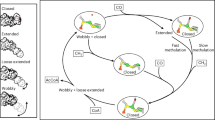
Abstract
Carbon monoxide dehydrogenases (CODHs) selectively catalyse the reversible reduction of CO2 to CO and water. The catalytic centre of CODHs contains a unique [NiFe4(OH)(µ3-S)4] cluster whose role in activating and converting CO2 is poorly understood. Here we reveal the structures of all catalytically relevant oxidation states with and without substrates and products bound. We show that the Ni–Fe cluster combines a rigid Fe–S core with a dynamic Ni(I/II)–Fe(II) dyad. The redox-active element is the Ni ion, cycling between square-planar Ni(II) and T-shaped Ni(I) states with metalloradical character, the latter serving as the nucleophile for CO2 activation. The Fe(II) ion switches between two positions, the one preceding CO2 activation is close to Ni(I) with a potential Ni(I)–Fe(II) interaction and the other binds the substrates CO2 and water. We demonstrate how the Ni–Fe cluster creates an efficient CO2 reduction catalyst and provides a blueprint for the design of novel catalysts based on abundant transition metals.

Similar content being viewed by others
Main
Catalysts that selectively bind and efficiently reduce CO2 are required for a sustainable economic future, but their development is challenged by high activation energies, product inhibition and lack of substrate selectivity1,2. Carbon monoxide dehydrogenases (CODHs) are ancient enzymes found in anaerobic bacteria and archaea that catalyse the reversible reduction of CO2 with two protons and two electrons to CO and water2,3,4,5. CODHs are among the few enzymes in nature that contain nickel, here integrated within a [NiFe4(OH)(µ3-S)4] cluster, known as the C cluster, which can be described as a distorted NiFe3S4 cubane connected to an Fe(II) in an exo position6,7,8. The C cluster transfers electrons to and from a [4Fe4S] cluster, known as the B cluster6,7. According to the current model of the catalytic cycle, the C cluster alternates between three different electronic states, referred to as Cred1, Cint and Cred2, which have been distinguished by means of their spectroscopic properties9,10,11,12. In its most reduced paramagnetic state (Cred2), the C cluster reduces CO2, while in the two-electron more oxidized paramagnetic Cred1 state, it binds and activates CO (refs. 9,10). The diamagnetic Cint state is one-electron more reduced than Cred1 and one-electron more oxidized than Cred2 (ref. 11).
Despite numerous spectroscopic and crystallographic investigations, the catalytic mechanism of the reduction of CO2 by CODHs remains poorly understood as we lack the structures of the spectroscopically defined C states and the electronic states of the determined structures13,14. Thus, key mechanistic questions remain unanswered, such as the structures of the three catalytic redox states, the catalytic role and electronic state of the Ni ion, the structure of the CO2-activating nucleophile and the reaction steps following CO2 binding, including the generation and release of the reaction product CO (ref. 14).
In this study, we resolved the structures of the catalytically relevant Cred1, Cint and Cred2 states and followed the reaction from CO2 activation to the CO-bound state by combining atomic resolution crystallography with infrared and electron paramagnetic resonance spectroscopy to assign electronic states to the structures. We show that the structural plasticity of the Ni–Fe dyad is essential for CO2 activation and that the Ni ion is the redox-active unit under turnover conditions.
Results
Parallel structural and spectroscopic studies
We used CODH-II from Carboxydothermus hydrogenoformans as a model CODH15. All investigated cluster states were prepared in crystal form and treated identically for analysis by X-ray diffraction, infrared (IR) and electron paramagnetic resonance (EPR) spectroscopy (Extended Data Fig. 1). The nine crystal structures were resolved at resolutions between 1.0 and 1.3 Å (Supplementary Table 1).
EPR analysis of the Cred1, Cint and Cred2 states
The crystals of all three states were incubated with 6 mM Ti(III)–ethylenediaminetetraacetate (Ti(III)–EDTA) and exhibited a dominant EPR signal in the range of g = 2.02 to g = 1.84 (Fig. 1 and Extended Data Fig. 2). Using the pH dependence of the midpoint potentials and by adding substrates, we were able to quantitatively accumulate all three catalytically relevant redox states of the C cluster in the crystalline state (Fig. 1a). While we accumulated the Cred1 state by treating crystals at pH 6.0, we observed the Cint state at pH 8.0 and the addition of 45 mM NaHCO3 at pH 8 quantitatively enriched the Cred2 state (Fig. 1a). The Cred1 state was assigned at pH 6, based on the typical g values at g = 1.82 and 1.64 (Supplementary Table 4), while at pH 8, we only observed a signal at g = 2.03 (Supplementary Table 4), characteristic of the gx value of the reduced B cluster (Bred)15. No species were detected in the spectral region typical of the C cluster, which indicates a diamagnetic Cint state. This intermediate converted to the Cred2 state (gy = 1.82 and gz = 1.74) when NaHCO3 was added. Although the EPR samples consisted of hundreds of crystals, the signals showed only slight anisotropy (Extended Data Fig. 3).
a, EPR spectra of the three electronic states of the C cluster in crystal suspensions, recorded at 10 K and 0.25 mW. The intense signal from Ti(III) (dashed lines) dominates the centre of the spectra (Extended Data Fig. 2). The rhombic signals from Cred1 (red) and Cred2 (blue) were simulated, while species indicative of the Ni-deficient C cluster state CredΔNi and Bred are highlighted in purple and dark cyan, respectively. The absence of any paramagnetic C cluster signal is indicative for the diamagnetic Cint state (orange) in the mddle EPR spectrum. The asterisk labels a deviation from the simulation, which could originate from an additional minor species or relate to the slight angle dependence of the EPR spectra (Extended Data Fig. 3). b–d, Crystal structures of the EPR-identified states determined from crystals treated in the same way as those used for EPR spectroscopy. For each state, the σA-weighted 2Fobserved − Fcalculated (2Fo − Fc) maps (Fobserved, observed structure factor amplitudes; Fcalculated, calculated structure factor amplitudes) in blue mesh, contour level = 1.0σ, and blue isosurface, contour level = 2.0σ) of Ni–Fe1 with the bound OH or CO2 ligand are shown separately to the right of the refined crystal structure. Two alternative positions of Fe1 are shown as ‘i’ for the ‘in’ conformation (denoted Fe1,i) and ‘o’ for the ‘out’ conformation (denoted Fe1,o), with the alternative conformation of His261 coordinating Fe not shown for clarity. The occupancies of the OH ligands in b and c and of CO2 in d were refined to 0.67, 0.52 and 0.66, respectively. Each state was chemically poised at stable redox potentials (Cred1 = −409 mV, Cint = −530 mV and Cred2 = −530 mV versus the standard hydrogen electrode (SHE)) using Ti(III)–EDTA (see Methods).
Structures of the Cred1, Cint and Cred2 states
In the Cred1 state, the C cluster clearly shows the electron density of a ligand between Ni and Fe1 (Fig. 1b and Extended Data Fig. 4). Bond lengths of 2.24 Å (Ni–OH) and 2.05 Å (Fe1–OH) are consistent with a bridging µ-OH ligand (Fig. 1b) and agree with model calculations predicting a Ni2+–Fe2+-bridging hydroxo ligand in the Cred1 state16. The Ni ligands have a slightly distorted square-planar arrangement with the bridging OH ligand lying below the plane defined by Ni and its three S ligands (Fig. 1b).
In contrast to the Cred1 state, the Ni in the C cluster in the Cint state has only three ligands in a slightly distorted T-shaped coordination (Fig. 1c). The hydroxy ligand is still part of the C cluster, but is further away from Ni (Ni–OH distance of 2.66 Å) and becomes a terminal ligand for Fe1 (Fe1–OH bond length of 1.96 Å; Fig. 1c). The structure is very similar to previously reported structures8 that are likely to represent the same EPR-silent state of the C cluster.
Adding Ti(III)–EDTA alone was not sufficient to generate the Cred2 state, which accumulated when we added NaHCO3. The conditions were very similar to those previously used to enrich the CO2-bound state, and the crystal structure of Cred2 is indistinguishable from the CO2-bound structures reported previously8,17, with CO2 bound by Ni and Fe1 (Fig. 1d) and all three having equivalent occupancies (Supplementary Table 2). The bond lengths and angles are consistent with CO2 being two-electron reduced and unprotonated, corresponding to a carbonite ligand17,18. The short Ni–C bond is consistent with a potential carbene character of the ligand17,19. The cooperative activation of CO2 by Ni and Fe1 is supported by the presence of amino acids within hydrogen-bonding distances of the oxygen atoms.
A pH jump splits the CO2 ligand and generates CO
We wondered why the C–O cleavage reaction in our crystals did not proceed from the Cred2 state because CO2 is sufficiently reduced. However, the carbonite ligand is unprotonated and C–O bond splitting requires contact with an electrophile (proton)2,20. It is therefore likely that at the slightly basic pH 8.0, the reduction of CO2 is less favourable than the oxidation of CO (ref. 4) and protonation may become rate-limiting. To increase the proton concentration in the solution, we lowered the pH by immersing crystals in the Cred2 state in a buffer at pH 4.0 for 20–30 s before cryo-cooling (hereafter referred to as the pH jump).
The structure after the pH jump shows a weaker electron density for the bound CO2 and additional features around the Ni ion (Fig. 2a). First, the electron density of Ni loses its spherical shape and becomes elongated. This elongation is also observed in anomalous difference maps, revealing two positions for the Ni ion. In one position, Ni is in the plane defined by its three sulfur-containing ligands, which is the position in the CO2-bound Cred2 state. The second position is 0.7 Å above the square-planar Ni ion, with similar Ni–S distances as before, but with a distorted tetrahedral ligand arrangement (Fig. 2a).
a, Crystal structure (left) and electron density map (right) of the CO- and CO2-bound state (0.15 and 0.60 occupancies, respectively) of the C cluster after the pH jump. In addition to the carbonite ligand, we observe electron density for a diatomic ligand above Ni, which is attributed to CO. Two alternative positions of Ni are shown: one with square-planar coordination (labelled A) and one with a distorted tetrahedral coordination (labelled B). b, IR spectra of identically treated crystals after the pH jump and incubation in a 12C- and 13C-containing NaHCO3 solution. The optical density (OD) is a measure of how much light is absorbed. c, The crystal structure (left) and electron density map (right) after incubation of crystals with CO in the presence of Ti(III)–EDTA (pH 8.0) reveals electron density for CO and CO2 bound to Ni (0.38 and 0.34 occupancies, respectively). d, IR spectra of crystals incubated with 12CO or 13CO in the presence of Ti(III)–EDTA (pH 8.0). Bands from Ni(I)–CO and Ni(II)–CO species are highlighted in green and pink, respectively. Additional signals between 1,850 and 1,950 cm−1 may also represent vibrations from metal carbonyls, but they could not be unambiguously assigned due to the absence of an isotopic shift. In these mixed-state structures, the major species are shown in dark colours, while the minor species are in light colours. In a and c, the electron density maps are shown as a mesh: σA-weighted 2Fo − Fc in blue with a contour of 1.0σ and Fo − Fc omit in green with a contour of 4.0σ. With Ti(III)–EDTA at pH 8.0, the crystals were chemically poised at −530 mV versus SHE.
Above this second Ni ion position, we observe an additional electron density with the typical elongated shape of a diatomic ligand. Modelling this density with water resulted in a residual electron density, indicating a terminally bound diatomic molecule, such as CO. The occupancies of the tetrahedral Ni ion and CO match (Supplementary Table 2), consistent with the CO being bound in the coordination sphere of the tetrahedrally coordinated Ni ion. The refined CO ligand has a Ni–C bond length of 1.83 Å and is tilted out of the expected linear Ni–C–O arrangement (Fig. 2a). To obtain further information on the identity of the ligand bound to Ni, we recorded IR spectra of the crystals following a pH jump. The IR spectrum reveals prominent bands at 1,971 and 1,980 cm−1 (Fig. 2b), which exhibited the expected isotopic shift to lower wavenumbers (1,925 and 1,935 cm−1) when the crystals were incubated with 13C-containing NaHCO3 solution.
Finally, to confirm the presence of a CO ligand bound to Ni, we incubated crystals under a CO/N2 atmosphere in the presence of Ti(III)–EDTA (pH 8.0, Cint state) at an overpressure of 83 kPa for 1 h. The resulting structure is very similar to the pH jump structure, with the same two Ni positions but a stronger electron density for the apical CO ligand and a higher occupancy of the tetrahedrally coordinated Ni (Fig. 2c and Extended Data Fig. 6). Electron density corresponding to a CO2 ligand bound to the square-planar coordinated Ni position shows that some of the CO has turned over. Compared with the pH jump structure, we observe more CO-bound and less CO2-bound molecules. The IR spectrum of the CO-incubated crystals shows the same dominant bands as in the pH jump experiments (Fig. 2d). In addition, some weaker bands between 2,020 and 2,060 cm−1 are also detected. When incubated with 13CO, all of the bands were shifted by −44 to −48 cm−1 (Fig. 2d). Without the pH jump or in the absence of CO, we observed no absorptions from metal-bound CO in the IR spectra of the crystals (Extended Data Fig. 7).
Thus, the spectroscopic data and structures are consistent with the conversion of CO2 to CO after the pH jump and a conversion of CO to the CO2-bound Cred2 state.
A Ni(I)–CO complex has formed
The main signals observed in the IR spectra between 1,950 and 2,000 cm−1 indicate the presence of CO bound to a Ni(I) ion, in line with the spectroscopic data from a recently reported CODH-mimicking Ni–Fe ferredoxin and other Ni(I)–CO complexes (Supplementary Table 3)21. Both CO-bound structures show that the Ni ion moves out of the sulfur plane to bind CO, adopting a distorted tetrahedral coordination that would be consistent with a Ni(I) ion (Fig. 2). The weaker signals above 2,020 cm−1 can be related to a smaller amount of CO bound to a Ni(II) ion.
The Cred2 state contains a carbonite ligand
While we observed the Cred2 signature with Ti(III)–EDTA only in the presence of CO2, we wondered whether we might be able to generate the Cred2 signal at more negative reduction potentials than those obtainable with Ti(III)–EDTA. We therefore treated CODH crystals at pH 8.0 with Eu(II)–diethylenetriaminepentaacetate (Eu(II)–DTPA)22 and checked for the generation of Cred2 by EPR spectroscopy. Crystal suspensions showed clear EPR signals for Bred, and we tentatively assigned additional signals slightly shifted from those of the B cluster to a minor amount of a Ni-deficient reduced C cluster (Fig. 3a)23. However, we observed only a very weak signal from Cred2, with most of the C cluster being EPR-silent (Extended Data Fig. 8). As a Ni–hydride has been proposed for the Cred2 state16, whose formation may be limited by available protons at pH 8, we increased the proton concentration, incubating CODH crystals in 8 mM Eu(II)–DTPA at pH 6.0, but still did not observe a quantitative paramagnetic signal of the C cluster (Fig. 3a). Only when we added NaHCO3 to the incubation solution at pH 8 did we observe a prominent signal from Cred2 with gx = 1.98, gy = 1.82 and gz = 1.74 (Fig. 3a and Supplementary Table 4). While this species was partially obscured in the Ti(III)–EDTA sample, we can fully define it in the presence of Eu(II)–DTPA, where an almost quantitative conversion to the Cred2 state is indicated (Extended Data Fig. 8). Significant line broadening of the Bred signal was observed, which suggests spin coupling between the paramagnetic B and C cluster states. As with Ti(III)–EDTA, CO2 is clearly defined in this state in the electron density map (Extended Data Fig. 4i) and has accumulated to an occupancy of 79% relative to the occupancy of Ni in the C cluster. In contrast to the structure obtained with Ti(III)–EDTA, some electron density of the CO ligand is visible. The presence of bound CO in the crystals is corroborated by the corresponding IR spectra that show a clear signal at 1,967 cm−1 (Extended Data Fig. 7), originating from Ni(I)-bound CO, as observed after a pH jump and incubation with CO (Fig. 2). The presence of CO in the NaHCO3-treated crystals is unlikely to be due to turnover at the C cluster, but to the ability of Eu(II)–DTPA to directly reduce CO2 (or hydrogen carbonate) to CO, which we observed in solution (Extended Data Fig. 9).
a, EPR spectra of crystals treated with Eu(II)–DTPA under different conditions. The EPR spectra were recorded at 10 K and 0.063 mW. The rhombic signals from Cred2 (blue), Cred2′ (light blue), Bred (dark cyan) and CredΔNi (violet) were simulated. The deconvoluted spectra are presented in Extended Data Fig. 8. b, Structure (left) and electron density map (right) of Eu(II)–DTPA-treated crystals at pH 6.0. The σA-weighted 2Fo − Fc map (blue mesh) is contoured at 3.0σ. Multiple crystals treated in the same way reproduced the shown structure, which has occupancies of 0.78 for both Ni,A and Fe1,i. c, Scheme showing the interconversion of the three observed Cint states, referred to as Cint,o for the OH-bound Cint state, Cint,i for the state with a short Ni–Fe1 distance devoid of an OH ligand and Cint,o–CO for the CO-bound state. d, The inward movement of Fe1 and its coordinating histidine (His261) on transitioning between its ‘in’ and ‘out’ positions. The Ti(III)-reduced pH 8.0 sample (Cint,o state) is shown in violet and the Eu(II)-reduced pH 6.0 sample (Cint,i state) is shown in blue. The σA-weighted 2Fo − Fc maps (blue mesh, Ti(III)-reduced pH 8.0; orange mesh, Eu(II)-reduced pH 6.0) are contoured at 1.0σ. For Eu(II)–DTPA at pH 8.0 and pH 6.0, the crystals were chemically poised at −660 and −632 mV versus SHE, respectively.
The Cred2 state is central to our understanding of the reaction mechanism of CODHs, but its structure remains uncertain, mostly because the similar spectral properties of Cred1 and Cred2 give no indication of where the two additional electrons of the Cred2 state may reside and different possible sites have been proposed, such as a Ni–hydride ligand, a Ni–Fe bond or a formal Ni(0) state16,24. Based on our data alone, we cannot rule out the existence of different Cred2 states. However, as the formation of Cred2 has frequently been reported to depend on the presence or absence of CO2, CO or cyanate9,12,25, the presence of a carbonite ligand seems to be the feature distinguishing the Cred1 and Cred2 states. It also explains why Cint accumulates in the presence of Ti(III)–citrate under N2/Ar atmosphere but is rapidly converted to Cred2 in a CO2 atmosphere12.
Although Cred2 can be described as containing a Ni(II) ion with its characteristic square-planar coordination, the short Ni–CO2 bond and calculated charges of around 1− for the ligand18 would be consistent with a greater charge delocalization between the carbonite ligand and the C cluster. This shift of charge from the carbonite ligand to the C cluster is probably also responsible for the slightly longer Ni–S bond length in the Cred2 state compared with in the Cred1 state (Extended Data Fig. 5). In summary, a carbonite ligand bound to the C cluster seems to agree with the spectroscopic data available for the Cred2 state that indicate small changes in charge and conserved spin states for Cred1 and Cred2, and the absence of an OH ligand at Fe1 in Cred2 (ref. 26).
A Ni(I) ion is characteristic of the Cint states
We observed the C cluster to be in EPR-silent Cint states under different conditions, namely, with Ti(III)–EDTA at pH 8.0, Eu(II)–DTPA at pH 8.0 and pH 6.0, and when we added CO to the reduced states. We therefore wondered whether all these EPR-silent states have something in common and compared their structures.
The structure of CODH incubated with 8 mM Eu(II)–DTPA at pH 8.0 (Extended Data Fig. 4h) is very similar to that incubated with Ti(III)–EDTA and CO, where the majority of the C cluster has a tetrahedrally coordinated Ni bound to CO and a minor fraction containing CO2 bound to square-planar Ni, explaining the observed traces of the Cred2 signature. The CO is likely to have been generated from and by Eu(II)–DTPA (Extended Data Fig. 9). In all of the structures described above, Fe1 is found in two positions: the dominant position is labelled ‘o’ for the ‘out’ position and the alternative position is labelled ‘i’ for the ‘in’ position (Extended Data Fig. 4). In the ‘out’ position, Fe1 carries an OH ligand, whereas in the ‘in’ position it loses its OH ligand, with Cys526 now completing its tetrahedral coordination. The in/out movement also leads to changes in the Ni–Fe distances, which in the ‘out’ position of Fe1 are 2.8 Å to the T-shaped Ni and 3.5 Å to the tetrahedral CO-bound Ni, whereas in the ‘in’ position of Fe1 the Ni–Fe1 distance shrinks to 2.6 Å. When we incubated crystals with 8 mM Eu(II)–DTPA at pH 6.0, the occupancy of the ‘out’ position of Fe1 strongly decreased and that of the ‘in’ position increased (Supplementary Table 2). Surprisingly, the Eu(II)–DTPA-reduced crystal structure at pH 6.0 shares the same structural properties previously proposed for Cred2, such as two unusually short Ni–Fe distances (Ni–Fe1: 2.60 Å and Ni–Fe3: 2.58 Å), which may indicate Ni–Fe bonds24 or the presence of a Ni–Fe-bridging hydride, similar to that in Ni–Fe hydrogenase, in which a Ni–Fe distance of 2.57 Å was found27. However, the spectroscopic data showed no indication of a Cred2 state when we lowered the pH and the C cluster remained EPR-silent. Incubating Eu(II)–DTPA-treated crystals at pH 6.0 with CO generated about 50% of the CO-bound state described above (equivalent to 67% of all Ni having CO bound) with Fe1 in the ‘out’ position with an OH ligand (Supplementary Table 2). This is in line with the corresponding IR spectra, which exhibit an intense band at 1,970 cm−1 (Extended Data Fig. 7i). Thus, the binding of CO to Ni results in its movement away from Fe1 and Fe3 and affects the in–out equilibrium of Fe1 (Fig. 3c), consistent with some Ni–Fe1 interaction when both metals are close. Notably, the related EPR spectrum shows a substoichiometric resonance at gx = 1.98, gy = 1.82 and gz = 1.79, similar but not identical to Cred2 (Fig. 3a). The intensity of the signal (Extended Data Fig. 8) coincides with the occupancy of the bound CO2 (Supplementary Table 2); therefore, we assign the structure to a Cred2′ state enriched at low pH. Despite the large movements of Fe1 and Ni, the Fe3S4 core of the C cluster remains unchanged in all of the observed structures (Fig. 4).
Superimposition of the structures of the C clusters in the five states determined in this study, namely, Cred1, Cred2, Cint,i, Cint,o and Cint–CO, showing that the Fe3S4 core remains unchanged despite the movements of the Ni and Fe1 ions. The five states are colour-coded and the positions of Ni and Fe1 are indicated.
In summary, we have observed three structurally distinct EPR-silent states of the C cluster, with different coordination spheres for Fe1, with an OH or a second cysteine ligand, and Ni, with a T-shaped coordination or tetrahedral geometry with a CO ligand (Fig. 3c). For the CO-bound state, the CO stretching frequency indicates the presence of Ni(I). In the other Cint structures, Ni has a slightly distorted T-shaped coordination. We argue that all of these Cint states contain a Ni(I) ion.
In the Cred1 state, the Ni ion has a distorted square-planar coordination with a Ni–Fe-bridging OH ligand, consistent with a Ni(II) ion. The one-electron reduction of the Cred1 state to the Cint state is concomitant with the dissociation of the OH ligand from the Ni ion, indicating its reduction to Ni(I) by populating the antibonding \(d_{x^2-y^2}\) orbital28. The formation of Ni(I) has chemical precedent as T-shaped complexes are well documented for Ni(I), but are atypical for Ni(II) (refs. 29,30). T-Shaped Ni(I) complexes, similar to other low-valent T-shaped 3d metals, harbour unusual reactivities, reflected in their designation as metalloradicals28,30. The reactivity of these complexes is determined by the unpaired electron directed towards the open coordination site and by their tendency to undergo one-electron oxidation to square-planar Ni(II) complexes.
The unsaturated threefold coordination of Ni in an Fe–S scaffold found in the Cint states has recently been mimicked in a synthetic model in which three-coordinate Ni was integrated into a WFe2S4 cluster31. Notably, the three-coordinate Ni was found to be Ni(I) in the oxidized and reduced states of the synthetic C cluster model with a Ni–W bond that is structurally analogous to the Ni–Fe3 interaction in the C cluster31. Finally, a Ni(I) ion would explain the observed diamagnetism of the C cluster in the Cint states, arising from antiferromagnetic coupling of the spin of the unpaired electron in Ni(I) with the S = 1/2 spin of the Fe3S4 unit.
Discussion
The CODH mechanism can be modified and refined in light of our results. First, we have revised the electronic structure of the C cluster and the role of Ni (Fig. 5a–h). During the catalytic cycle, only one electron is directly taken up by the C cluster and the only metal ion that changes its oxidation state during the catalytic cycle is Ni, cycling between Ni(I) and Ni(II). As CO2 is reduced by two electrons, the second electron must be delivered by the B cluster. The oxidation state of the B cluster is likely to determine whether CO oxidation (with Box) or CO2 reduction (with Bred) is the prevalent reaction, which is supported by the more negative reduction potential of the B cluster in the presence of CO2 (ref. 12). The [Fe3S4] unit of the C cluster serves to integrate and hold the Ni ion, creating its T-shaped S coordination with electron buffering capacity. The two electrons that distinguish Cred1 (Fig. 5d) and Cred2 (Fig. 5a) are stored in a carbonite ligand. The Ni–OH bond observed in the Cred1 state would indicate that CO reacts by inserting into the Ni–OH bond (Fig. 5c), forming a Ni–C(O)O(H)–Fe unit (Fig. 5b), whose deprotonation generates the observed Cred2 state (Fig. 5a).
a, In the Cred2 state (PDB: 9FPH), the C cluster carries an unprotonated carbonite ligand. b,c, Hydrogen bonds from His93 and Lys563, together with the covalency of the Ni–C bond, maintain negative charges on the O atoms and foster the protonation of the Fe-bound O of CO2 (b), destabilizing the C–O bond and splitting the COOH ligand (c). d, The Cred1–CO state is likely to be short-lived because Ni(II) is an atypical CO binder51. Under oxidizing conditions, CO will diffuse out of the active site and generate the Cred1 state (PDB: 9FPG). e,f, A two-electron reduction will generate the Cint,o state (PDB: 9FPF) (e), whose protonation gives the Cint,i state (PDB: 9FPN) (f). g, The Cint,i state is ready to activate CO2, giving the Cint,i–CO2 state, and regenerate the Cred2 state in a, which requires two internal electron transfers to CO2, one from Ni(I) and the second from Bred via the C cluster, marked by a hash (#). h, Under strong reducing conditions, the B cluster (Bred) may be reduced so that Ni(II)–CO (Cred1–CO) can be reduced to Ni(I)–CO (Cint,o–CO state; PDB: 9FPI, 9FPJ and 9FPO). The electron transfer is probably facilitated by a shift of the Ni(I)/Ni(II) reduction potential caused by the π-acidic CO ligand. After reduction, Ni(I) moves out of the S plane, which aligns CO with the hydrophobic gas tunnel. This state can react after a one-electron reduction, either by reacting with CO2, which would support the dissociation of CO, or CO can dissociate first, giving the Cint,o state. The potentially slower pathway along Cint,o–CO may be prevented in the presence of CO2 by the more negative redox potential of the B cluster12. When the external CO concentration is high under reducing conditions, the formation of the Cint,o–CO state may be responsible for the observed inhibitory effect of CO (ref. 25). We propose the reverse cycle including Cred1 for the CO oxidation reaction. The asterisks indicate that Box can be reduced to the Bred state under reducing conditions.
The revised electronic structure of the C cluster in the different states also has direct implications for the activation of CO2. Notably, the Cred2 state is not the state that activates and reacts with CO2, as it already contains a carbonite ligand (Fig. 5a). It is therefore the Cint state that is responsible for the initial binding and activation of CO2, indicating that the Ni(I) ion is the nucleophile that reacts with CO2. Of the three different Cint states (Cint,o, Cint,o–CO and Cint,i in Fig. 5e,h,f) that we observed, it is likely that it is the Cint,i state that reacts with CO2 (Fig. 5f). The T-shaped Ni(I) ion, with its metalloradical character exposing its singly occupied molecular orbital at the binding site of CO2, is most probably a central element of the CO2 activation complex.
Synthetic and computational models of the CO2 reduction cycle suggest that the Ni(I)–CO complex may persist until CO2 is activated at the Ni ion32,33. However, the affinity of the Cint states for CO seems to be only moderate, in line with T-shaped Ni(I)–CO complexes that, despite being sufficiently π-basic, are not strong CO binders29. The distortion from linearity along the Ni–C–O axis, enforced by the protein matrix, probably lowers the CO affinity even further (Fig. 5h). At low CO concentrations and neutral to slightly acidic conditions, where CODHs reduce CO2 more rapidly4, Cint,i would be the dominant species, together contributing to a reductive activation of CO2. CO2 would encounter Ni(I) and Fe1 with open coordination sites pointing towards the dynamic CO2 channel (Fig. 5f,g)34. How the proximity and potential metal–metal interaction between Ni(I) and Fe1 in the Cint,i state further modulates the reactivity with CO2 needs to be investigated in the future.
Our study supports a role for Ni as the redox-active, nucleophilic unit in the C cluster: it accepts one electron and supports the storage of two electrons in its carbonite ligand. The reactivity of the T-shaped Ni(I) metalloradical state and its dynamic interplay with an Fe(II) ion provide a blueprint to develop CO2-converting catalysts.
Methods
Production of carbon monoxide dehydrogenase II from Carboxydothermus hydrogenoformans
The cooS-II gene encoding carbon monoxide dehydrogenase II from Carboxydothermus hydrogenoformans (CODH-IICh) was amplified by PCR from the genomic DNA of C. hydrogenoformans Z-2901 (DSM 6008) and cloned into a linearized pET28a vector (Novagen) to generate the expression plasmid pPKS2. This plasmid was transformed into Escherichia coli Rosetta (DE3) cells co-harbouring pRKISC (ref. 8). Cultures were grown at 30 °C in a modified terrific broth medium supplemented with 1.5% (v/v) glycerol, 0.04 mM NiCl2, 0.45 mM Na2S, 0.6 mM FeSO4 and 2 mM cysteine under a continuous nitrogen flux. Protein expression was induced with 0.2 mM isopropyl-β-d-thiogalactoside at an optical density of 0.6–0.7 at 600 nm. After 1 h, 1 mM NiCl2, 0.75 mM Na2S, 1 mM FeSO4 and 50 mM KNO3 were added. Cells were collected aerobically by centrifugation and frozen in liquid nitrogen.
All subsequent purification steps were conducted inside an anoxic glove box (Model B, COY Laboratory Products) under an atmosphere of 95% N2 and 5% H2 at 20 °C. Cells were lysed by sonication on ice and the lysate was solubilized with 0.9% (v/v) deoxycholic acid for 60 min. After ultracentrifugation, the supernatant was applied to a Ni2+-immobilized metal affinity chromatography column (Ni Sepharose, GE Healthcare). All of the buffers used for purification contained 2 mM Na dithionite. Imidazole was removed from the eluted protein by gel filtration (G-25 column, 10 mm × 60 mm) and the protein was concentrated using a centrifugal filter unit with a molecular weight cut-off of 30 kDa (Vivaspin 500, Vivascience). Purified CODH-IICh was stored at −80 °C.
Enzyme activity
The specific CO oxidation activity of CODH-IICh was determined by monitoring the CO-dependent reduction of oxidized methyl viologen (MVox) at 578 nm (reduced MV, molar absorption coefficient at 578 nm = 9.7 mM−1 cm−1) with a CO headspace at 25 °C. The protein batches used for crystallization had a specific activity of 1,226–1,279 U mg−1 (corresponding to 11,529–12,028 U mg−1 at 70 °C (ref. 17)), where 1 U can be defined as reducing 2 µmol MVox per minute.
Crystallization and crystal treatment
Crystallization and soaking experiments were carried out in an anoxic glove box. Crystals of CODH-IICh were grown by hanging drop vapour diffusion using 16–18 mg ml−1 of protein in 14–18% (w/v) polyethylene glycol (PEG) 2000 monomethyl ether (MME), 100 mM sodium 4-(2-hydroxyethyl)-1-piperazineethanesulfonate (HEPES; pH 7.5) and 2 mM Na dithionite by mixing equal volumes of protein solution and reservoir (2 µl each). All crystals were collected in a solution containing 25% (w/v) PEG 2000 MME, 50 mM HEPES (pH 7.5) and 2 mM Na dithionite and subjected to post-crystallization treatment as described below.
The crystal treatment procedures are schematically depicted in Extended Data Fig. 1.
We previously used the Ti(III)–citrate complex (−600 mV versus SHE at pH 8.0) to reduce CODH-IICh in kinetic and crystallographic investigations8,17,35. In this study, we used the more stable Ti(III)–EDTA (−530 mV versus SHE at pH 8.0) complex as reducing agent, mixing 20% (w/v) Ti(III) chloride with a 25% molar excess of EDTA to a final concentration of 100 mM Ti(III)–EDTA in 0.2 M MOPS buffer (pH 7.2)36. The structures of the reduced and CO2-bound samples determined in this work using Ti(III)–EDTA superimpose without obvious structural differences on the previously published atomic resolution structures17. In addition, a stock solution of Eu(II)–DTPA (ref. 22) was prepared by mixing equimolar amounts of Eu(II) chloride and DTPA to a final concentration of 100 mM in 1 M Tris-HCl buffer (pH 8.0)37, which was also used as a reducing agent for its highly negative reduction potential and significantly lower background in the spectral region of interest in EPR spectroscopy.
Multiple Ti(III)-reduced samples of the C cluster were prepared (Extended Data Fig. 4). For the initial Ti(III)-reduced pH 8.0 sample, single crystals of CODH-IICh were incubated in buffer A1 (100 mM Tris-HCl (pH 8.0), 25% (w/v) PEG 2000 MME and 6 mM Ti(III)–EDTA) for 3 h. For buffer A1, we measured a redox potential of −530 mV versus SHE. The Ti(III)-reduced CO-bound sample was prepared by placing the Ti(III)-reduced crystals in buffer A1 in a homemade gas chamber pressurized with 83 kPa CO for 1 h (Ti(III)-reduced pH 8.0 + CO sample). The Ti(III)-reduced pH 6.0 sample was obtained by incubating single crystals of CODH-IICh in buffer A2 (100 mM Bis-Tris (pH 6.0), 25% (w/v) PEG 2000 MME and 6 mM Ti(III)–EDTA) for 3 h. The Ti(III)-reduced CO2-bound sample was prepared by soaking the Ti(III)-reduced pH 8.0 crystals in buffer A1 in the presence of 45 mM NaHCO3 (pH 7.9 buffered by 100 mM Tris-Cl) for 30 min (Ti(III)-reduced pH 8.0 + CO2 sample)8. The rapid pH drop (referred to as pH jump) of the Ti(III)-reduced CO2-bound crystals was achieved by incubating the crystals in buffer A3 (100 mM Na acetate buffer (pH 4.0), 25% PEG 2000 MME and 6 mM Ti(III)–EDTA) for 20–30 s (Ti(III)-reduced pH 8.0 + CO2 pH jump sample). CODH-IICh crystals with isotopes of 13C for the Ti(III)-reduced pH 8.0 13CO- and 13CO2-bound samples were prepared in the same way as their 12CO and 12CO2 counterparts.
Two control experiments were performed on the Ti(III)-reduced pH 8 + CO2 pH jump sample (Extended Data Fig. 4):
-
1.
A pseudo-pH jump was achieved by incubating the Ti(III)-reduced pH 8.0 + CO2 sample in buffer A1 without CO2 (pH 8.0) for 10 min.
-
2.
A potential jump of the Ti(III)-reduced pH 8.0 + CO2 sample from −530 mV to −191 mV versus SHE maintaining pH 8.0 was carried out by incubating the crystals in buffer B1 (100 mM Tris-HCl (pH 8.0), 25 % PEG 2000 MME and 2 mM anthraquinone-2,6-disulfonate) for 20–30 s.
To obtain the Eu(II)–DTPA-reduced samples (Extended Data Fig. 4) at pH 8.0 (Eu(II)-reduced pH 8.0 sample) and pH 6.0 (Eu(II)-reduced pH 6.0 sample), single crystals were incubated in buffer C1 (100 mM Tris-HCl (pH 8.0), 25% (w/v) PEG 2000 MME and 8 mM Eu(II)–DTPA) and buffer C2 (100 mM Bi-Tris-HCl (pH 6.0), 25% (w/v) PEG 2000 MME and 8 mM Eu(II)–DTPA), respectively, for 3 h. The Eu(II)-reduced CO2-bound pH 8.0 state was prepared by soaking the Eu(II)-reduced pH 8.0 crystals in buffer C1 in the presence of 45 mM NaHCO3 (pH 7.9 buffered by 100 mM Tris-HCl) for 30 min (Eu(II)-reduced pH 8.0 + CO2 sample). The Eu(II)-reduced CO-bound sample at pH 6.0 was prepared by placing the Eu(II)-reduced pH 6.0 crystals in buffer C2 in a homemade gas chamber pressurized with 83 kPa CO for 1 h (Eu(II)-reduced pH 6.0 + CO sample).
Crystals of the final samples were shock-cooled directly in liquid nitrogen, where the 25% (w/v) PEG 2000 MME in the final soaking solution acted as cryo-protectant.
The redox potentials of the soaking solutions containing Ti(III)–EDTA and Eu(II)-DTPA were measured using a PT 5900 BNC redox electrode with an integrated Ag/AgCl reference (Xylem Analytics). The redox potential of each solution was measured in a glove box (model B, COY Laboratory Products) under an atmosphere of 95% N2 and 5% H2. Each measurement was carried out using 2 ml of the respective solution, allowing 15–20 min under stirring for the stabilization of the redox potential readings.
Sample preparation for spectroscopic analysis
All samples for crystal IR and EPR spectroscopic analysis were prepared by the same treatments as the crystals prepared for X-ray diffraction analysis described as illustrated in Extended Data Fig. 1. For IR spectroscopy, four to eight larger crystals were transferred to a custom-made IR plate and frozen in liquid nitrogen immediately38. For EPR spectroscopy, several hundred crystals from 6 to 24 crystallization drops were collected for each sample, transferred to an EPR tube and stored in liquid nitrogen until further use.
The reduction of CO2 to CO or ligand degradation (for example, by decarboxylation) was monitored by IR spectroscopy (Extended Data Fig. 9), analysing the formation of carboxyhaem (Fe(II)–CO). The assay contained 10 mM haemoglobin from bovine blood, the reducing agent (10 mM Na thionite, Ti(III)–EDTA or Eu(II)–DTPA) and, if indicated, NaHCO3 (70 mM) as the CO2 source, using the same buffers as in the crystal assays.
Data collection, structure determination and refinement
Diffraction data were collected on beamlines 14.1 and 14.2 operated by Helmholtz-Zentrum Berlin at the BESSY-II storage ring39 and beamlines P13 and P14 operated by EMBL Hamburg at the PETRA-III storage ring (DESY)40 at 0.9184 Å and 100 K. Diffraction data were integrated and scaled using XDSAPP41,42, autoPROC43 or the EDNA framework Fast Processing System44.
Initially, a rigid body refinement using model 3B51.pdb8 was followed by two rounds of simulated annealing refinement using Phenix45 to minimize model bias. Restraints of metal clusters of CODH-IICh were generated using REFMAC5 from CCP446 and further adjusted in the course of the refinement.
We began modelling each sample using the published model with [NiFe4S4] as the C cluster with two alternative exo-Fe1 positions (Fe1,i and Fe1,o). All structures were initially refined without modelling the non-protein ligands of the C cluster. After global refinement of the protein structure, refinement of the C cluster followed by refinement of Ni and ligand modelling were carried out. In general, looser restraints were used for the refinement of all of the metal clusters because of sufficiently high resolution (dmin ≥ 1.31 Å). All ligand Fo − Fc omit maps were calculated after removal of the ligand. After iterative manual model building using Coot47, several cycles of iterative refinements were routinely performed using phenix.refine from Phenix.
After refining the C cluster in each sample, including Ni and Fe1, we followed the developing strong positive densities in the Fo − Fc omit maps to reveal several ligands around the C cluster and then modelled these accordingly as CO, CO2, OH and/or H2O (see below for details).
In several of the samples (Extended Data Fig. 4d,e,h,i,k), after the first refinement using [NiFe4S4] as the C cluster, the Ni would automatically be refined by phenix.refine using relaxed restraints to a position (referred to as Ni,X in Extended Data Fig. 6a). The refined Ni,X would be placed above that of Ni,A observed in other samples, with a higher B factor and clear positive density above and below Ni,X even after anisotropic refinement (Extended Data Fig. 6a). When we placed Ni at the position Ni,A with an occupancy of Ni,X, we observed a positive density above and a negative density at Ni,A after B-factor refinement (Extended Data Fig. 6c). After occupancy and B-factor refinement of Ni,A, the negative density was resolved; however, the positive density above it persisted (Extended Data Fig. 6d). This strongly suggests that Ni has two alternative positions: Ni,A and Ni,B (Extended Data Fig. 6e). Ni,B and Ni,A were refined using loose bond distance and angle restraints, which allow clear refinement of both of the Ni positions. For ligand refinement in subsequent steps, the positional and occupancy refinement of the two alternative Ni ions and the rest of the C cluster is switched off.
The initial refinements of the C cluster in many samples (Extended Data Fig. 4c–i,k) revealed a triatomic density bridging Ni and Fe1,o, also observed in the −600 mV + CO2 state of CODH-IICh (Protein Data Bank (PDB): 4UDX)8,17. The triatomic density was modelled well as CO2, resulting in a structure indistinguishable from the published −600 mV + CO2 structure.
After modelling and refinement of the C cluster, a density observed in the Fo − Fc omit maps between Fe1 and Ni was modelled as an OH ligand (Extended Data Fig. 4a,b), first as Fe1,o–OH (Fig. 1c), as in previously published structures of CODH-IICh (PDB: 3B51)8, and then as a µ2-bridging OH (Fig. 1b)
In cases where simultaneous states existed, once CO2 was refined, a positive density persisted next to the O2 of CO2, bound to Fe1,o, which overlaps with the position of the OH. This indicates the presence of a terminal OH ligand bound to Fe1,o in the C cluster, where the product CO2 was not formed. The modelling of OH resolved the positive density (Extended Data Fig. 4d,h,k).
After the initial refinement of the C cluster, the Ni positions in several of the samples (Extended Data Fig. 4d,e,h,i,k) exhibited a strong positive electron density in the Fo – Fc omit map at the apical space of the Ni ion of the C cluster (Extended Data Fig. 6) that was visibly elongated and diatomic in character. When a water molecule was modelled as a control in the apical density and refined with tight distance restraints, a strong positive density was observed between the water and the Ni,B atom, indicating the presence of a diatomic molecule. As these samples were incubated with CO under high pressure or CO2 under turnover conditions, we modelled the observed density with CO. The Ni,A–CO distance of 2.39 Å is longer than the typical metal–carbonyl bond length of 1.80 Å (ref. 48), while the Ni,B–CO distance of 1.81 Å aligns well with the typical bond length. Therefore, the observed positive density above the Ni ion was modelled with CO completing the tetrahedral geometry of Ni,B (Extended Data Fig. 4d).
For all of the structures containing bound CO2, a weak Fo − Fc omit electron density was revealed above the Ni ion, identical to the water molecule modelled in the previously published −600 mV + CO2 state (PDB: 4UDX)17. We therefore modelled it as a water molecule as well (Extended Data Fig. 4c,f,g) and always found it to be within the distance of a hydrogen-bonding interaction and with a low occupancy. In samples with mixed states (Extended Data Fig. 4d,e,h,i,k), a slight deformation of the 2Fo – Fc map and a weak positive density of the Fo – Fc map, even after CO modelling, suggested the presence of a small amount of water. The observed positive density was resolved after several cycles of refinement with the water molecule.
After the final refinements of all of the reported structures, 96.35–96.68% of the residues were in the Ramachandran-favoured region and 0.16–0.47% were outliers. Other data collection and refinement statistics for the reported structures are provided in Supplementary Table 1.
IR spectroscopy
The IR spectra of protein crystals were recorded on a Bruker Tensor 27 Fourier transform (FT) IR spectrometer linked to a Hyperion 3000 IR microscope equipped with a ×20 IR transmission objective and a mercury cadmium telluride detector38. The temperature was adjusted to 80 K using a liquid N2-cooled cryo-stage (Linkam Scientific). The IR spectra of protein solutions were recorded using a Bruker Vertex 80v FTIR spectrometer at 283 K. The protein solution was injected into a homemade, gas-tight IR transmission cell incorporating two CaF2 windows separated by a 50-µm Teflon spacer. All IR spectroscopic data were accumulated and processed using the OPUS software (versions 6.5 and 7.5) from Bruker.
EPR spectroscopy
EPR spectra were recorded on a Bruker EMX plus X-Band spectrometer equipped with an ER 4122 super-high Q resonator using the Xenon software (version 1.1b.58) from Bruker. The temperature of the Oxford ESR900 helium flow cryostat was controlled by an Oxford ITC4 temperature controller. As references, an EPR spectrum of buffer solution recorded with identical experimental parameters was subtracted from the spectra of the samples, unless indicated otherwise. A spline function was used to correct broad baseline drifts, for example, from Eu(II)–DTPA. To monitor the anisotropic spectral changes in the crystal suspension (Extended Data Fig. 3), EPR tubes were rotated clockwise within the resonator while recording spectra for each respective rotation angle relative to the starting position. The following experimental parameters were used: microwave frequency, 9.3 GHz; modulation amplitude, 10 G; modulation frequency, 100 kHz. The EPR spectra were simulated using MATLAB toolbox EasySpin (version 5.2.36)49.
Control experiments
Pseudo-jump control experiments were performed to check whether the observed changes in the position of CO2 or its occupancy during the pH jump could be attributed to the dissociation of CO2 from the C cluster when soaked in a buffer without a CO2 source. Therefore, we soaked the CO2-bound crystals in buffer A1 without NaHCO3 instead of in buffer A3 as used in the pH jump. The crystals were soaked for 10 min in buffer A1, compared with 20 s in buffer A3 for the pH jump, to ascertain whether ligand dissociation took place over a longer period of time. The CO2 ligand of the pseudo-jump was indistinguishable in structure and occupancy (Extended Data Fig. 4f) from that in the other CO2-bound samples.
The original pH jump buffer used Ti(III)–EDTA as the reducing agent. However, the reduction potential for Ti(III)–EDTA is pH-dependent and drops to −234 mV versus SHE at pH 4.0 (buffer A3), while it is −530 mV versus SHE at pH 8 (buffer A1). Therefore, we simulated a similar potential drop by soaking Ti(III)-reduced pH 8.0 + CO2 crystals in buffer A3 without a change in pH to make sure that our observations were inherently due to a change in pH alone and not to a change in the reduction potential. We did not observe any structural change in the potential jump structure compared with the Ti(III)-reduced pH 8.0 + CO2 sample (Extended Data Fig. 4g).
Reproducibility
To test the reproducibility of the CO-bound structures of the Ti(III)-reduced pH 8.0 + CO and Ti(III)-reduced pH 8.0 + CO2 pH jump samples, following the same treatment procedures, we determined several more crystal structures for both samples over the course of several months and multiple synchrotron measurements. These crystal structures were less clearly resolved at the apical and equatorial positions in their respective 2Fo − Fc and Fo − Fc omit maps and/or were of lower resolution. However, their electron densities showed the same changes that were modelled and refined in the same manner as the structures detailed in this Article.
Due to the unique structure of the Eu(II)-reduced pH 6.0 sample, with the exo-Fe being in the Fe1, i position, we determined several more structures of multiple crystals using the same treatment procedure independently of each other at different time points. Nearly all of them indicated the same observed structural changes and were modelled and refined in the same manner as the structure elucidated in this study.
The samples used for the IR and EPR measurements were reproduced simultaneously with those used for the crystal structure determinations.
Other software
The figures of structures presented in this Article were prepared using UCSF Chimera (version 1.16)50, PerkinElmer ChemDraw Prime 19.1 and Adobe Illustrator 2020. The figures showing spectra were prepared using Origin 2024, Origin 2025 and Inkscape (1.3.2). The graphical abstract was prepared using PyMol (2.5.8) and Powerpoint 365 (version 2504).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and reflection files of all structures have been deposited with the Protein Data Bank at http://www.pdb.org with accession numbers 9FPF (Ti(III)-reduced, pH 8.0 crystal, Cint,o state), 9FPG (Ti(III)-reduced pH 6.0 crystal, Cred1 state), 9FPH (Ti(III)-reduced, pH 8.0 + CO2 crystal, Cred2 state), 9FPI (Ti(III)-reduced, pH 8.0 + CO crystal, Cint,o–CO state), 9FPJ (Ti(III)-reduced + CO2 pH jump crystal, Cred2 and Cint,o–CO states), 9FPK (Eu(II)-reduced, pH 8.0 crystal), 9FPL (Eu(II)-reduced, pH 8.0 + CO2 crystal), 9FPN (Eu(II)-reduced, pH 6.0 crystal, Cint,i state) and 9FPO (Eu(II)-reduced, pH 6.0 + CO crystal, Cint,o + CO state). Other crystallographic structures and data used in this study that have already been published are also deposited with the Protein Data Bank (3B51 and 4UDX). Source data are provided with this paper.
References
Hansen, H. A., Varley, J. B., Peterson, A. A. & Nørskov, J. K. Understanding trends in the electrocatalytic activity of metals and enzymes for CO2 reduction to CO. J. Phys. Chem. Lett. 4, 388–392 (2013).
Appel, A. M. et al. Frontiers, opportunities, and challenges in biochemical and chemical catalysis of CO2 fixation. Chem. Rev. 113, 6621–6658 (2013).
Shin, W., Lee, S. H., Shin, J. W., Lee, S. P. & Kim, Y. Highly selective electrocatalytic conversion of CO2 to CO at −0.57 V (NHE) by carbon monoxide dehydrogenase from Moorella thermoacetica. J. Am. Chem. Soc. 125, 14688–14689 (2003).
Parkin, A., Seravalli, J., Vincent, K. A., Ragsdale, S. W. & Armstrong, F. A. Rapid and efficient electrocatalytic CO2/CO interconversions by Carboxydothermus hydrogenoformans CO dehydrogenase I on an electrode. J. Am. Chem. Soc. 129, 10328–10329 (2007).
Adam, P. S., Borrel, G. & Gribaldo, S. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc. Natl Acad. Sci. USA 115, E1166–E1173 (2018).
Dobbek, H., Svetlitchnyi, V., Gremer, L., Huber, R. & Meyer, O. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293, 1281–1285 (2001).
Drennan, C. L., Heo, J., Sintchak, M. D., Schreiter, E. & Ludden, P. W. Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase. Proc. Natl Acad. Sci. USA 98, 11973–11978 (2001).
Jeoung, J.-H. & Dobbek, H. Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase. Science 318, 1461–1464 (2007).
Lindahl, P. A., Munck, E. & Ragsdale, S. W. CO dehydrogenase from Clostridium thermoaceticum. EPR and electrochemical studies in CO2 and argon atmospheres. J. Biol. Chem. 265, 3873–3879 (1990).
Hu, Z. et al. Nature of the C-cluster in Ni-containing carbon monoxide dehydrogenases. J. Am. Chem. Soc. 118, 830–844 (1996).
Anderson, M. E. & Lindahl, P. A. Spectroscopic states of the CO oxidation/CO2 reduction active site of carbon monoxide dehydrogenase and mechanistic implications. Biochemistry 35, 8371–8380 (1996).
Fraser, D. M. & Lindahl, P. A. Evidence for a proposed intermediate redox state in the CO/CO2 active site of acetyl-CoA synthase (carbon monoxide dehydrogenase) from Clostridium thermoaceticum. Biochemistry 38, 15706–15711 (1999).
Lindahl, P. A. Implications of a carboxylate-bound C-cluster structure of carbon monoxide dehydrogenase. Angew. Chem. Int. Ed. 47, 4054–4056 (2008).
Newman-Stonebraker, S. H., Gerard, T. J. & Holland, P. L. Opportunities for insight into the mechanism of efficient CO2/CO interconversion at a nickel-iron cluster in CO dehydrogenase. Chem 10, 1655–1667 (2024).
Svetlitchnyi, V., Peschel, C., Acker, G. & Meyer, O. Two membrane-associated NiFeS-carbon monoxide dehydrogenases from the anaerobic carbon-monoxide-utilizing eubacterium Carboxydothermus hydrogenoformans. J. Bacteriol. 183, 5134–5144 (2001).
Amara, P., Mouesca, J.-M., Volbeda, A. & Fontecilla-Camps, J. C. Carbon monoxide dehydrogenase reaction mechanism: a likely case of abnormal CO2 insertion to a Ni–H− bond. Inorg. Chem. 50, 1868–1878 (2011).
Fesseler, J., Jeoung, J.-H. & Dobbek, H. How the [NiFe4S4] cluster of CO dehydrogenase activates CO2 and NCO−. Angew. Chem. Int. Ed. 54, 8560–8564 (2015).
Breglia, R. et al. First-principles calculations on Ni,Fe-containing carbon monoxide dehydrogenases reveal key stereoelectronic features for binding and release of CO2 to/from the C-cluster. Inorg. Chem. 60, 387–402 (2021).
Zimmermann, P., Hoof, S., Braun-Cula, B., Herwig, C. & Limberg, C. A biomimetic nickel complex with a reduced CO2 ligand generated by formate deprotonation and its behaviour towards CO2. Angew. Chem. Int. Ed. 57, 7230–7233 (2018).
Shafaat, H. S. & Yang, J. Y. Uniting biological and chemical strategies for selective CO2 reduction. Nat. Catal. 4, 928–933 (2021).
Lewis, L. C., Sanabria-Gracia, J. A., Lee, Y., Jenkins, A. J. & Shafaat, H. S. Electronic isomerism in a heterometallic nickel–iron–sulfur cluster models substrate binding and cyanide inhibition of carbon monoxide dehydrogenase. Chem. Sci. 15, 5916–5928 (2024).
Vincent, K. A. et al. Instantaneous, stoichiometric generation of powerfully reducing states of protein active sites using Eu(II) and polyaminocarboxylate ligands. Chem. Commun. 2590–2591 (2003).
Spangler, N. J., Lindahl, P. A., Bandarian, V. & Ludden, P. W. Spectroelectrochemical characterization of the metal centers in carbon monoxide dehydrogenase (CODH) and nickel-deficient CODH from Rhodospirillum rubrum. J. Biol. Chem. 271, 7973–7977 (1996).
Lindahl, P. A. Metal–metal bonds in biology. J. Inorg. Biochem. 106, 172–178 (2012).
Wang, V. C. C., Can, M., Pierce, E., Ragsdale, S. W. & Armstrong, F. A. A unified electrocatalytic description of the action of inhibitors of nickel carbon monoxide dehydrogenase. J. Am. Chem. Soc. 135, 2198–2206 (2013).
DeRose, V. J., Telser, J., Anderson, M. E., Lindahl, P. A. & Hoffman, B. M. A multinuclear ENDOR study of the C-cluster in CO dehydrogenase from Clostridium thermoaceticum: evidence for HxO and histidine coordination to the [Fe4S4] center. J. Am. Chem. Soc. 120, 8767–8776 (1998).
Ogata, H., Nishikawa, K. & Lubitz, W. Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase. Nature 520, 571–574 (2015).
Yoo, C. & Lee, Y. A T‐shaped nickel(I) metalloradical species. Angew. Chem. Int. Ed. 129, 9630–9634 (2017).
Ingleson, M. J. et al. Influence of the d-electron count on CO binding by three-coordinate [(tBu2PCH2SiMe2)2N]Fe, -Co, and -Ni. Inorg. Chem. 47, 407–409 (2008).
Ott, J. C., Bürgy, D., Guan, H. & Gade, L. H. 3d metal complexes in T-shaped geometry as a gateway to metalloradical reactivity. Acc. Chem. Res. 55, 857–868 (2022).
Wilson, D. W. N. et al. Three-coordinate nickel and metal–metal interactions in a heterometallic iron–sulfur cluster. J. Am. Chem. Soc. 146, 4013–4025 (2024).
Sahoo, D., Yoo, C. & Lee, Y. Direct CO2 addition to a Ni(0)–CO species allows the selective generation of a nickel(II) carboxylate with expulsion of CO. J. Am. Chem. Soc. 140, 2179–2185 (2018).
Liao, R. Z. & Siegbahn, P. E. M. Energetics for the mechanism of nickel-containing carbon monoxide dehydrogenase. Inorg. Chem. 58, 7931–7938 (2019).
Wang, P.-h, Bruschi, M., De Gioia, L. & Blumberger, J. Uncovering a dynamically formed substrate access tunnel in carbon monoxide dehydrogenase/acetyl-CoA synthase. J. Am. Chem. Soc. 135, 9493–9502 (2013).
Basak, Y., Jeoung, J.-H., Domnik, L., Ruickoldt, J. & Dobbek, H. Substrate activation at the Ni,Fe cluster of CO dehydrogenases: the influence of the protein matrix. ACS Catal. 12, 12711–12719 (2022).
Ruickoldt, J., Basak, Y., Domnik, L., Jeoung, J.-H. & Dobbek, H. On the kinetics of CO2 reduction by Ni, Fe-CO dehydrogenases. ACS Catal. 12, 13131–13142 (2022).
Lee, C. C., Hu, Y. & Ribbe, M. W. Reduction and condensation of aldehydes by the isolated cofactor of nitrogenase. ACS Cent. Sci. 4, 1430–1435 (2018).
Lorent, C. et al. Exploring structure and function of redox intermediates in [NiFe]-hydrogenases by an advanced experimental approach for solvated, lyophilized and crystallized metalloenzymes. Angew. Chem. Int. Ed. 60, 15854–15862 (2021).
Mueller, U. et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: current status and perspectives. Eur. Phys. J. 130, 233 (2015).
Cianci, M. et al. P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Radiat. 24, 323–332 (2017).
Sparta, K. M., Krug, M., Heinemann, U., Mueller, U. & Weiss, M. S. XDSAPP2.0. J. Appl. Crystallogr. 49, 1085–1092 (2016).
Krug, M., Weiss, M. S., Heinemann, U. & Mueller, U. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Crystallogr. 45, 568–572 (2012).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 67, 293–302 (2011).
Incardona, M. F. et al. EDNA: a framework for plugin-based applications applied to X-ray experiment online data analysis. J. Synchrotron Radiat. 16, 872–879 (2009).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Hocking, R. K. & Hambley, T. W. Database analysis of transition metal carbonyl bond lengths: insight into the periodicity of π back-bonding, σ donation, and the factors affecting the electronic structure of the TM−C⋮O moiety. Organometallics 26, 2815–2823 (2007).
Stoll, S. & Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 178, 42–55 (2006).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Macgregor, S. A., Lu, Z., Eisenstein, O. & Crabtree, R. H. Why nickel(II) binds CO best in trigonal bipyramidal and square pyramidal geometries and possible consequences for CO dehydrogenase. Inorg. Chem. 33, 3616–3618 (1994).
Ragsdale, S. W. & Kumar, M. Nickel-containing carbon monoxide dehydrogenase/acetyl-CoA synthase. Chem. Rev. 96, 2515–2540 (1996).
Heo, J., Staples, C. R., Telser, J. & Ludden, P. W. Rhodospirillum rubrum CO-dehydrogenase. Part 2. Spectroscopic investigation and assignment of spin−spin coupling signals. J. Am. Chem. Soc. 121, 11045–11057 (1999).
Falk, M. & Miller, A. G. Infrared spectrum of carbon dioxide in aqueous solution. Vib. Spectrosc. 4, 105–108 (1992).
Choc, M. G. & Caughey, W. S. Evidence from infrared and 13C NMR spectra for discrete rapidly interconverting conformers at the carbon monoxide binding sites of hemoglobins A and Zurich. J. Biol. Chem. 256, 1831–1838 (1981).
Yan, Z.-H. et al. Photo-generated dinuclear {Eu(II)}2 active sites for selective CO2 reduction in a photosensitizing metal-organic framework. Nat. Commun. 9, 199 (2018).
Matsumoto, A. & Azuma, N. Photodecomposition of europium(III) acetate and formate in aqueous solutions. J. Phys. Chem. 92, 1830–1835 (1988).
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; grant no. DO785/6-2, H.D.) and Germany’s Excellence Strategy (EXC 2008–390540038–UniSysCat, I.Z. and H.D.). We thank C. Limberg (Humboldt-Universität zu Berlin, Germany) for reading of the manuscript and C. Teutloff (Freie Universität Berlin) for helpful discussions. We acknowledge access to the PETRA III storage ring (DESY, Hamburg, Germany) and the BESSY II storage ring (Berlin) through the Joint Berlin MX-Laboratory sponsored by Helmholtz-Zentrum Berlin für Materialien und Energie, Freie Universität Berlin, Humboldt-Universität zu Berlin, Max-Delbrück-Centrum and the Leibniz-Institut für Molekulare Pharmakologie.
Funding
Open access funding provided by Humboldt-Universität zu Berlin.
Author information
Authors and Affiliations
Contributions
Y.B., C.L. and H.D. designed the experiments. Y.B. and C.L. carried out the experiments and processed the data. Y.B., C.L., J.-H.J., I.Z. and H.D. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Catalysis thanks Maria Romao, Itzel Vaca and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 XRD, IR and EPR measurements.
The final conditions of the crystal samples and the associated experiments are indicated in a square box with a distinct colour (pink for the Ti(III)-EDTA-based treatment and yellow for the Eu(II)-DTPA-based treatment).
Extended Data Fig. 2 EPR spectra of Ti(III)-EDTA and Eu(II)-DTPA at pH 8.0 recorded at 10 K with 1 mW microwave power.
(a) Broad range scan of Ti(III)-EDTA and Eu(II)-DTPA. (b) Narrow range scan of Ti(III)-EDTA, Ti(III)-EDTA + CO2 and Eu(II)-DTPA. Whereas the signal of the paramagnetic Ti(III) dominates central parts of the EPR spectrum, spectra of Eu(II)-DTPA show significantly less background in the CODH-relevant spectral region between g-values of 1.6 and 2.4. The S = ½ signal from Ti(III) (gx = 1.97, gy = 1.95, gz = 1.90) is broadened and slightly shifted to larger g-values, if NaHCO3 is added to the crystallisation buffer.
Extended Data Fig. 3 Angle-dependent EPR spectra of CODH crystal suspensions recorded at 10 K and 1 mW microwave power, reduced with Eu(II)-DTPA or Ti(III)-EDTA.
(a) Ti(III)-reduced pH 6.0 sample. (b) Ti(III)-reduced pH 8.0 + CO2 sample. (c) Eu(II)-reduced pH 8.0 sample. (d) Eu(II)-reduced pH 8.0 + CO2 sample. (e) Eu(II)-reduced pH 6.0 sample. (f) Eu(II)-reduced pH 6.0 + CO sample. The different paramagnetic species have the following color code: Bred (dark cyan), CredΔNi (violet), Cred1 (red), Cred2 (blue) and Cred2’ (light blue). For the displayed spectra the resonator background was not subtracted and no baseline correction was performed to identify small anisotropic changes upon rotation of the sample.
Extended Data Fig. 4 Crystallographic view of all Ti(III)- and Eu(II)-reduced structures.
(a) Ti(III)-reduced pH 8.0 (Cint,o state). (b) Ti(III)-reduced pH 6.0 (Cred1 state). (c) Ti(III)-reduced pH 8.0 + CO2 (Cred2 state). (d) Ti(III)-reduced pH 8.0 + CO (Cint,o-CO state). (e) Ti(III)-reduced + CO2 pH jump (Cint,o-CO state). (f) Ti(III)-reduced pH 8.0 + CO2 pseudo-jump sample. (g) Ti(III)-reduced pH 8.0 + CO2 potential jump sample. (h) Eu(II)-reduced pH 8.0. (i) Eu(II)-reduced pH 8.0 + CO2. (j) Eu(II)-reduced pH 6.0 (Cint,i state). (k) Eu(II)-reduced pH 6.0 + CO (Cint,o-CO state). Sigma-A weighted 2Fo – Fc maps are contoured at 1.0 σ (blue mesh) and omit Fo – Fc maps (green mesh) are contoured at 4.0 σ for the ligands after the first round of refinements using simulated annealing without corresponding models.
Extended Data Fig. 5 Schematic drawing of cluster C in different Ti(III)-EDTA and Eu(II)-DTPA reduced structures.
(a) Ti(III)-reduced pH 8.0 (Cint,o state). (b) Ti(III)-reduced pH 6.0 (Cred1 state). (c) Ti(III)-reduced pH 8.0 + CO2 (Cred2 state). (d) Ti(III)-reduced pH 8.0 + CO (Cint,o-CO state). (e) Ti(III)-reduced + CO2 pH jump (Cint,o-CO state). (f) Eu(II)-reduced pH 8.0. (g) Eu(II)-reduced + CO2. (h) Eu(II)-reduced pH 6.0 (Cint,i state). (i) Eu(II)-reduced pH 6.0 + CO (Cint,o-CO state). Distances are depicted in Ångstrom (Å). Other than (h) Fe1,i has not been shown for clarity.
Extended Data Fig. 6 Modelling of the two Ni positions in the C-cluster of the Ti(III)-reduced pH 8.0 + CO sample.
(a) Ni in the Ti(III)-reduced pH 8.0 sample when refined initially at Ni,X. (b) Depiction of anomalous difference Fourier maps for Ni and Fe atoms depicted. (c) Ni refined as a single position (Ni,A), with the same occupancy as Ni,X (69%). (d) Ni,A refined at 31% occupancy. (e) Final occupancy refinement: Ni,A (31%) and Ni,B (38%). Sigma-A weighted 2Fo – Fc maps (blue mesh) are contoured at 1.0 σ. Fo – Fc omit maps are contoured at + (green mesh) and - (red mesh) 5.0 σ. Anomalous difference Fourier maps in (b) for Fe atoms (orange mesh) is contoured at 7.0 σ and for Ni ions (violet mesh) is contoured at 5.0 σ. Fe1,i has not been shown for clarity.
Extended Data Fig. 7 Reproducibility of IR spectra of crystals soaked in Ti(III)-EDTA or Eu(II)-DTPA recorded at 80 K.
(a) Ti(III)-reduced pH 8.0 sample. (b) Ti(III)-reduced pH 8.0 + CO sample (The inset shows the dark-minus-light difference spectrum after illumination with 530 nm LED for 0.5 hour). (c) Ti(III)-reduced pH 8.0 + CO2 sample. (d) Ti(III)-reduced + CO2 pH jump sample. (e) Ti(III)-reduced pH 6.0 sample. (f) Eu(II)-reduced pH 8.0 sample. (g) Eu(II)-reduced pH 8.0 + CO2 sample. (h) Eu(II)-reduced pH 6.0 sample. (i) Eu(II)-reduced pH 6.0 + CO sample. The integrated intensity of the CO bands of the Eu(II)-reduced pH 8.0 sample is approximately 3 times less, and the intensity of the Eu(II)-reduced pH 6.0 sample 20 times less, respectively, compared to the intensity of the Eu(II)-reduced pH 8.0 + CO2 or Eu(II)-reduced pH 6.0 + CO sample, for crystals of similar size. Additional minor signals between 1850 and 1950 cm−1 may also represent metal carbonyl vibrations, but could not be unambiguously assigned due to the absence of an isotopic shift (Fig. 2)
Extended Data Fig. 8 EPR experimental spectra and simulations of CODH crystal suspensions recorded at 10 K, reduced with Eu(II)-DTPA.
(a) Eu(II)-reduced pH 6.0 sample and (b) Eu(II)-reduced pH 6.0 + CO sample, both recorded at 0.063 mW microwave power. (c) Eu(II)-reduced pH 8.0 sample and (d) Eu(II)-reduced pH 8.0 + CO2 sample, both recorded at 0.25 mW microwave power. The black solid line represents the experimental spectrum and the grey dotted line the simulated spectrum. The simulated species have the following colour code: Bred (dark cyan), CredΔNi (violet), Cred2 (blue), Cred2’ (light blue) and Ni-signal (brown). The difference between the experimental and the simulated data likely corresponds to the observed minor orientation dependence of the EPR spectra (Extended Data Fig. 3). The Ni-signal (g = 2.35, 2.25, 2.21; gav = 2.27) is presumably related to a weak population of Ni from a degraded C-cluster or non-physiological nickel complex. A similar type of signal with a gav significantly larger than 2 (gav of Cred1 and Cred2 signals are clearly below 2, see Table S3) was observed before for the CODH from Moorella thermoacetica (assigned to a Cred3 or C* state52) or the CODH from Rhodospirillum rubrum53. For these states a catalytic relevance has not yet been shown. The Bred signal was used as an internal standard for quantification, since the B-cluster is fully occupied in the crystal structure and should be fully reduced at the applied redox potential.
Extended Data Fig. 9 Utilising IR spectroscopy at 283 K to monitor the CO2 content and carboxyheme (Fe(II)-CO) formation in haemoglobin solution for different chemical reductants.
(a) IR absorbance spectra of Ti(III)-EDTA + CO2 at pH 6, Eu(II)-DTPA + CO2 at pH 8, Ti(III)-EDTA + CO2 at pH 8, sodium dithionite (NaDT) at pH 8 + CO2, Eu(II)-DTPA at pH 6, Eu(II)-DTPA at pH 8 and Ti(III)-EDTA at pH 8 (from top to bottom) after 1 hour incubation with the reductant. The displayed spectral range is characteristic for the antisymmetric stretching vibration of CO2 in aqueous solution at 2343 cm−154. At lower pH values the CO2/HCO3− equilibrium is shifted towards CO2. (b) IR spectra of NaDT at pH 8 + CO2, Ti(III)-EDTA + CO2 at pH 8, Ti(III)-EDTA + CO2 at pH 6 and Ti(III)-EDTA + CO2 at pH 8 (from top to bottom). The spectra are displayed as second derivatives due to the weak intensity of the Fe(II)-CO band. The displayed spectral range is characteristic for the CO stretching vibration of carboxyheme at 1950 cm−155. (c) IR spectra of Eu(II)-DTPA at pH 6, Eu(II)-DTPA at pH 8 and Eu(II)-DTPA + CO2 at pH 8 (from top to bottom). The dashed and solid lines represent the spectra after 1 hour and 13 hours incubation, respectively. (d) Time-dependent evolution of the integrated signal intensity of the Fe(II)-CO band at 1950 cm−1 of Eu(II)-DTPA at pH 8 (red) and Eu(II)-DTPA + CO2 at pH 8 (blue). The inset shows the absorbance spectra after 13 hours. The carboxyheme formation for Eu(II)-DTPA at pH 8 and Eu(II)-DTPA + CO2 follows different kinetics. The direct CO2 reduction by the europium ions, but also a degradation of the carboxylate-containing DTPA ligands, might be contributing to the observed reactivity56,57.
Supplementary information
Supplementary Information
Supplementary Tables 1-4.
Source data
Source Data Fig. 1
Source data for Fig. 1a.
Source Data Fig. 2
Source data for Fig. 2b,d.
Source Data Fig. 3
Source data for Fig. 3a.
Source Data Extended Data Fig. 2
Source data for Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Source data for Extended Data Fig. 3.
Source Data Extended Data Fig. 7
Source data for Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Source data for Extended Data Fig. 8.
Source Data Extended Data Fig. 9
Source data for Extended Data Fig. 9.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Basak, Y., Lorent, C., Jeoung, JH. et al. Metalloradical-driven enzymatic CO2 reduction by a dynamic Ni–Fe cluster. Nat Catal 8, 794–803 (2025). https://doi.org/10.1038/s41929-025-01388-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41929-025-01388-5
This article is cited by
-
Unsymmetric bimetallic centres in metalloproteins and synthetic models
Nature Reviews Chemistry (2025)