinical insights by generating valuable information sequentially through its two-stage process. The initial Stage 1 classification identifies children likely needing further evaluation (non-TD), after which Stage 2 differentiates between a higher likelihood of ASD versus being High-Risk. This process enables reliable severity stratification, which in turn facilitates several key clinical applications. It supports early identification and prioritization by helping to identify children who warrant comprehensive diagnostic evaluation, a crucial benefit given extended waiting periods for specialized care¹. Furthermore, the objective outputs serve as valuable supplementary information for enhanced assessment, especially for individuals with borderline or ambiguous clinical presentations. Finally, the framework directly supports informing personalized intervention, as the detailed stratification allows clinicians to tailor the intensity and focus of early intervention plans more effectively based on the child’s specific assessed needs⁸.
In any screening paradigm, misclassifications are inevitable, and understanding their downstream effects is crucial. The core workflow presented here utilizes two sequential binary models—Stage 1 (TD vs. [High-Risk + ASD]) and Stage 2 (High-Risk vs. ASD)—followed by calibrated risk stratification correlated with ADOS-2 scores. While misclassifying TD children as false positives can increase parental anxiety and follow-up burden, the two‐stage design cushions this impact. A TD child mistakenly flagged in Stage 1 is highly unlikely to receive a High‐Risk score in Stage 2, thereby reducing unnecessary diagnostic referrals. Clinicians can further use model confidence scores; for instance, children with borderline probabilities in Stage 2 may be monitored over time rather than being sent directly for a full evaluation. Conversely, missing an ASD diagnosis (a false negative) delays critical early intervention. Although the pipeline achieves >90% ASD sensitivity, the small number of misses in Stage 2 (1/11 in the external cohort) underscores that no automated tool is infallible, reinforcing the need for continued developmental surveillance by primary care providers and the option for repeat screening if concerns persist.
To build clinical trust and facilitate informed decision-making, micro-level interpretability features like SHAP values and attention maps are critical. These tools allow clinicians to peer into the model’s “reasoning” by revealing which specific phrases from questionnaires or structured task outcomes most strongly influenced a prediction. For example, if the model heavily weighted a phrase like “not attending to social approaches from adults” towards an ASD classification, it directly aligns with clinical diagnostic criteria, allowing the clinician to cross-reference this with their own observations. This transparency transforms the model from a black box into a collaborative partner, enabling clinicians to discuss complex or borderline cases by integrating the model’s evidence with their own clinical expertise.
This study directly addresses several limitations of prior research. The multimodal approach, incorporating text and audio data, overcomes the constraints of unimodal methods that have dominated previous ASD classification studies13,20,41. By harnessing the power of deep learning models like RoBERTa33 and Whisper34, the framework moves beyond traditional machine learning techniques that often struggle with the complexity and heterogeneity of ASD presentations. Furthermore, the focus on model calibration, drawing on principles from uncertainty quantification research42,43,44 enhances the reliability and interpretability of the model’s predictions, addressing a crucial gap often overlooked in earlier work20,39. These findings align with the global shift toward scalable, accessible, and efficient digital health solutions, particularly in under-resourced settings. The app-based approach for data collection, coupled with automated AI-driven analysis, has the potential to significantly reduce the time and resources required for ASD screening, which is relevant given the increasing prevalence of ASD1 and the development of other AI-based diagnostic tools10.
Despite these advancements, the study has limitations. The initial sample sizes (n ≈ 35 per group) limit the precision of low-frequency error estimates, and the findings may not be generalizable to all populations, as the study was conducted in a specific Korean clinical setting. While the text-based component used English ASD terms for semantic processing of questionnaire data, the cultural and linguistic specificity of the context must be considered. The audio component, however, focused on more language-agnostic acoustic features, which may enhance transferability. A second limitation involves data collection and ground truth. The uncontrolled nature of audio recording, including ambient noise and variable child participation, impacted the quality of child vocalizations. Additionally, ADOS-2 scores were used as the ground truth for risk stratification; however, these scores do not always correspond with clinicians’ final judgments and represent only one of several diagnostic tools.
Future research should focus on validating the framework in larger, more diverse populations with different linguistic, cultural, and socioeconomic backgrounds to assess fairness and robustness, including its performance in children with co-occurring developmental conditions. Further model optimization could be achieved by incorporating additional data modalities, such as video or physiological measures, and exploring adaptive thresholds to trade off false positives versus false negatives. Implementing standardized protocols for audio collection could also improve performance. Finally, longitudinal studies are needed to evaluate the long-term impact of this AI-driven approach on developmental trajectories, and investigating its integration with electronic health records could streamline clinical workflows.
In sum, this study successfully developed and validated a novel multimodal AI framework that integrates voice data and semantically analyzed questionnaire responses for effective ASD screening and severity stratification in toddlers. The primary clinical implication lies in its potential to address critical bottlenecks in current care pathways by offering objective support for prioritizing assessments, enhancing clinical judgment, and informing the tailoring of early intervention intensity based on the initial severity level identified by the model3. By leveraging readily available data through a mobile application combined with advanced AI, this framework represents a scalable solution. Its implementation, following rigorous validation, could substantially enhance current ASD care pathways, reduce the burden on specialized services, and ultimately improve developmental outcomes for children with ASD and their families.
Methods
This study employed a two-stage AI framework (Fig. 8) for ASD risk stratification, integrating multimodal data from surveys, parent-child interactions, and clinical assessments.
The framework shows a sequential process beginning with initial classification and culminating in actionable risk stratification. This diagram illustrates how Stage 1 (Model 1) uses voice and questionnaire data to classify individuals as Typically Developing (TD) or at-risk (High+ASD). Individuals flagged as at-risk proceed to Stage 2 (Model 2), which uses semantic features and task outcomes to further classify them as High-Risk or diagnosed ASD, leading to stratification into three clinical risk categories with recommended actions.
Study design and participants
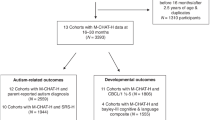
This study recruited 1242 children aged 18–48 months from clinical settings across 9 hospitals in the Republic of Korea. Data collection was primarily conducted using a mobile application, which gathered responses to key ASD screening questionnaires (including the M-CHAT-R/F, SCQ-L, and SRS) —with the M-CHAT-R/F administered for children aged 18–30 months and the SCQ-L for those aged 24–48 months— and recorded voice data from parent-child interaction videos during standardized tasks. Participants were categorized into three groups: TD, high-risk for ASD, and confirmed ASD. This categorization was based on results from these screening tools and clinical diagnoses performed according to DSM-5 criteria. Figure 9 details the participant flow through the study, illustrating the inclusion and exclusion criteria applied based on data availability and clearly delineating the sample sizes and specific data inputs for both Stage 1 and Stage 2 of the two-stage AI model analysis.
This flowchart details participant recruitment, exclusion criteria, and data allocation for the two-stage AI model. It shows how participants were selected based on the availability of mobile application data, including screening tool responses and voice recordings. Stage 1 included 818 participants to differentiate TD from non-TD individuals, while Stage 2 included 515 participants from the non-TD group to differentiate between High-Risk and ASD individuals. (MCHAT: Modified Checklist for Autism in Toddlers; SCQ-L: Social Communication Questionnaire-Lifetime; SRS: Social Responsiveness Scale; ASD: Autism Spectrum Disorder; TD: Typically Developing; non-TD: Non-Typically Developing).
Participants in the study were categorized into three distinct groups: ASD, High-Risk (HR), and TD (TD). The ASD group included toddlers with a confirmed diagnosis based on a combination of screening tools and clinical diagnoses using DSM-5 criteria6. These diagnoses were further supported by comprehensive clinical evaluations utilizing standardized assessment tools, including the Autism Diagnostic Observation Schedule, Second Edition (ADOS-2)45, and the Korean Childhood Autism Rating Scale, Second Edition (K-CARS-2)5. The High-Risk (HR) group comprised toddlers identified through primary screening tools such as the Modified Checklist for Autism in Toddlers, Revised with Follow-Up (M-CHAT-R/F), Q-CHAT, SCQ, SRR, and Bedevel46,47. Toddlers with delayed language development, a family history of ASD, or preterm birth before 36 weeks were also classified into this group. These children exhibited potential ASD traits requiring follow-up evaluations but did not meet the full criteria for an ASD diagnosis. Finally, the TD group consisted of toddlers exhibiting typical developmental patterns without any indications of ASD traits, as determined by the M-CHAT-R/F47, and who had no family history of developmental disorders.
Ethical considerations
This study was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the Institutional Review Board (IRB) of Severance Hospital, Yonsei University College of Medicine (IRB No. 4-2022-1468), and the IRBs of all participating hospitals in this multi-center study: Seoul National University Hospital (IRB No. 2209-096-1360), Eunpyeong St. Mary’s Hospital (IRB No. 2022-3419-0002), Wonkwang University Hospital (IRB No. 2022-12-023-001), Seoul National University Bundang Hospital (IRB No. 2305-829-401), Hanyang University Hospital (IRB No. 2022-12-007-001), Chungbuk National University Hospital (IRB No. 2023-04-034), and Seoul St. Mary’s Hospital (IRB No. KC24ENDI0198). Written informed consent was obtained from the legal guardian of every participant before any data collection.
Procedures for data protection were implemented to uphold the highest ethical standards for collecting data from minors. Upon enrollment, each child was assigned a unique, non-identifiable “Study ID,” and no direct identifiers appear in analysis files. All data were stored on encrypted servers at Yonsei University, using AES-256 and HASH-256 encryption for sensitive information. Access to the database was restricted to authorized personnel through role-based permissions. Data management complied with the Personal Information Protection Act and the Bioethics and Safety Act. Raw audio recordings were deleted after feature extraction, and pseudonymized data will be stored for 10 years from the study’s conclusion before being securely destroyed, with any extension requiring further IRB review. All methods and procedures are described here in the main manuscript, not in Supplementary Information.
Recruitment
The study employed a multi-pronged recruitment strategy, partnering with clinical institutions, community childcare centers, and pediatric clinics specializing in developmental delays to reach a diverse population. Targeted outreach to high-risk groups was conducted through inclusive daycare centers and by distributing brochures in specialized clinics. Participant engagement was fostered by offering incentives like detailed developmental assessment reports.
Data collection
Data collection for this study involved three primary sources: survey data, digital phenotyping data, and clinical data.
Survey data was acquired through standardized questionnaires completed by parents, including the Modified Checklist for Autism in Toddlers (MCHAT)12,46, the Social Communication Questionnaire Lifetime (SCQ-L)12,46, and the Social Responsiveness Scale second edition (SRS-2)20,23. These surveys provided text-based data for the AI model. The initial sample based on MCHAT and SCQ-L scores consisted of 1242 children, including 434 TD, 331 High-Risk, and 477 with ASD. For the Stage 2 analysis requiring SRS and 5-task audio data, 32 participants were excluded, resulting in a sample of 162 High-Risk and 353 ASD children (Total = 515).
Digital phenotyping data was collected from 897 children via a custom-developed mobile application. This included audio data from parent-child interactions, where the app guided parents through a series of standardized tasks designed to elicit specific behaviors relevant to ASD diagnosis (Table 4). The number of interaction tasks varied by age, with children aged 18–23 months completing four tasks, those aged 24–35 months completing five, and those aged 36–48 months completing six. These tasks included Responding to Name, Imitation (one-step for 18–23 months, two-step for 24–48 months), Ball Play, Symbolic Play, Requesting Help, and Free Play. Audio from the Free Play task was analyzed for the Stage 1 model using data from 294 TD, 214 High-Risk, and 389 ASD children. Audio from 547 children was used in the Stage 2 model. Additionally, task success/failure data was collected for each of the five structured tasks (excluding Free Play), where caregivers evaluated and recorded the child’s performance based on predefined criteria; this binary information was used directly in the Stage 2 model.
Finally, clinical data was collected, including assessments from ADOS-2 Module 1 (ADOS-2 Mod1), ADOS-2 Module 2 (ADOS-2 Mod2), and the ADOS-2 Toddler Module (ADOS-2 ModT). To ensure consistency across modules, a composite score—referred to as the ADOS-2 total(T) score—was calculated by summing the ADOS social affect (ADOS_SA) score and the ADOS restricted and repetitive behavior (ADOS_RRB) score. This ADOS-2 total(T) score served as the ground truth for model validation and risk stratification.
Language delay definition and calculation
In this study, language delay was defined as a delay of 7 months or more in either receptive or expressive language age compared to the child’s chronological age. Language development was assessed using either the Preschool Receptive-Expressive Language Scale (PRES) or the Sequenced Language Scale for Infants (SELSI). For each child, the obtained receptive and expressive language ages were subtracted from their chronological age. If the receptive or expressive language delay was 7 months or greater, the child was classified as having a language delay.
Stage 1 model development
The development of the AI model employed a two-stage approach. This section details the methodology used for Stage 1, focusing on distinguishing TD children from those at High-Risk (HR) for or diagnosed with ASD.
The initial dataset comprised data from 1242 participants. Following the exclusion of 61 participants due to missing free-play audio data or the presence of specific confounding factors (high-risk ASD with family history or high-risk ASD with premature birth), the final dataset for Stage 1 analysis consisted of 818 children. This cohort included 273 TD children, 175 children identified as High-Risk for ASD, and 370 children diagnosed with ASD.
Data preprocessing
Text-based analysis utilized responses from the Modified Checklist for Autism in Toddlers (MCHAT) and the Social Communication Questionnaire - Lifetime (SCQ-L). We employed a mapping strategy to enhance the extraction of ASD-relevant information from these questionnaires. Specifically, 1943 medical concepts derived from Zhao et al. (2022)13 were used to map the survey items to a broader set of 3336 ASD-related terms, creating a richer textual representation for identifying high-risk individuals. Subsequently, a Sentence Transformer model (Reimers & Gurevych, 2019) was implemented to calculate cosine similarity scores between each survey item and the expanded set of ASD terms. The top five matching terms for each item, based on cosine similarity, were identified. From this subset, 1–2 clinically relevant terms were carefully selected as keywords, guided by expert clinical judgment. These selected keywords served as the foundation for training a RoBERTa-based binary classification model. This model was designed to differentiate between TD children and the combined group of high-risk and ASD children, leveraging the textual information derived from the MCHAT and SCQ-L.
The audio data consisted of recordings from parent-child interaction tasks. Each recording, with a total duration of 3 min, was segmented into 30-s intervals. These segments were then processed using a Multiple Instance Learning (MIL) framework (Ilse et al.)48, with the Whisper model (Radford et al.)34 serving as the feature extraction backbone. To address the presence of language delay, children’s receptive and expressive language development ages were assessed. Children exhibiting a delay of 7 months or more compared to their chronological age were labeled as having a language delay. This language delay status was then predicted using the Whisper model (Radford et al.)34 within the MIL framework (Ilse et al.)48, enabling effective feature extraction from the variable-length audio inputs.
Model architecture
A multi-modal neural network was constructed to integrate both text and audio data. This network was based on the pre-trained ‘RoBERTa-large’ and Whisper Encoder models. The ‘RoBERTa-large’ model served as the text pathway, extracting features from the preprocessed textual data derived from the questionnaires. Concurrently, a pre-trained whisper Encoder model, which was fine-tuned as part of this study, formed the audio pathway, extracting features from the segmented audio data. The extracted text and audio features were then concatenated to form a unified representation. This combined feature vector was fed into a final classification layer, which produced logits for classifying children into either the TD group or the combined HR + ASD group. As an auxiliary task, a separate output layer was included to predict the presence or absence of language delay (Yes/No) based on the fused features. Finally, a hard ensemble approach was implemented to combine the predictions generated by the MCHAT/SCQ-L-based binary classification model and the language delay prediction model, ultimately classifying children into the TD or combined HR + ASD groups.
The overall loss function for Stage 1 was a composite of losses from the main classification task and the auxiliary language delay prediction task. The total loss function, which equally weighted the classification loss and the language-delay prediction loss, is defined in Eq. (1).
Here, \({{\rm{L}}}_{{\rm{CE}}}\) and \({L}_{{LD}}\) represent the cross-entropy losses for the main classification task and the auxiliary language delay task, respectively.
The classification error for the primary task (classifying TD vs. High-Risk/ASD) is computed using Eq. (2):
In this equation, N is the total number of samples, \({y}_{i}\) is the true class label for sample \(i\) (0 for TD, 1 for High-Risk/ASD), and \({p}_{i}\) is the predicted probability of the positive class (High-Risk/ASD) for sample \(i\).
The classification error for predicting language delay is computed using Eq. (3):
Here, \(N\) represents the total number of samples, \({z}_{i}\) is the true binary label for language delay (1 for delay, 0 for no delay), and \({q}_{i}\) is the predicted probability of language delay for sample \(i\).
Model training and evaluation
The dataset used for training and evaluation for Stage 1 comprised data from 273 TD, 175 high-risk, and 370 ASD participants. Model training was conducted using 5-fold stratified cross-validation. For each fold, the data was split into ~70% training, 10% validation, and 20% held-out test set. The splits were stratified by class (TD, HR, ASD) to maintain class balance across all subsets. The same partitioning was used consistently across both training stages.
The ‘RoBERTa-large’ model was initialized with pre-trained weights, serving as the text pathway for feature extraction from the preprocessed textual data. Concurrently, the Whisper model encoder was fine-tuned across all its layers on our dataset, rather than being used as a frozen feature extractor, to form the audio pathway. In addition to the primary classification task (distinguishing typically developing from high-risk/ASD children), an auxiliary classifier was included to predict language delay status, leveraging the model’s ability to capture clinically relevant vocal features. The model was trained using the cross-entropy loss function for both the main classification task and the auxiliary language delay task. No explicit data augmentation was applied to the audio recordings; this decision was based on the natural variability and ambient noise present in the real-world clinical and home environments, as preliminary experiments showed that artificial augmentation degraded performance.
Features from the audio (Whisper encoder hidden states) and text (RoBERTa embeddings) models were concatenated at the feature level before being fed into a fully connected neural network for classification. This multimodal fusion allowed the model to jointly leverage both vocal and linguistic information. No ensemble methods were employed during inference; the final predictions were generated by this single integrated multimodal model.
Hyperparameters were set as follows: a batch size of 4, trained for 10 epochs, a fixed learning rate of 1e-5, and a weight decay of 0.01. The AdamW optimizer was employed, and no learning rate scheduler was used. While explicit early stopping with patience was not applied, the model checkpoint with the lowest validation loss during training was selected and saved for evaluation. During inference, the trained model was set to evaluation mode and processed test data in batches of 64, without shuffling. Model performance was evaluated using AUC, accuracy, and loss, calculated on the held-out test set for each fold.
Stage 2 model development
This stage integrated data from two sources: the outcomes of parent-child interaction tasks and responses from the Social Responsiveness Scale-2 (SRS-2). The success or failure of each of the six parent-child interaction tasks was recorded. The criteria for determining success or failure were based on clinically relevant behavioral markers. Examples of these markers include responding to a name within two attempts, correctly imitating at least one action, and engaging in symbolic play. The initial task success/failure labels were primarily provided by parents during the assessment. To ensure validity and quality control, these parent-reported labels for a stratified sample of 50 subjects (comprising 227 total task items) were rigorously validated against video-reviewed ground truth by experienced clinicians. A subset of 10 subjects (46 tasks) underwent re-review by clinicians to ensure clarity and consistency. Consistency was further ensured through a comprehensive inter-rater reliability analysis; Cohen’s kappa (κ) was calculated to quantify the agreement between parent-reported labels and clinician video reviews, yielding κ = 0.885 (95% CI: 0.85–0.92). This indicates “almost perfect agreement” according to Landis and Koch (1977)49 benchmarks. The raw observed agreement was 94.27% (214 agreed items out of 227 total items), with only 13 discrepancies identified, representing an exceptionally low mismatch rate of 5.7% for clinical and behavioral studies. This robust reliability supports the validity and quality of the parent-reported task outcomes used in our study. Like the text preprocessing in Stage 1, SRS survey responses were mapped to ASD-related terms to enhance the extraction of relevant information. The task success/failure data, represented as binary outcomes for each task, and the mapped SRS terms were combined to train a RoBERTa-Large model using fine-tuning. The model was trained for 10 epochs with a batch size of 8 and a learning rate of 2e-5. A cross-entropy loss function and the AdamW optimizer were employed for training. To prevent overfitting, early stopping based on validation loss was implemented. This trained model achieved a promising AUC of 0.93.
Stage 2 model training and evaluation
For Stage 2, model training and evaluation involved a dataset of 162 High-Risk and 353 ASD children. We employed StratifiedGroupKFold for all splits to ensure consistency in labels and subject grouping across the dataset. An initial approximate 80:10:10 (train:validation: test) split was performed, which was later updated following the inclusion of new subjects, resulting in final sample counts of Train (455), Validation (58), and Test (57). Fixed train/validation/test sets were used for evaluation, rather than explicit cross-validation folds in this stage.
The RoBERTa-Large model was fine-tuned for 10 epochs. An effective batch size of 32 was used, achieved with a per-device train batch size of 8 and a gradient accumulation of 4 steps. The learning rate was fixed at 2e-5, and a weight decay of 1e-8 was used for regularization, with 0 warmup steps. Evaluation and model saving occurred per epoch, and the best model was loaded based on the lowest evaluation loss on the validation set (load_best_model_at_end = True, metric_for_best_model = “eval_loss”). No explicit hyperparameter search was performed, with the listed values being those used for fine-tuning. A cross-entropy loss function and the AdamW optimizer were employed for training.
Model performance was comprehensively evaluated using several metrics. The Area Under the Receiver Operating Characteristic curve (AUROC) was used to assess the model’s ability to discriminate between classes. The F1-score, the harmonic mean of precision and recall, provided a balanced measure of accuracy, while overall accuracy measured the correctness of the model’s predictions. Precision, the proportion of accurate positive predictions among all positive predictions, and recall, the proportion of accurate positive predictions among all actual positive cases, were also calculated.
Model calibration and correlation analysis
Furthermore, the ECE was computed to quantify the alignment between predicted probabilities and observed frequencies across multiple bins. The ECE is calculated using the formula shown in Eq. (4):
In this equation, \({B}_{i}\) represents the i-th bin containing predicted probabilities, \({{|B}}_{i}|\) is the number of predictions within that bin, and N is the total number of predictions. The term \({accuracy}\left({B}_{i}\right)\) refers to the fraction of true positives in bin i, while \({confidence}\left({B}_{i}\right)\) is the mean predicted probability for the predictions in that same bin.
A lower ECE indicates better calibration, meaning the predicted probabilities more accurately reflect the actual likelihood of the outcome.
Isotonic regression was employed for calibration to ensure that the model’s predicted probabilities aligned with clinically defined ADOS-2 risk categories. Isotonic regression is a non-parametric method that adjusts the predicted probabilities to better match the observed outcomes while preserving the original ranking of predictions (monotonicity).
The calibrated probabilities \({p}_{{cal}}\) are obtained by solving the optimization problem shown in Eq. (5):
In this equation, \({P}_{{iso}}\) represents the isotonic probabilities, \({y}_{i}\) is the true label for the \({i}_{{th}}\) sample, and N is the total number of samples.
We used 10 equal-width bins (edges at 0.0, 0.1, …, 1.0) to compute ECE (Eq. 4), and we fitted a scikit-learn Isotonic Regression (PAV algorithm, out_of_bounds = “clip”) to obtain calibrated probabilities under the monotonicity constraint (Eq. 5).
To investigate the relationship between the model’s predicted probabilities and ADOS scores, both Pearson and Spearman correlation analyses were conducted. The Pearson correlation coefficient (“r”), which measures the linear relationship between two variables, was calculated using the following formula.
The Pearson correlation coefficient, which measures the linear relationship between two variables, is calculated using Eq. (6):
In this equation, \({{\rm{x}}}_{{\rm{i}}}\) and \({{\rm{y}}}_{{\rm{i}}}\) are the values of the first variable (Mean Calibrated Probabilities) and the second variable (ADOS-2 total(T) score) respectively, while \(\bar{{\rm{x}}}\) and \(\bar{{\rm{y}}}\) are the means of each variable. The term n represents the total number of data points.
The Spearman rank correlation coefficient ρ, a non-parametric measure of rank correlation, was also calculated using Eq. (7):
Here, \({d}_{i}\) is the difference in ranks between the two variables for each data point \(i\) (i.e., \(\text{Rank}\left({x}_{i}\right)-\text{Rank}\left({y}_{i}\right)\) and n is the number of data points.
ADOS scores were scaled to a range of 0–1 for these analyses. For risk stratification, a data-driven threshold optimization procedure was employed to define thresholds for classifying individuals into “Low Risk,” “Moderate Risk,” and “High Risk” categories based on the calibrated predicted probabilities. The thresholds were optimized to maximize agreement (measured by accuracy) with ADOS-2 based risk categories. These ADOS-2 based risk categories were defined as follows: Low Risk: ADOS-2 TOTAL(T) < 7; Moderate Risk: 7 ≤ ADOS-2 TOTAL(T) ≤ 13; and High Risk: ADOS-2 TOTAL(T) > 13. The optimization process involved a grid search over a range of threshold values. The lower threshold (t1) varied from 0.1 to 0.45 in increments of 0.05, and the upper threshold (t2) ranged from t1 + 0.1 to 0.85 in increments of 0.05.
Risk stratification and threshold optimization
As shown in Eq. (8), the predicted risk \({R}_{p}\) is classified based on the calibrated probability (\(P\)) using two thresholds \({T}_{1}\) and \({T}_{2}\):
In this equation, \(P\) represents the predicted probability (e.g., the mean calibrated probability for an individual), while \({T}_{1}\) and \({T}_{2}\) T2 are the thresholds that separate the risk groups.
As defined in Eq. (9), the ADOS risk \({R}_{a}\) is determined using the ADOS-2 TOTAL(T) score(\(S\)) with fixed thresholds S1 and S2:
In this equation, \(S\) is the ADOS-2 TOTAL(T) score for an individual, while S1 and S2 are the thresholds for the ADOS risk categories (e.g.,\({S}_{1}\) = 7, \({S}_{2}\) = 13).
The combined risk \({R}_{c}\) is fusion of \({R}_{p}\) and \({R}_{a}\), is defined as shown in Eq. (10):
The agreement metric (A) measures the similarity between \({{\rm{R}}}_{{\rm{p}}}\) and \({{\rm{R}}}_{{\rm{a}}}\), can be computed using accuracy as defined in Eq. (11):
Alternatively, Eq. (12) defines the agreement using a weighted F1 score formulation:
In this formula, \(T{P}_{i}\), \(F{P}_{i}\), and \(F{N}_{i}\) are the true positives, false positives, and false negatives for class \(i\), respectively. The term \({W}_{i}\) represents the weight for class \(i\), which is proportional to its representation in the dataset.
The optimal thresholds \(\left({T}_{1}^{* },{T}_{2}^{* }\right)\), which maximize the agreement metric, are determined by solving the optimization problem shown in Eq. (13):
This optimization is performed by iterating the thresholds \({T}_{1}{and}{T}_{2}\) over a defined range (e.g., \({T}_{1}\in \left[\mathrm{0.1,0.5}\right],{T}_{2}\in \left[{T}_{1}+\mathrm{0.1,0.9}\right]\)), where \(A\left({T}_{1},{T}_{2}\right)\) is the agreement metric computed for each pair of thresholds.
A combined risk assessment strategy was implemented by integrating both the model-predicted risk and the ADOS-2-based risk. This combined risk was determined according to the following criteria: An individual was classified as “High Risk” if either the model-predicted risk or the ADOS-2 risk was high. “Moderate Risk” was assigned if one of the risks was moderate and the other was high. Finally, an individual was classified as “Low Risk” if both the model-predicted risk and the ADOS-2 risk were either low or moderate. This combined approach aimed to leverage the strengths of the AI model and the established clinical assessment.
Statistical analysis
Descriptive statistics summarized participant characteristics. Chi-square tests with post-hoc Bonferroni correction examined associations between categorical variables. Statistical significance was set at p < 0.05. Analyses used Python (version 3.9) with pandas, NumPy, SciPy, scikit-learn, statsmodels, Matplotlib, torch, torchaudio, transformers, and Seaborn libraries. We trained the model using a Quadro RTX 8000. The model was implemented using PyTorch, utilizing a GPU with fixed random seeds for ensuring reproducibility.
Data availability
The raw datasets generated and analyzed during the current study are not publicly available due to their sensitive nature, which includes identifiable clinical information and audio recordings of child participants. Public dissemination of this data would breach patient privacy and violate the terms of the Institutional Review Board (IRB) approval under which it was collected. To facilitate reproducibility, a comprehensive data schema defining the structure and variables of the minimal dataset is provided in Supplementary Data 1. This schema details all variables derived from clinical assessments (ADOS-2, PRES, SELSI), screening questionnaires (MCHAT, SCQ-L, SRS-2), and audio feature extraction, as used in the final analyses. The processed data itself cannot be shared. However, the provided schema allows other researchers to structure their own datasets in the same format, enabling them to replicate our methodology and analyses.
Code availability
The complete source code used for data preprocessing, model development, training, and analysis is openly available in a GitHub repository: https://github.com/skwgbobf/Multimodal-AI-ASD-Risk-ScreeningF. The repository includes the Jupyter notebooks used to generate the final datasets for both stages of the model (e.g., Stage1_model/notebooks/1.Preprocess_git.ipynb and stage2_model/notebooks/1_Data_Preprocessing/model2_data preprocess_Final.ipynb), the implementation of the two-stage deep learning framework, and all scripts required to reproduce the results presented in the manuscript.
Change history
15 September 2025
In the Acknowledgements section of this article the grant number "DMHR25E01" relating to "National Center for Mental Health and the Digital Healthcare Center at Severance Hospital was incorrectly given as "MHER22A01". The article has been corrected.
References
Perochon, S. et al. Early detection of autism using digital behavioral phenotyping. Nat. Med. 29, 2489–2497 (2023).
Stevens, E. et al. Identification and analysis of behavioral phenotypes in autism spectrum disorder via unsupervised machine learning. Int. J. Med. Inform. 129, 29–36 (2019).
Qin, L. et al. New advances in the diagnosis and treatment of autism spectrum disorders. Eur. J. Med. Res. 29, 322 (2024).
Ilias, L., Mouzakitis, S. & Askounis, D. Calibration of transformer-based models for identifying stress and depression in social media. IEEE Trans. Comput. Soc. Syst. 1, 12 (2023).
Kim, S. W. & Yoo, H. J. Korean Childhood Autism Rating Scale 2nd edn (K-CARS-2) (Hakjisa Publisher, 2020).
Christiansz, J. A. et al. Autism spectrum disorder in the DSM-5: diagnostic sensitivity and specificity in early childhood. J. Autism Dev. Disord. 46, 2054–2063 (2016).
Kohli, M., Kar, A. K. & Sinha, S. The role of intelligent technologies in early detection of autism spectrum disorder (ASD): a scoping review. IEEE Access 10, 104887–104913 (2022).
Zhu, F. L. et al. A multimodal machine learning system in early screening for toddlers with autism spectrum disorders based on the response to name. Front. Psychiatry 14, 1039293 (2023).
Anagnostopoulou, P. et al. Artificial intelligence in autism assessment. Int. J. Emerg. Technol. Learn. 15, 95–107 (2020).
Megerian, J. T. et al. An artificial intelligence-based medical device to aid the diagnosis of autism spectrum disorder. npj Digit. Med. 5, 57 (2022).
Zhu, X. et al. RMER-DT: robust multimodal emotion recognition in conversational contexts based on diffusion and transformers. Inf. Fusion 123, 103268 (2025)
Rubio-Martín, S. et al. Enhancing ASD detection accuracy: a combined approach of machine learning and deep learning models with natural language processing. Health Inf. Sci. Syst. 12, 20 (2024).
Zhao, M. et al. Development of a phenotype ontology for autism spectrum disorder by natural language processing on electronic health records. J. Neurodev. Disord. 14, 32 (2022).
Sariyanidi, E. et al. Comparison of human experts and AI in predicting autism from facial behavior. CEUR Workshop Proc. 3359, 48–57 (2023).
Moridian, P. et al. Automatic autism spectrum disorder detection using artificial intelligence methods with MRI neuroimaging: a review. Front. Mol. Neurosci. 15, 999605 (2022).
Helmy, E. et al. Role of artificial intelligence for autism diagnosis using DTI and fMRI: a survey. Biomedicines 11, 1858 (2023).
Sohl, K. et al. Feasibility and impact of integrating an artificial intelligence–based diagnosis aid for autism into the extension for community health outcomes autism primary care model: protocol for a prospective observational study. JMIR Res. Protoc. 11, e37576 (2022).
Xu, A. et al. Understanding spoken language development of children with ASD using pre-trained speech embeddings. arXiv https://arxiv.org/abs/2305.14117 (2023).
Eni, M. et al. Estimating autism severity in young children from speech signals using a deep neural network. IEEE Access 8, 139489–139500 (2020).
Rahman, M. M. et al. A review of machine learning methods of feature selection and classification for autism spectrum disorder. Brain Sci. 10, 949 (2020).
Han, J., Jiang, G., Ouyang, G. & Li, X. A multimodal approach for identifying autism spectrum disorders in children. IEEE Trans. Neural Syst. Rehabil. Eng. 30, 2003–2011 (2022).
Dcouto, S. S. & Pradeepkandhasamy, J. Multimodal deep learning in early autism detection—recent advances and challenges. Eng. Proc. 59, 205 (2023).
Tamizhmalar, D., Subbiah, S. & Premkumar, R. A multimodal diagnostic framework for autism spectrum disorder using deep learning: an in-depth exploration. In Proc. 2024 International Conference on. Power, Energy, Control and Transmission Systems (ICPECTS) 1–5 (IEEE, 2024).
Luo, J., Phan, H. & Reiss, J. Cross-modal fusion techniques for utterance-level emotion recognition from text and speech. In Proc. 2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 1–5 (IEEE, 2023).
Hazarika, D., Poria, S., Majumder, N., & Cambria, E. CIME: contextual interaction-based multimodal emotion analysis with enhanced semantic information. In Proc. 2018 Conference on Empirical Methods in Natural Language Processing 4467–4477 (2018).
Wang, R., Wang, Y., Cambria, E., Zhu, X. & Liu, Z. Contrastive-based removal of negative information in multimodal emotion analysis. Cogn. Comput. 17, 107–118 (2025).
Milling, M. et al. Evaluating the impact of voice activity detection on speech emotion recognition for autistic children. Front. Comput. Sci. 4, 837269 (2022).
Lee, J. H. et al. Deep-learning-based detection of infants with autism spectrum disorder using auto-encoder feature representation. Sensors 20, 6762 (2020).
Korkmaz, C., Cirakman, E., Tekalp, A. M., & Dogan, Z. Trustworthy SR: Resolving Ambiguity in Image Super-resolution via Diffusion Models and Human Feedback. arXiv preprint arXiv:2402.07597 (2024).
Zhu, X. et al. A client-server based recognition system: non-contact single/multiple emotional and behavioral state assessment methods. Comput. Biol. Med. 260, 108564 (2025).
Zhang, Y., Wang, X., Wen, J. & Zhu, X. WiFi-based non-contact human presence detection technology. Sci. Rep. 14, 3605 (2024).
Zheng, J. et al. Dynamic spectral graph anomaly detection. In Proc. AAAI Conference on Artificial Intelligence Vol. 39, 13410–13418 (2025).
Liu, Y. et al. RoBERTa: a robustly optimized BERT pretraining approach. arXiv preprint arXiv:1907.11692 364 (2019).
Radford, A. et al. Robust speech recognition via large-scale weak supervision. In Proc. 40th International Conference on Machine Learning (ICML) Vol. 202, 28492–28518 (PMLR, 2023).
Gong, Y. et al. AST: audio spectrogram transformer. https://arxiv.org/abs/2104.01778 (2021).
Nixon, J. et al. Measuring calibration in deep learning. In Proc. CVPR Workshops Vol. 2, 7 (2019).
Buddenkotte, T. et al. Calibrating ensembles for scalable uncertainty quantification in deep learning-based medical image segmentation. Comput. Biol. Med. 163, 107096 (2023).
Choi, E. S. et al. Applying artificial intelligence for diagnostic classification of Korean autism spectrum disorder. Psychiatry Investig. 17, 1090–1095 (2020).
Abdar, M. et al. A review of uncertainty quantification in deep learning: techniques, applications and challenges. Inf. Fusion 76, 243–297 (2021).
Chang, M. C. et al. The use of artificial intelligence to predict the prognosis of patients undergoing central nervous system rehabilitation: a narrative review. Healthcare 11, 2687 (2023).
Huang, S. C. et al. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. npj Digit. Med. 3, 136 (2020).
Lakshminarayanan, B., Pritzel, A. & Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. Advances in Neural Information Processing Systems 30 (2017).
Wilson, A. G. & Izmailov, P. Bayesian deep learning and a probabilistic perspective of generalization. Adv. Neural Inf. Process. Syst. 33, 4697–4708 (2020).
Hüllermeier, E. & Waegeman, W. Aleatoric and epistemic uncertainty in machine learning: an introduction to concepts and methods. Mach. Learn 110, 457–506 (2021).
Western Psychological Services. Autism Diagnostic Observation Schedule 2nd edn (ADOS-2) (Western Psychological Services, 2013).
Mujeeb Rahman, K. K. & Subashini, M. A deep neural network-based model for screening autism spectrum disorder using the quantitative checklist for autism in toddlers (QCHAT). J. Autism Dev. Disord. 52, 1–15 (2022).
Robins, D. L., Deborah Fein, and Marianne Barton. Modified checklist for autism in toddlers, revised, with follow-up (M-CHAT-R/F) TM. LineageN (2009).
Ilse, M. et al. Attention-based deep multiple instance learning. In Proc. 35th International conference on machine (ICML) Vol. 80, 2127–2136 (PMLR, 2018).
Landis, J. R. & Koch, G. G. The measurement of observer agreement for categorical data. Biometrics 33, 159–174 (1977).
Acknowledgements
This study was supported by funding from the National Center for Mental Health (grant number: DMHR25E01) and the Digital Healthcare Center at Severance Hospital, Yonsei University College of Medicine. We thank the clinicians and research staff at the nine participating hospitals for their assistance with data collection and validation. We are also grateful to the children and families who participated in this study.
Author information
Authors and Affiliations
Contributions
Concept and Design: S.B., J.H., H.K. Data Acquisition, Preprocessing, Cleaning and Validation: S.H., J.Y., H.C., J.L., R.D., H.S., H.K., H.L., M.P., E.K., C.Y., D.L., H.Y., Y.L., G.B., J.I.K., H.S., H.K., E.J., S.C., J.S., J.H.Y., S.J. Model Development and Analysis: S.B., J.H. Data Interpretation and Clinical Inference: S.H., R.D., B.K., K.C.•Drafting of the Manuscript: S.B. Critical Revision for Intellectual Content: J.H., S.H., J.M., H.K., R.D. •Statistical Analysis: S.B., J.H., J.M. Supervision: H.K., B.K., K.C. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bae, S., Hong, J., Ha, S. et al. Multimodal AI for risk stratification in autism spectrum disorder: integrating voice and screening tools. npj Digit. Med. 8, 538 (2025). https://doi.org/10.1038/s41746-025-01914-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-01914-6
This article is cited by
-
Alzheimer’s diagnosis from EEG with reliable probabilities: subject-wise, leakage-free evaluation and isotonic calibration
Journal of Engineering and Applied Science (2025)