Abstract
The deep biosphere is an energy constrained ecosystem yet fosters diverse microbial communities that are key in biogeochemical cycling. Whether microbial communities in deep biosphere groundwaters are shaped by infiltration of allochthonous surface microorganisms or the evolution of autochthonous species remains unresolved. In this study, 16S rRNA gene amplicon analyses showed that few groups of surface microbes infiltrated deep biosphere groundwaters at the Äspö Hard Rock Laboratory, Sweden, but that such populations constituted up to 49% of the microbial abundance. The dominant persisting phyla included Patescibacteria, Proteobacteria, and Epsilonbacteraeota. Despite the hydrological connection of the Baltic Sea with the studied groundwaters, infiltrating microbes predominantly originated from deep soil groundwater. Most deep biosphere groundwater populations lacked surface representatives, suggesting that they have evolved from ancient autochthonous populations. We propose that deep biosphere groundwater communities in the Fennoscandian Shield consist of selected infiltrated and indigenous populations adapted to the prevailing conditions.
Similar content being viewed by others
Introduction
The terrestrial deep biosphere is temporally and spatially separated from the Earth’s photosynthesis-driven surface, yet accommodates a diverse microbial community with an estimated number of 2 to 6 × 1029 cells1, that is key in driving the Earth’s biogeochemical cycles2,3,4,5. Despite limitations imposed by the predominantly extreme low carbon and energy conditions, subsurface microbial communities are alive6, active7,8,9, and metabolically versatile; exhibiting energy conservation strategies that include nitrate and sulfate reduction, fermentation, methanogenesis, and acetogenesis10,11,12. Life in the terrestrial subsurface can be dictated by infiltration of organic carbon13, while communities at greater depths are largely separated from surface input and are sustained by alternative sources of energy such as the abiotically produced “geogases” carbon dioxide and hydrogen13,14,15 and iron and manganese-rich minerals16. However, the limited study sites compared to the vast volume of the deep biosphere biome results in largely undescribed communities including how the microbial assemblages are shaped.
Deep subsurface microbial communities exhibit distinct traits to cope with low energy flux, including small cell size10, streamlined genomes17,18, and a high functional interactivity such as syntrophy3,19,20. For instance, Patescibacteria are suggested to be ubiquitous in shallow groundwaters due to their ease of mobilization from soils, small genomes, and ability to survive in low carbon and energy conditions21,22. In contrast to the paucity of deep terrestrial biosphere studies, the marine sub-seafloor sediment microbial diversity has been shown to decrease with depth and consequently age of the sediment23,24,25 in response to gradients in both temperature and the availability of organic carbon26. Starnawski et al.27 also identified a sub-seafloor community derived by selection of a small group of surface sediment taxa with the ability to persist under severe energy limitation for >5000 years. In addition, analysis of marine sulfate reducing microbes has shown that the sediment surface community influences sub-seafloor populations through the process of species sorting whereby the geochemical conditions shape the microbial communities by favoring distinct populations28. Additional work on Indian Ocean and up to 1.3 Ma old Bering Sea sediments obtained from depths down to 332 m below the seafloor shows community dependence on the relative abundance of the population in the near-seafloor sediment29. Yet, for the terrestrial deep subsurface, the question remains whether surface microbes infiltrate the deep biosphere and possess traits that allow them to survive in this predominantly low energy and nutrient poor ecosystem or if an indigenous deep groundwater community has developed that is independent of the surface world.
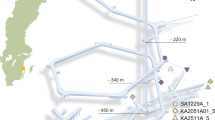
Previous studies have explored the microbial diversity of surface waters30, and benthic waters plus sediments31,32,33 in the south-western part of the Baltic Sea region. Microbial communities in the deep biosphere were investigated in fractures intersected by boreholes emanating from the Äspö Hard Rock Laboratory (HRL) that is built partly under the land and partly under the sea. Äspö HRL, operated by the Swedish Nuclear Fuel and Waste Management Company, is a 3.6 km long tunnel extending 460 m below sea level into ~1.8 Ga years old fractured bedrock consisting of Fennoscandian Shield Paleoproterozoic granitoids34,35,36 that bear groundwaters with contrasting depths, chemical compositions, and residence times10,13,37. Water from the terrestrial landscape and the Baltic Sea are transported deep into the bedrock via vertical to subvertical fractures and thus, provide an ideal situation to investigate how the microbial communities in the deep biosphere groundwaters are influenced by different surface communities37,38,39. The groundwaters are distinguished according to their origin with “meteoric groundwater” derived from the land surface, “modern marine groundwater” from the overlying Baltic Sea, and “old saline groundwater” thought to be completely separated from surface energy influx10,13,39,40,41. Many microbiological studies have been carried out at Äspö HRL including ‘omics’ investigations into community structure and dynamics10,13,37. These microbial communities survive in the deep biosphere by syntrophy and symbiotic associations that alleviates the ‘tragedy of the commons’42 that is aided by biofilm/aggregate formation43.
Here we set out to elucidate the potential role of infiltration into deep biosphere groundwaters of surface and near-surface microbes for the structuring of the deep microbiota. 16S rRNA gene amplicon datasets from nearby aquatic, sediment, soil, and terrestrial deep biosphere environments were collected to unravel potential similarities between their microbial communities. We hypothesized that the fixed niches harboring a common core microbial deep biosphere biome42 drive species sorting of surface microbes, thereby resulting in reduced diversities.
Results and discussion
Geochemistry and water flow
The meteoric and the deeper modern marine and old saline groundwaters have different characteristics including the concentration of dissolved organic carbon (DOC), chloride concentration, and δ18O values (Fig. 1 plus S1 for additional parameters). The shallowest sampled bedrock fracture carried a groundwater below the surface of the Äspö Island that was classified as containing 80% meteoric water based upon δ18O values, chloride concentration, low magnesium plus sulfate concentrations, and high DOC content. This groundwater constituted precipitation that originally soaked into the soil groundwater and was transported downward to reach the meteoric groundwater after eight months to a year39,44. Chloride concentrations and δ18O values of the modern marine groundwater ranged from 2.06 to 4.10 g L−1 and −7.3 to −9.2‰ relative to Standard Mean Ocean Water, respectively, that were similar to the 3.38 g L−1 and −6‰ values, respectively reported in the Baltic Sea by Mathurin et al.39. These data supported the modern marine groundwater being infiltrated by Baltic Sea water with a residence time of several years up to 20 years39. Additional evidence for the infiltration is that the modern marine groundwater was connected with the overlying Baltic Sea via extensive vertical to subvertical fractures, leading to drawdown of marine water45. Finally, old saline groundwater had chloride concentrations in the range of 12.0 to 16.2 g L−1 and δ18O values in the range of −11.3 to −12.4‰. The low δ18O values in particular revealed a high degree of separation from other groundwater types and hence, residence times that would extend to thousands of years or more45. The old saline groundwater likely originated as a marine water that underwent substantial changes in chemistry over geological time scales when in contact with bedrock surfaces and secondary minerals on the fracture walls38. However, it cannot be ruled out that marginal mixing between the sampled fractures occurred, such that the old saline groundwater could contain a minor fraction of modern marine groundwater and vice versa. This can be regarded as a minor disturbance that does not substantially affect the overall results and interpretations.
DOC; dissolved organic carbon. δ18O is the 18O/16O ratio relative to Standard Mean Ocean Water, expressed in parts per thousand (‰). The boxes are formed by the first and third quartiles while the whiskers extend to 1.5 times the inter-quartile range. Additional chemical parameters including strontium and manganese concentrations are shown in Fig. S1.
16S rRNA gene amplicon sequencing
16S rRNA gene amplicon sequences (totaling 214 samples) were analyzed from a variety of environments including surface and benthic seawater; upper (depth 0–1 cm) and lower sediments (6 and 20 cm); terrestrial upper (2–3 m) and lower (5 m) soil groundwaters; and meteoric, modern marine, and old saline deep groundwaters residing in bedrock fractures at depths between 70 and 460 m below the surface. The samples from the surface seawater, lower sediment, upper and lower soil groundwaters, and meteoric groundwaters were sequenced for this study while the data from the remaining environments have been previously published. An overview figure of the various sampling sites is provided in Fig. 2, with sequencing data repository references in Table S1, and sampling details in Table S2. In total 18 million sequencing reads were included in this study, encompassing 48.7 thousand amplicon sequence variants (ASVs). On average, each sample contained 83 ± 72 thousand sequencing reads (mean ± standard deviation). The upper sediment samples had the lowest sequencing depth with 34 ± 19 thousand reads on average per sample (n = 19) while the old saline groundwater samples had 108 ± 72 thousand reads (n = 15; details provided in Table S3). The rarefaction curves depicting the relationship between sequencing depth and ASV count are asymptotic for nearly all samples, indicating a sufficient sequencing effort for microbial diversity estimates (Fig. S2).
a Map of south-east Sweden with the sampling sites marked as a star. b Bar plot depicting the 11 most abundant phyla over all the environments with the remaining phyla grouped as “Other”. The number of replicates within each environment is displayed underneath each bar label. The environments are ordered according to increasing depth although they do not represent a physical continuum due to the multiple sampling sites as shown in (a).
Microbial community description
On phylum level, the Baltic surface seawater community had a similar composition to the Baltic benthic seawater community. Likewise, the composition of sediment communities (upper and lower) resembled each other along with the soil versus deep biosphere groundwaters communities (Fig. 2). The Actinobacteria and Cyanobacteria were the most abundant phyla in the Baltic surface seawater community with an abundance of 30 and 23%, respectively. In contrast, both upper and lower sediment communities were characterized by a high abundance of Proteobacteria (47 and 37%, respectively), Bacteroidetes (15 and 8%), and Chloroflexi (5 and 16%). Finally, both soil and deep biosphere groundwater communities were characterized by a high abundance of Proteobacteria and Patescibacteria with a combined abundance above 50% that increased up to 74% in the upper soil groundwater. The high abundance of the Patescibacteria clearly distinguished the soil plus deep biosphere groundwaters from the sediment plus seawater communities (Fig. 2) and is consistent with the Patescibacteria being abundant in shallow aquifers21,46. The Patescibacteria dominated the upper soil and meteoric groundwaters (49 and 43%) after which the abundance of this clade decreased with depth in the modern marine (28%) and old saline (21%) groundwaters. Abundant representatives from the Patescibacteria included “Candidatus Falkowbacteria” and “Ca. Kaiserbacteria” (Fig. S3) that both constituted more than 10% of the shallow soil and modern marine groundwater communities. The persistence of this clade in subsurface groundwaters was likely due to its capacity to maintain growth under low energy conditions21,46. Within the Proteobacteria, the Betaproteobacteriales and the Campylobacterales orders were mainly responsible for the abundance of this phylum in the modern marine and old saline groundwaters (Fig. S3). On genus level, Sulfurimonas and Thiobacillus were abundant representatives of the Proteobacteria, together comprising 27 and 17% of the microbial abundance in the modern marine and old saline groundwaters, respectively. Although Cyanobacteria are reported to be present in the deep biosphere47,48, this phylum was only scarcely present in the meteoric (0.4%) and modern marine (0.02%) groundwaters and was not detected in the old saline groundwater.
Alpha diversity, according to the Shannon index, peaked in the lower sediment (6.7 ± 0.60; Fig. 3) and upper soil groundwater (6.6 ± 1.1) before decreasing with depth to 3.9 ± 0.64 in the old saline groundwater community. For example, the alpha diversity in the lower sediment community was statistically higher than in the modern marine groundwater community (Tukey HSD’s p value = 6.3e−3, all pairwise tests in Table S4). However, the lower sediment community alpha diversity was higher compared to the upper sediment (albeit insignificant, p = 0.22) that contrasts with the general notion that diversity decreases with sediment depth27,49. This incongruence was potentially caused by sampling a larger part of the deeper sediment column (i.e. top 1 cm for upper sediment compared to 6 plus 20 cm depth for lower sediment), thereby capturing more fine-scale variation e.g., local diversity hotspots in redox transition zones50, that would positively affect the overall diversity. Within the groundwaters, a negative correlation between alpha diversity and depth was observed (Pearson’s r = −0.73, p = 2.7e−13; Fig. 3) and also between diversity and chloride content (r = −0.33, p = 0.005) while the correlation of alpha diversity with DOC was positive (r = 0.60, p = 1.1e−7). In addition, depth correlated with DOC (r = −0.63, p = 1.1e−3) that in general showed the deeper groundwaters contained less DOC and have reduced diversities. These correlations help explain why the old saline groundwater had a reduced diversity compared to e.g., the meteoric groundwater as the former is at greater depth and has a very low organic carbon content (Fig. 1). The ordination plot (Fig. 4) revealed a high dissimilarity between deep biosphere groundwater communities and Baltic surface and benthic, upper and lower soil, plus upper and lower sediment microbial communities, which was confirmed by statistical testing (Table S5). The modern marine groundwater clusters with the old saline groundwater (Fig. S4) while in contrast, the meteoric groundwater sat alone between the other deep biosphere groundwaters and the lower soil groundwater.
a Boxplot combined with a dot plot. The box is formed by the first and third quartiles while the whiskers extend to 1.5 times the inter-quartile range. The data points in the Baltic surface seawater left of the whisker indicates outliers. The environments are ordered according to increasing depth although they do not represent a physical continuum due to the multiple sampling sites as shown in Fig. 2a. Scatterplots depicting the relation of alpha diversity with depth (b) and with dissolved organic carbon (c). Shading in b, c represents a 95% confidence interval.
The beta diversity was estimated according to the Bray-Curtis dissimilarity index. The clustering was verified by multivariate statistical testing (Table S4).
The results showed that the deep biosphere groundwaters had a lower alpha diversity than soil groundwaters and sediments. This diversity decreased with depth, retention time from a few years in meteoric to thousands of years in old saline groundwater, and dissolved organic carbon content (Fig. 3). Despite infiltration of the Baltic Sea, the microbial community in the modern marine groundwater did not resemble the community in the Baltic surface seawater, as illustrated by the composition on phylum level (Fig. 2) and beta diversity analysis (Fig. 4).
Connectivity of surface microbes with the deep biosphere
All deep biosphere groundwaters shared at least 8% of their ASVs with the surface and near-surface environments (Fig. 5), supporting the concept that “Everything is everywhere, but, the environment selects”51. For example, of the 8567 ASVs in the modern marine groundwater, 748 ASVs had a (near) surface representative and these taxa accounted for 49% of the abundance in the modern marine groundwater community. 572 out of 2836 ASVs in the old saline groundwater had a (near) surface representative that accounted for 36% of this groundwater community (Fig. 5). Despite seawater infiltration of bedrock fractures37, the number and the abundance of persisting ASVs between the Baltic surface seawater and modern marine groundwater was low (Table S6). Pairwise comparison of the various communities revealed that the lower soil groundwater was the most important source of surface microbes to the deep biosphere groundwaters (Table S6). For example, 227 out of a total of 1660 meteoric ASVs persisted between lower soil and meteoric groundwaters that constituted 22% of the abundance in the meteoric groundwater community. This overlap was dominated by the Patescibacteria, demonstrating that the capacity of ultra-small cells to infiltrate shallow groundwaters from soils21 also extends to the deep terrestrial biosphere22. Potentially, the dominance of this group among the persisting ASVs is facilitated by their episymbiosis on bacterial or archaeal hosts and this lifestyle may also be an adaptation to low-energy environments such as the deep biosphere52. In addition to the Patescibacteria, the Proteobacteria and Epsilonbacteraeota phyla were also abundant in the overlapping community (Fig. 5) and interestingly, the Epsilonbacteraeota phylum was only represented by the genera Sulfurimonas and Sulfuricurvum. The most abundant genera affiliated with the Proteobacteria were Syntrophus and Hydrogenophaga. Most Proteobacteria and Epsilonbacteraeota likely survived due to the prevalence and importance of sulfate reduction in oligotrophic deep biosphere groundwaters53,54.
a Venn diagram depicting the number of shared ASVs between each of the groundwaters and the surface plus near-surface communities with their relative abundance in parentheses. Surface and near-surface is a reference to the seawater, sediment, and soil groundwater samples. b The taxonomy of the shared ASVs sorted at the taxonomic level of phylum. c The taxonomy of the ASVs that lack a surface or near-surface representative, sorted at the taxonomic level of phylum. For each groundwater, the relative abundance of b, c sums up to 1. The overlap among the deep biosphere groundwater communities can be found in Table S5.
The modern marine groundwater shared 2186 out of its 9315 ASVs with the old saline groundwater and these ASVs comprised 92 and 98% of the abundance in these groundwaters, respectively. This high degree of overlap between both communities is also depicted in Fig. 4 wherein the samples of both environments form a cluster. That the old saline groundwater community was predominantly a subset of the modern marine groundwater community suggested that its diversity was constrained by its geochemistry such as a very low organic carbon content (1–1.4 mg L−1), long retention time (up to thousands of years), and very high chloride content (12–16 g L−1). In contrast, the modern marine groundwater community shared relatively few taxa with e.g., the Baltic benthic seawater and upper sediment communities (Table S6a) but these shared ASVs comprised 34 and 33% of this groundwater’s microbial abundance, respectively (Table S6b). Likewise, the old saline groundwater community shared few taxa with e.g., the upper sediment community but these shared ASVs comprise 23% of this groundwater’s microbial abundance. Starnawski et al.27 found a similar pattern with 79 OTUs persisting throughout the marine sediment column that made up to as much as 40–50% of the microbial community in deeper sediment layers while these OTUs represented less than 10% of the total diversity. Although a significant proportion of the diversity in the deep biosphere groundwaters originated from surface infiltration, it is not entirely a subset. This is illustrated in Fig. 5 where the majority of deep biosphere ASVs lack a (near) surface representative and suggests that part of the diversity in the deep biosphere likely constituted a long-term community separated from the surface for many thousands of years. However, it should be noted that these missing surface representatives might not have been captured in the (near) surface samples due to fine-grained spatial heterogeneity in e.g., the soil groundwaters. That only a small number of ASVs from the Baltic surface seawater persisted in deep biosphere groundwaters, despite infiltration of seawater37, shows that surface microbes unable to survive in the challenging low energy conditions were outcompeted and the cellular components rapidly consumed during nutrient recycling6.
Core and accessory deep biosphere taxa
The deep biosphere groundwaters shared 154 out of a total of 9025 ASVs that were present in all three groundwater types. These ASVs, referred to as the core community, constituted 24, 24, and 14% of the microbial community in the meteoric, modern marine, and old saline groundwaters, respectively of which 52 were affiliated with the Patescibacteria (Fig. S5). Abundant taxa within the Patescibacteria included “Ca. Falkowbacteria” in the modern marine groundwater (12%) along with “Ca. Levybacteria” (5.7%) and “Ca. Kaiserbacteria” (2.1%) in the meteoric groundwater. Nine of these core ASVs were affiliated with the genus Syntrophus (class Deltaproteobacteria) of which representatives have been identified in groundwater from a 1.34 km deep former mine55. In addition to the core community, the meteoric, modern marine, and old saline groundwaters harbored an accessory community of 1324 (67% of the community), 4895 (6.0%), and 978 (2.1%) unique ASVs, respectively that were not retrieved in any other environment. 29% of the ASVs unique to the meteoric groundwater were affiliated with the Patescibacteria (Fig. S5). The contrast between the three groundwater types, especially between meteoric groundwater and modern marine plus old saline groundwater, suggested a role of the groundwater’s differing depth, retention time, and chemistry that favors different microbial populations. Finally, this analysis of deep biosphere groundwaters confirmed the previously reported presence of a core microbiome42.
Conclusions
The results reveal connectivity of surface and near-surface environments with the deep biosphere at the Äspö HRL, allowing infiltration of microbes. Consequently, the deep biosphere is potentially prone to alterations in surface biome microbial communities, such as climate change. Although the number of ASVs persisting among various environments was relatively low, these taxa comprise up to 49% of the deep biosphere microbial community. Despite hydrological connectivity of the Baltic Sea with the majority of the studied deep biosphere groundwaters, the most important source of microbes from the (near) surface was the lower soil groundwater. This supported the concept of species sorting whereby in this case, microbial populations migrate, but the restrictive geochemical conditions in the deep biosphere selects. The abundant persistent ASVs aligned with the phyla Patescibacteria, Proteobacteria, and Epsilonbacteraeota. Finally, the absence of a surface representative for the majority of abundant deep biosphere taxa suggested that a large portion of the deep biosphere microbiome were from resident autochthonous populations that live isolated from the photosynthetic driven surface.
Methods
Published datasets used in this study
A total of 29 previously published 16S rRNA gene amplicon (V3–V4 region) samples, each sampled in replicates were included in this study (Table S1). Baltic benthic seawater and upper sediment samples were collected using a gravity corer, sampling the top 1 cm layer of sediment and the overlying benthic water31,32,33. Modern marine and old saline groundwaters from the Äspö HRL were collected under in situ pressure using a 0.1 μm filter connected to the respective boreholes as previously described13. The microbial communities in Baltic benthic seawater plus upper sediment31,32,33 and modern marine plus old saline groundwaters13 have been previously described.
Sampling sites and cell capture
In addition to the published amplicon data, a further 122 samples were collected from various environments (overview provided in Fig. 1 and sampling details in Table S2). Baltic surface seawater samples (n = 93) were captured as part of a time-series between October 2011 and December 2013 at the Linnaeus Microbial Observatory (LMO), located 11 km off the northeast coast of Öland using a Ruttner sampler at a depth of 2 m below the water surface56. The lower sediment (n = 14) was sampled in June 2018 with a gravity corer in a bay near Äspö HRL (Borholmsfjärden) according to Broman et al.33 except that the sediment core was cut at 6 and 20 cm below the sediment-water interface. To account for spatial heterogeneity, seven sites within Borholmsfjärden were sampled at two depths, yielding 14 samples. With salinity as the major driver for microbial community composition in the Baltic Sea57, salinity concentrations were measured for the surface seawater samples in LMO (range 6.6–7.6%), the benthic water in Loftahammar (6.5%)32, and the water overlying the sediment in Borholmsfjärden (range 6.1–6.5%). A one-way ANOVA showed no significant differences in salinity regimes among the three environments (p = 0.15). Furthermore, soil groundwaters were sampled in October 2019 using soil tubes available on Äspö Island designated as upper (2–3 m below the surface; three tubes; SSM42, SSM215, & SSM268, n = 9) and lower (5 m depth; one tube; SSM22, n = 3) soil groundwater. Finally, meteoric groundwater (borehole KR0015B at 70 m depth, n = 3) was sampled in October 2019. Both soil and meteoric groundwaters were sampled in triplicates as described for the modern marine and old saline groundwaters in Lopez-Fernandez et al.13. Briefly, to avoid contamination from stagnant water in the sampling connections and boreholes, three section volumes were flushed and discarded before connecting a high-pressure filter holder (Millipore) containing a sterile 0.1 µm pore size polyvinylidene fluoride Durapore Membrane filter and the water allowed to pass through the filter. The filters were aseptically collected in cryotubes and immediately frozen in liquid nitrogen for transport to the laboratory where the samples were kept at −80 °C.
The chemistry data was part of SKB’s extensive geochemical monitoring program that is publicly available as the Sicada database. For the modern marine and old saline groundwaters, this data has been published in Lopez-Fernandez, et al.13. For the meteoric and soil groundwaters, chemistry data was retrieved for this study with the sampling dates (November 2019) as close as possible to the microbiology sampling effort during October 2019.
DNA extraction, PCR amplification, and sequencing
DNA was extracted using a phenol-chloroform based method58 for the Baltic surface seawater samples; the Qiagen DNeasy PowerSoil Kit for the upper and lower sediment samples; and the Qiagen DNeasy PowerWater Kit for both soil and meteoric groundwater samples. Regarding the soil and meteoric groundwater samples, the manufacturer’s protocol was followed apart from re-suspending the extracted DNA in 50 μL instead of 100 μL elution buffer. 16S rRNA gene fragment sequences (V3–V4 region) were PCR amplified using the primer set 341F and 805R57 according to the protocol described in Hugerth et al.59. DNA concentration was analyzed using a Qubit 2.0 Fluorometer (Life Technologies) and amplicon specificity by gel electrophoresis. Sequencing was carried out at the Science for Life Laboratory, Sweden on the Illumina MiSeq platform, producing 2 × 300 bp paired-end reads13.
Bioinformatic analyses
Raw sequencing reads were processed using the Ampliseq pipeline (v1.2.0)60 that relied on FastQC (v0.11.8), Cutadapt (v2.8)61, MultiQC (v1.9)62, QIIME2 (v2019.10.0)63, DADA2 (v1.14.1)64, and the SILVA reference database (v138.1)65. Samples with less than 1000 reads were excluded from downstream analysis, thereby removing three from the 96 samples from the Baltic surface seawater environment. To test for DNA extraction kit contaminants that can be an issue in low biomass environments, four negative control DNA extracts were sequenced and processed using identical parameters. One sample yielded a total of 86 ASVs that were removed from the dataset prior to downstream analysis.
Statistics and reproducibility
Absolute counts were standardized according to relative abundance by dividing an ASV’s count by the total number of counts within a sample as this has been reported to be more accurate than rarefying microbiome data66. Alpha diversity was estimated using the Shannon-Weaver index, taking the mean over replicates, followed by statistical testing of the diversity between environments by a one-way ANOVA and post-hoc Tukey’s HSD test while correcting for multiple comparisons. Correlation between alpha diversity, depth, and DOC was quantified according to Pearson correlation and tested for significance using the Pearson’s product moment correlation coefficient. Normality was checked prior to statistical testing with the Shapiro-Wilk test, Levene’s test, and quantile-quantile plots. Beta diversity was estimated by the Bray-Curtis dissimilarity index and tested for significance between groups using a permutational analysis of variance (PERMANOVA). Prior to PERMANOVA, the homogeneity of within-group variation was assessed using PERMDISP67. The null hypothesis tested with this procedure was that the average within-group variation was equivalent among groups. Beta diversity was estimated according to Bray-Curtis dissimilarities and visualized using the nonmetric multidimensional scaling (NMDS) function on default settings in the R Vegan (v2.5)68 package. Statistics and plot generation were performed in R Studio (version 3.6.3.)69. A compiled version of the R script, generated using knitr70 (v1.33), is uploaded to a public repository with the link provided in the code availability statement below.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data are available for the samples from nucleic acid sequencing repositories as detailed in Table S1.
Code availability
The R Markdown document is provided in GitHub at: https://github.com/geweaa/connectivity/.
References
Magnabosco, C. et al. The biomass and biodiversity of the continental subsurface. Nat. Geosci. 11, 707–717 (2018).
Pedersen, K., Bengtsson, A. F., Edlund, J. S. & Eriksson, L. C. Sulphate-controlled diversity of subterranean microbial communities over depth in deep groundwater with opposing gradients of sulphate and methane. Geomicrobiol. J. 31, 617–631 (2014).
Bell, E. et al. Active sulfur cycling in the terrestrial deep subsurface. ISME J. 14, 1260–1272 (2020).
Kotelnikova, S. Microbial production and oxidation of methane in deep subsurface. Earth-Sci. Rev. 58, 367–395 (2002).
Onstott, T. C. et al. In Enigmatic Microorganisms and Life in Extreme Environments (Joseph Seckbach ed) 487–500 (Springer Netherlands, 1999).
Lopez-Fernandez, M. et al. Investigation of viable taxa in the deep terrestrial biosphere suggests high rates of nutrient recycling. FEMS Microbiol. Ecol. 94, fiy121 (2018).
Lopez-Fernandez, M. et al. Metatranscriptomes reveal all three domains of life are active, but are dominated by bacteria in the Fennoscandian crystalline granitic continental deep biosphere. mBio 9, e01792–01718 (2018).
Baker, B. J. et al. Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. ISME J. 7, 1962–1973 (2013).
Lau, M. C. et al. An oligotrophic deep-subsurface community dependent on syntrophy is dominated by sulfur-driven autotrophic denitrifiers. Proc. Natl Acad. Sci. USA 113, E7927–E7936 (2016).
Wu, X. et al. Microbial metagenomes from three aquifers in the Fennoscandian shield terrestrial deep biosphere reveal metabolic partitioning among populations. ISME J. 10, 1192–1203 (2016).
Simkus, D. N. et al. Variations in microbial carbon sources and cycling in the deep continental subsurface. Geochim. Cosmochim. Acta 173, 264–283 (2016).
Jones, R. E., Beeman, R. E. & Suflita, J. M. Anaerobic metabolic processes in the deep terrestrial subsurface. Geomicrobiol. J. 7, 117–130 (1989).
Lopez-Fernandez, M., Astrom, M., Bertilsson, S. & Dopson, M. Depth and dissolved organic carbon shape microbial communities in surface influenced but not ancient saline terrestrial aquifers. Front. Microbiol. 9, 2880 (2018).
Holm, N. G., Oze, C., Mousis, O., Waite, J. & Guilbert-Lepoutre, A. Serpentinization and the formation of H2 and CH4 on celestial bodies (planets, moons, comets). Astrobiology 15, 587–600 (2015).
Parkes, R. J. et al. Rock-crushing derived hydrogen directly supports a methanogenic community: significance for the deep biosphere. Environ. Microbiol. Rep. 11, 165–172 (2019).
Casar, C. P. et al. Mineral-hosted biofilm communities in the continental deep subsurface, deep mine microbial observatory, SD, USA. Geobiology 18, 508–522 (2020).
Hug, L. A. et al. A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016).
Nelson, W. C. & Stegen, J. C. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 6, 713 (2015).
Kietäväinen, R. & Purkamo, L. The origin, source, and cycling of methane in deep crystalline rock biosphere. Front. Microbiol. 6, 725 (2015).
Schwank, K. et al. An archaeal symbiont-host association from the deep terrestrial subsurface. ISME J. 13, 2135–2139 (2019).
Herrmann, M. et al. Predominance of Cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 10, 1407 (2019).
Probst, A. J. et al. Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat. Microbiol. 3, 328–336 (2018).
Lomstein, B. A., Langerhuus, A. T., D’Hondt, S., Jørgensen, B. B. & Spivack, A. J. Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484, 101–104 (2012).
Walsh, E. A. et al. Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 10, 979–989 (2016).
Nunoura, T. et al. Variance and potential niche separation of microbial communities in subseafloor sediments off Shimokita Peninsula, Japan. Environ. Microbiol. 18, 1889–1906 (2016).
Malinverno, A. & Martinez, E. A. The effect of temperature on organic carbon degradation in marine sediments. Sci. Rep. 5, 17861 (2015).
Starnawski, P. et al. Microbial community assembly and evolution in subseafloor sediment. Proc. Natl Acad. Sci. USA 114, 2940–2945 (2017).
Marshall, I. P. G. et al. Environmental filtering determines family-level structure of sulfate-reducing microbial communities in subsurface marine sediments. ISME J. 13, 1920–1932 (2019).
Kirkpatrick, J. B., Walsh, E. A. & D’Hondt, S. Microbial selection and survival in subseafloor sediment. Front. Microbiol. 10, 956 (2019).
Lindh, M. V. et al. Metapopulation theory identifies biogeographical patterns among core and satellite marine bacteria scaling from tens to thousands of kilometers. Environ. Microbiol. 19, 1222–1236 (2017).
Broman, E. et al. Spring and late summer phytoplankton biomass impact on the coastal sediment microbial community structure. Microb. Ecol. 77, 288–303 (2019).
Broman, E., Sachpazidou, V., Pinhassi, J. & Dopson, M. Oxygenation of hypoxic coastal baltic sea sediments impacts on chemistry, microbial community composition, and metabolism. Front. Microbiol. 8, 2453 (2017).
Broman, E., Sjostedt, J., Pinhassi, J. & Dopson, M. Shifts in coastal sediment oxygenation cause pronounced changes in microbial community composition and associated metabolism. Microbiome 5, 96 (2017).
Alakangas, L. J., Mathurin, F. A. & Åström, M. E. Diverse fractionation patterns of rare earth elements in deep fracture groundwater in the baltic shield–progress from utilisation of diffusive gradients in thin-films (DGT) at the Äspö Hard Rock Laboratory. Geochim. Cosmochim. Acta 269, 15–38 (2020).
Drake, H., Åström, M. E., Tullborg, E.-L., Whitehouse, M. & Fallick, A. E. Variability of sulphur isotope ratios in pyrite and dissolved sulphate in granitoid fractures down to 1 km depth–evidence for widespread activity of sulphur reducing bacteria. Geochim. Cosmochim. Acta 102, 143–161 (2013).
Drake, H., Tullborg, E.-L., Hogmalm, K. J. & Åström, M. E. Trace metal distribution and isotope variations in low-temperature calcite and groundwater in granitoid fractures down to 1 km depth. Geochim. Cosmochim. Acta 84, 217–238 (2012).
Hubalek, V. et al. Connectivity to the surface determines diversity patterns in subsurface aquifers of the Fennoscandian shield. ISME J. 10, 2447–2458 (2016).
Laaksoharju, M. et al. Hydrogeochemical evaluation and modelling performed within the Swedish site investigation programme. Appl. Geochem. 23, 1761–1795 (2008).
Mathurin, F. A., Åström, M. E., Laaksoharju, M., Kalinowski, B. E. & Tullborg, E.-L. Effect of tunnel excavation on source and mixing of groundwater in a coastal granitoidic fracture network. Environ. Sci. Technol. 46, 12779–12786 (2012).
Laaksoharju, M., Gascoyne, M. & Gurban, I. Understanding groundwater chemistry using mixing models. Appl. Geochem. 23, 1921–1940 (2008).
Smellie, J. A. T., Laaksoharju, M. & Wikberg, P. Äspö, SE Sweden: a natural groundwater flow model derived from hydrogeochemical observations. J. Hydrol. 172, 147–169 (1995).
Mehrshad, M. et al. Energy efficiency and biological interactions define the core microbiome of deep oligotrophic groundwater. Nat. Commun. 12, 1–12 (2021).
Wu, X. et al. Potential for hydrogen-oxidizing chemolithoautotrophic and diazotrophic populations to initiate biofilm formation in oligotrophic, deep terrestrial subsurface waters. Microbiome 5, 37 (2017).
Banwart, S. et al. Organic carbon oxidation induced by large-scale shallow water intrusion into a vertical fracture zone at the Äspö Hard Rock Laboratory (Sweden). J. Contaminant Hydrol. 21, 115–125 (1996).
Louvat, D., Michelot, J. L. & Aranyossy, J. F. Origin and residence time of salinity in the Äspö groundwater system. Appl. Geochem. 14, 917–925 (1999).
Tian, R. et al. Small and mighty: adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 8, 51 (2020).
Puente-Sánchez, F. et al. Viable cyanobacteria in the deep continental subsurface. Proc. Natl Acad. Sci. USA 115, 10702–10707 (2018).
Miettinen, H. et al. Microbiome composition and geochemical characteristics of deep subsurface high-pressure environment, Pyhäsalmi mine Finland. Front. Microbiol. 6, 1203 (2015).
Hoshino, T. et al. Global diversity of microbial communities in marine sediment. Proc. Natl Acad. Sci. USA 117, 27587–27597 (2020).
Zhao, R., Hannisdal, B., Mogollon, J. M. & Jørgensen, S. L. Nitrifier abundance and diversity peak at deep redox transition zones. Sci. Rep. 9, 8633 (2019).
O’Malley, M. A. ‘Everything is everywhere: but the environment selects’: ubiquitous distribution and ecological determinism in microbial biogeography. Stud. Hist. Philos. Sci. Part C: Stud. Hist. Philos. Biol. Biomed. Sci. 39, 314–325 (2008).
Castelle, C. J. et al. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat. Rev. Microbiol. 16, 629–645 (2018).
Pedersen, K. Subterranean microbial populations metabolize hydrogen and acetate under in situ conditions in granitic groundwater at 450 m depth in the Äspö Hard Rock Laboratory, Sweden. FEMS Microbiol. Ecol. 81, 217–229 (2012).
Pedersen, K. Metabolic activity of subterranean microbial communities in deep granitic groundwater supplemented with methane and H2. ISME J. 7, 839–849 (2013).
Rastogi, G. et al. Microbial and mineralogical characterizations of soils collected from the deep biosphere of the former homestake gold mine, South Dakota. Microb. Ecol. 60, 539–550 (2010).
Bunse, C. et al. High frequency multi-year variability in Baltic Sea microbial plankton stocks and activities. Front. Microbiol. 9, 3296 (2019).
Herlemann, D. P. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571 (2011).
Bunse, C. et al. Spatio-temporal interdependence of bacteria and phytoplankton during a Baltic Sea spring bloom. Front. Microbiol. 7, 517 (2016).
Hugerth, L. W. et al. DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl. Environ. Microbiol. 80, 5116–5123 (2014).
Straub, D. et al. Interpretations of environmental microbial community studies are biased by the selected 16S rRNA (Gene) amplicon sequencing pipeline. Front. Microbiol. 11, 2652 (2020).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
McMurdie, P. J. & Holmes, S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10, e1003531 (2014).
Anderson, M. J. Distance‐based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253 (2006).
Oksanen, J. et al. The vegan package. Community Ecol. package 10, 631–637 (2007).
Team, R. C. R: A language and environment for statistical computing. (2013).
Xie, Y. Dynamic Documents with R and knitr (Chapman and Hall/CRC, 2017).
Acknowledgements
The authors thank The Swedish Nuclear Fuel and Waste Management Co (SKB) for access to the Äspö HRL and Sicada database. The study was supported by The Swedish Research Council (contracts 2018-04311 and 2017-04422). M.D. thanks the Crafoord Foundation (contract 20180599). S.B. acknowledges financial support from the Swedish Research Council and Science for Life Laboratory. C.B. acknowledges funding by HIFMB, a collaboration between the Alfred-Wegener-Institute, Helmholtz-Center for Polar and Marine Research, and the Carl-von-Ossietzky University Oldenburg, initially funded by the Ministry for Science and Culture of Lower Saxony (MWK) and the Volkswagen Foundation through the “Niedersächsisches Vorab” grant program (grant number ZN3285). High-throughput sequencing was carried out at the National Genomics Infrastructure hosted by the Science for Life Laboratory. Bioinformatics analyses were carried out utilizing the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) at Uppsala University (projects SNIC 2021/22-628 and SNIC 2021/6-256). The computations were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX partially funded by the Swedish Research Council through grant agreement no. 2016-07213.
Funding
Open access funding provided by Linnaeus University.
Author information
Authors and Affiliations
Contributions
M.D. and S.B. conceived the study; M.D. and G.W. designed the research; G.W., S.T., M.M., L.A., V.S., C.B., J.P., M.K., M.Å., and S.B. produced and/or analyzed data; G.W., M.M., and M.D drafted the manuscript with comments from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review information
Communications Biology thanks Daniel Lipus, Rui Zhao, and Romy Chakraborty for their contribution to the peer review of this work. Primary Handling Editor: Luke R. Grinham. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Westmeijer, G., Mehrshad, M., Turner, S. et al. Connectivity of Fennoscandian Shield terrestrial deep biosphere microbiomes with surface communities. Commun Biol 5, 37 (2022). https://doi.org/10.1038/s42003-021-02980-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-021-02980-8
This article is cited by
-
Biosignatures of an ancient bedrock- and impact structure-hosted deep biosphere: current knowledge and future perspectives
Discover Geoscience (2025)
-
Candidatus Desulforudis audaxviator dominates a 975 m deep groundwater community in central Sweden
Communications Biology (2024)
-
Terrigenous dissolved organic matter persists in the energy-limited deep groundwaters of the Fennoscandian Shield
Nature Communications (2022)