Abstract
The processes of forming lineages undergoing widespread radiations remain a knowledge gap that is fundamental to our understanding of the geographic distributions of species. Although early studies emphasized the importance of dispersal ability and historical migration events, key innovations that promote rapid diversification and/or adaptation to new habitats may also strongly influence distribution ranges. Juniperus is the second largest genus of conifers and is widely distributed throughout the Northern Hemisphere. Here, we used phylogenetic, phenotypic, and climatic data to investigate the contributions of these processes to the wide distribution and rapid diversification of Juniperus. Combining a time-scaled phylogeny and macroevolutionary theory, we show that the key innovations of berry-like seed cones and dioecy promoted the rapid diversification of Juniperus and that increased dispersal ability promoted allopatric speciation. Ecological niches had significant divergence among different clades of Juniperus. Biogeographic results supported multiple long-distance dispersal events and niche variation that contributed to the modern range of Juniperus, while both phenotypic adaptation and ecological opportunity probably drove its distribution range. Our findings suggest that the current widespread distribution is likely the result of significant divergence driven by niche variation in which ecological opportunities from key innovation and phenotypic divergence.
Similar content being viewed by others
Introduction
Species richness and species diversity are not distributed randomly in space; instead generally exhibiting regular distribution patterns1,2,3,4. Species may be dispersed over different environmental gradients or may follow distinct geographic features (such as islands and mountains)5,6,7. Rapid radiation characterized by lineage diversification is one of the central processes responsible for species-rich clade diversity8. A species-rich clade has often been attributed to key innovations or novel traits conferring an evolutionary advantage9,10 in new environments or in situ habitat shifts, thereby obtaining new ecological niches11. Key innovations are characteristics that loosen the restrictions on adaptive evolution and/or boost competitiveness; these can lead to rapid bursts of diversification by facilitating the occupation of new adaptive zones12. Innovations such as the seed dispersal in the early divergence of Fagaceae13 or the fleshy fruits of plants14 exemplify how such novel traits can have dramatic effects on lineage diversification. Environmental factors such as the paleoclimate15, mountain heterogeneity16,17, islands18, and temperature changes19,20 may lead to the appearance of new and varied niches, creating novel ecological opportunities for living species. Meanwhile, key innovations can create new ecological niches and increase individual fitness, and thus promote rapid diversification and extend the current distribution21,22,23. To understand the evolutionary history of a rapid diversification group, we need to investigate both key innovations and ecological opportunities.
Juniperus is the second-largest genus of conifers and the largest genus within the living Cupressaceae, comprising about 75 species in three monophyletic sections24,25. Section Caryocedrus includes only one dioecious taxon, J. drupacea native to Greece, Turkey, Lebanon, and Syria26. Section Juniperus includes 14 species, with 12 species distributed in Eastern hemisphere, J. jackii endemic to North America, plus the circumboreal J. communis25. Section Sabina contains approximately 60 species distributed in Asia, Africa, southwestern North America, and the Mediterranean region. Recently, considering the agreement between the phylogenetic results and morphological characters, the genus Juniperus sensu lato was divided into three genera, Juniperus sensu stricto (sect. Juniperus), Sabina (sect. Sabina), and Arceuthos (sect. Caryocedrus)27.
The Juniperus likely originated in Eurasia during the Eocene and was a member of the south Eurasian Tethyan vegetation. The present range of Juniperus was presumably influenced by long-distance dispersal and migration across land bridges28. The living Juniperus are a major component of arid and semi-arid landscapes throughout the Northern Hemisphere, and many species are ecologically and economically important conifers24. Juniperus is one of the most diverse genera of the conifers; species are found from sea level to above timberline29. Most Juniperus species prefer to grow on limestone; however, some taxa grow on granite, sand dunes, and sandstone, and the habitats range from deserts to bogs24. Diversity of species distributions across habitats predicts ecological niche variation in Juniperus species. Whether ecological niche variation has contributed to the widespread distribution of Juniperus and the causal factors involved remain unclear.
A key element in the evolution of Juniperus was the evolution of a fleshy female cone, functionally resembling a berry, and dioecy, unlike other genera in the Cupressaceae30. Compared to other conifers, Juniperus had a relatively high number of polyploidization events and at least 15% of Juniperus taxa are tetraploids31. Furthermore, there are significant morphological differences between the three sections, including leaf shape, female cone size, texture, and color25,26. During a complex evolutionary history that included long generation times, and long-term climatic changes28, the influence of key innovations (such as the ‘berry-like’ seed cones, and dioecy) and morphological divergence in the diversification rate and species diversity of Juniperus remain unknown.
In this study, we investigated the importance of key innovations, phenotypic adaptation, evolutionary history, and ecological opportunity in the species richness and wide distribution of Juniperus. We first constructed a robust phylogeny using the whole chloroplast genomes among almost all living Juniperus species; i.e., 70 of the 75 species recognized by Adams24. Based on this phylogenetic framework, we estimated divergence times, assessed diversification rates, reconstructed ancestral ranges and ancestral niches, and inferred evolutionary shifts of habitat. We derived species diversity patterns and estimated the effects of environmental variables. We investigated whether key innovations, environmental changes, niche variation, and historical dispersal events have been associated with Juniperus diversification and range expansion.
Results
Phylogeny and divergence time of Juniperus
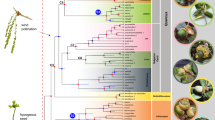
Using the chloroplast genome and adding species with several chloroplast gene sequences from GenBank, we constructed the most complete phylogenetic tree for Juniperus to date. The tree topologies from ML and BI analyses were congruent, and two-thirds of the nodes had the highest possible support values (ML = 100%) and posterior probability values (PP = 1.0), indicating highly resolved relationships among Juniperus species (Fig. 1, Supplementary Fig. 1 and Supplementary Fig. 2). Our results strongly supported Juniperus sensu lato as a sister group to Cupressus sensu lato, and the three sections were in well-supported clades (sect. Sabina, sect. Juniperus, and sect. Carycedrus), consistent with the results using morphology and with other molecular studies (Fig. 1)25,27,28,32.
a Phylogeny of Juniperus. BI tree inferred from the whole chloroplast genome along with chloroplast genes of several species downloaded from GenBank. The numbers attached to the branches show the Bayesian posterior probability (PP) and maximum likelihood bootstrap support (BS). The PP = 1/BS = 100 is not shown. b–d The global distribution patterns of species diversity (b), phylogenetic diversity (c), and weighted endemism (d) of Juniperus plotted in grid cells of 1° × 1°. The map was downloaded from the WorldClim (https://www.worldclim.org/).
The results for divergence time showed that the earliest ancestor of Juniperus appeared approximately 58.36 Ma (stem age, 95% highest posterior density (HPD): 47.91–71.23 Ma) during the middle of the Paleocene period, and the crown age was estimated as 35.92 Ma (95% HPD: 33.01–37.71 Ma), during the later Eocene period (Supplementary Fig. 3). The divergence time between sect. Juniperus and sect. Carycedrus was 28.27 Ma (95% HPD: 20.47–35.36 Ma), and the crown age of sect. Sabina was 31.35 Ma (95% HPD: 28.26–34.69 Ma), during the Oligocene period. Diversification of sect. Sabina into four clades occurred in the Oligocene, apparently in a relatively short time.
Global distribution patterns of Juniperus diversity
Finally, a total of 409,200 distribution sites were obtained, including all species of Juniperus, and the distribution data covered 4383 grid cells. The results showed that Juniperus was widely distributed in the Northern hemisphere (Fig. 1b), and only J. procera had spread into the Southern hemisphere along the East African Rift Valley region. The species J. communis was the most widely distributed, largely in high-latitude regions. The species richness and phylogenetic diversity patterns uncovered there were three distinct centers of species diversity, the Himalaya-Hengduan Mountains, the Mediterranean coast, and Western North America (Fig. 1b, c). The species endemism pattern coincided with the species richness (Fig. 1d), indicating different regions had specific species diversification. The Caribbean islands, Atlantic islands (Azores and the Canary Islands) of West Africa and the Japan and Taiwan islands in Asia were host to endemic species, including J. barbadensis, J. cedrus, J. morrisonicola and so on.
Driving factors of distribution patterns of Juniperus
According to the multiple regression analysis, eight variables (bio2, bio5, bio8, bio11, bio14, bio15, bio16, ME) were significantly correlated with the species richness of global Juniperus (R2 = 0.30), including three temperature variables, four precipitation variables, and the mean elevation. The species richness of Juniperus was positively correlated with the maximum temperature of the warmest month (bio5), precipitation of the wettest quarter (bio16), and the mean elevation (ME), while being negatively correlated with precipitation of the driest month (bio14) and precipitation seasonality (bio15) (Supplementary Fig. 4). These results indicated that both topography heterogeneity and temperature had significant effects on species diversity (Supplementary Fig. 4i).
Ecological niche divergence and ancestral niche reconstruction of Juniperus
Ecological niches showed significant divergence among different clades of Juniperus according to the 21 environmental variables (Supplementary Fig. 5). Section Sabina Clade I, sect. Sabina Clade IV, and sect. Carycedrus clade appeared to be more tolerant to heat, as the ecological niche values for maximum temperature of the warmest month (bio5) were higher than in other clades. The niche values of precipitation of the wettest quarter (bio16) in sect. Sabina Clade I, sect. Sabina Clade IV, and Sect. Juniperus had lower values, while sect. Sabina Clade II had the highest values. Section Sabina Clade I, sect. Carycedrus, and sect. Sabina Clade II were distributed at high altitudes; in particular, sect. Sabina Clade II was found at the highest altitudes, over 3000 m. Those results indicated that sect. Sabina Clade II was more suited to the humid and cold environment of the high altitude of the Himalaya-Hengduan Mountains, and sect. Sabina Clade I was better adapted to the high temperature and low rainfall of Western North America (Supplementary Fig. 5).
Based on the phylogenetic relationships and the ecological niches of the current species, we reconstructed the ancestral ecological niche of Juniperus by using RevBayes (Fig. 2 and Supplementary Fig. 6). Three environmental variables, bio5, bio16, and ME, representing the temperature, precipitation, and topography variables, respectively, were used to reconstruct the ancestral niche of Juniperus (Fig. 2). The results suggested that the ancestor of Juniperus evolved in a warm and semi-arid environment with the maximum temperature of the warmest month of ~28.9 °C and the precipitation of the wettest quarter of ~315 mm. The reconstruction of the ME suggested that the ancestor of Juniperus had inhabited a middle altitude at an elevation of ~883 m.
a–c Relationships between ecological variables (bio5, Max Temperature of the Warmest Month; bio16, Precipitation of the Wettest Quarter, and ME. Mean Elevation) and species richness. d–f Ancestral niche estimation. The point estimates and 95% credible intervals of traits reconstructed for common ancestors of section and clade are shown in numerical values and bars. g–i Ancestral niche estimation and niche differentiation among six clades of Juniperus. The shaded areas represent 95% credible intervals. The letters on the boxplots represent the results of multiple comparisons.
For the ancestor of the sister clades of sect. Sabina Clade I and sect. Sabina Clade II, the maximum temperature was ~28.8 °C in the warmest month, similar to the other four clades (Fig. 2d, g). However, the ancestral niches of sect. Sabina Clade I and sect. Sabina Clade II had significant divergence, while the ancestor of sect. Sabina Clade II had a maximum temperature of ~20.6 °C, and the sect. Sabina Clade I had a maximum temperature of ~29.9 °C in the warmest month. The results of the ancestral niche reconstruction for the sect. Sabina Clade I suggested that the value of bio5 had increased, and during the diversification of the group, sect. Sabina Clade I became adapted to high-temperature environments. Based on the time-scaled phylogeny, the results suggest that the ancestor of sect. Sabina Clade I adapted to high-temperature environments around 24 Ma.
For the precipitation of the wettest quarter, there was also a significant difference between the ancestors of sect. Sabina Clade I and sect. Sabina Clade II. The results of the ancestral niche reconstruction showed that the precipitation for the ancestor of sect. Sabina Clade II in the wettest quarter had increased to ~348.1 mm, a value that was higher than those for the ancestors of other clades. The ancestor of sect. Sabina Clade IV had lower precipitation ( ~ 254.2 mm) and needed to adapt to an arid environment. The results of the ancestral niche reconstruction in the sect. Sabina Clade I showed that the precipitation environments were very divergent during the diversification of the group (Fig. 2e, h).
The ancestral elevation distribution of Juniperus is shown in Fig. 2f, i, and the results were used to determine whether the ancestors of the different clades inhabited high-elevation environments. The results showed that the ancestors of the three sections were distributed at elevations below 1000 m, and different clades in the sect. Sabina had significantly diverged. The ancestors of sect. Sabina Clade I and sect. Sabina Clade II had likely already inhabited the high-elevation environments. The sect. Sabina Clade II inhabited the Qinghai-Tibet Plateau (QTP) at an elevation of ~2,787.8 m, an elevation that is within the range of many extant species. With the uplift of the QTP, there was a subsequent creation of variation in elevation during the diversification of this clade approximately 17 Ma.
Macroevolutionary rate and trait innovation in Juniperus
Based on the time-scaled phylogeny of the Cupressaceae (Supplementary Fig. 7 and Supplementary Fig. 8), we detected one shift in the net diversification rates (Fig. 3a). The net diversification rates in Cupressus sensu lato and Juniperus had significant increase. The results indicated a 4.66-fold increase in the diversification rate compared to the background rate. Furthermore, the speciation and net diversification rates were significantly increased at ~50.0 Ma (Fig. 3c), coinciding with the diversification of Cupressus sensu lato and Juniperus and including the maximum number of species in the Cupressaceae. The results indicated that Juniperus rapidly radiated and that the speciation rate had been maintained at a high level, with no shift among different sections (Supplementary Fig. 9).
a Phylogenetic relationships of Cupressaceae sensu lato based on the whole chloroplast genome, with branch colors representing the rates of diversification and branch length representing the times of differentiation as detected by BAMM and BEAST, respectively. The photos shows three birds feeding Juniperus cones: Passer montanus, Cyanopica cyanus, and Bombycilla japonica (photo credit Kangjia Liu). b Diversification analyses using BiSSE demonstrating the effect of two traits on speciation rates: dioecious (yellow) vs. monoecious (blue) and berry-like seed cones (orange) vs. normal seed cones (purple). The variable traits within Cupressaceae sensu lato are represented on the right side of the phylogenetic tree a. The four photos represent the specific traits of Juniperus (photo credit Kangjia Liu). c Rate-through-time plots for speciation (red lines), extinction (blue lines), and net diversification (green lines) with 95% confidence intervals indicated by shaded areas. The estimates were made using BAMM.
Based on the evolutionary tree results for the Cupressaceae, the BiSSE module analyses demonstrated that various clade-specific traits influenced the diversification rates within the Cupressaceae (Supplementary Table 1). For the four selected binary traits, the berry-like seed cones (vs. normal seed cones) and dioecious (vs. monoecious) traits were strongly correlated with the rapid diversification of Juniperus (Fig. 3b and Supplementary Fig. 10). Each of the two traits acted in concert on the two components (speciation rate and extinction rate) of diversification: the dioecious species with berry-like seed cones had higher speciation rates than monoecious species with normal seed cones. The shrubby life history and scale-like leaf type did not make significant contributions to the diversity of Juniperus, possibly as a result of the synapomorphy of other branches in Cupressaceae (Supplementary Fig. 10). In Juniperus, there were some unique morphological characteristics in each section such as the species of sect. Sabina Clade II that was largely distributed in the Himalaya-Hengduan Mountains and evolved one seed and thick seed coat to adapt to the alpine environment and red cones appeared in sect. Juniperus.
Historical biogeography of Juniperus
According to the ancestral range estimation (Fig. 4), the distribution area of the common ancestor of Juniperus and Cupressus sensu lato. was probably located in Asia. For Juniperus, the area of origin may have been Europe or Asia. The common ancestor of sect. Juniperus was inferred to have originated in Europe, whereas that of sect. Sabina was probably in Europe, Asia, or a combination of these two. The ancestral distribution areas of the common ancestors of sect. Sabina Clade I, Clade II, Clade III, and Clade IV were inferred to be Western North America, Asia, Asia, and the Mediterranean coast, respectively.
a The division of geographical areas (A. Western North America; B. Eastern North America; C. Central America; D. Europe plus the Mediterranean coast; E. Africa and southern Arabia; F. Asia; G. Japan and Taiwan Islands). The map was downloaded from the WorldClim (https://www.worldclim.org/). b Density plots of speciation, dispersal, and vicariance events. The density curves are total speciation events (green line), dispersal speciation events (red line), and vicariance speciation events (yellow line). Historical temperature change results deduced by Zachos are represented by brown lines. c The number of dispersal events among different geographical areas. d Ancestral range estimation using RASP with DEC + J model. Divergence times were estimated using BEAST based on the chloroplast genome sequences along with five fossils.
We identified 36 dispersal events and ten vicariance events. Most of the dispersal and vicariance events occurred about 17 Ma in the early Miocene and as recently as 5 Ma (Fig. 4b). Our results indicated that Juniperus diversified within Europe and Asia, and the first dispersal event from Asia to Western North America (A) occurred at 28.29 Ma in the middle Oligocene, leading to the common ancestor of sect. Sabina Clade I. The second dispersal from Europe to Eastern North America (B) occurred at 16.38 Ma. There was a transcontinental dispersal event occurring in the sect. Sabina Clade III at 10.68 Ma, resulting in migration from Asia to Central America. The largest number of dispersal events occurred between Western and Eastern North America, and between Asia (F) and Japan and Taiwan Islands (G) (nine times), followed by between Western North America and Central America (C) (seven times) and between Europe plus the Mediterranean coast (D) and Asia (six times) (Fig. 4c).
Discussion
The diversification of many plant groups has been influenced by the acquisition of new morphological traits such as the berry fleshy fruits in Calophyllum33, vertebrate pollination syndromes in Andean bellflowers34, life history types in Corydalis35, and polyploidy in Allium36. The species of Juniperus are strong candidates for increasing the species numbers compared to other Cupressaceae genera37. Among the traits we tested, state changes in cone types and/or sex types may explain accelerated net diversification in Juniperus and the related Cupressus sensu lato compared to other genera in the Cupressaceae. BiSSE analyses were used to discover the details concerning how the network of trait changes has been associated with the dynamics of diversification in the Cupressaceae (Fig. 3b).
The berry-like seed cones trait is a novel morphological characteristic that evolved at about the same time as the intensification of climate change and a steady decline in global temperatures in the later Eocene period (Fig. 3). The acquisition of a berry-like seed cones, as a key innovation, was a newly evolved beneficial trait that enabled the occupation of new niche space, increasing the ability for seed dispersibility and permitting rapid range shifts in response to climate change. This in turn led to an increased speciation rate in Juniperus.
Our results demonstrate that the hypothesis of transition from dry to fleshy fruits in angiosperms coincided with changes in diversification rates14,38,39. The evolution of fleshy fruits, including berries and drupes, is considered a key innovation in angiosperm lineages such as Bromeliaceae40 and Calophyllum of the Calophyllaceae33. Berries are primarily bird-dispersed, and the berry-like seed cones of Juniperus are also dispersed by birds41. Furthermore, fleshy-fruited plants and their associated frugivores are frequently generalists42, suggesting that seed dispersal does not depend on a particular species and that it can promote diversification43.
Dioecy, the sexual system in which male and female organs are found on separate individuals, is the norm in many animal clades, but rather rare in flowering plants, occurring in only about 6% of all species44. Previous studies have shown that dioecy is advantageous with higher specialization for sex-specific functions under various ecological and environmental conditions45,46,47,48. There are inconsistent results regarding the relative diversification rates of dioecious lineages compared to their non-dioecious counterparts based on different methods and datasets47,49,50. The BAMM and BiSSE results showed the dioecious trait in Juniperus was associated with an increase in the speciation rate (Fig. 3). It is noteworthy that few species of Juniperus are monoecious, and these species did not form a clade (Figs. 1 and 3), indicating that the common ancestor of Juniperus species may have been diecious, and the monoecious species are likely to represent local reversals from dioecious species.
Dioecy is thought to be positive for facilitating outcrossing, especially when other outcrossing mechanisms are absent, thereby minimizing inbreeding depression51. Population genetic analyses supported the hypothesis that there is high genetic diversity within populations and low to moderate population differentiation of Juniperus, in agreement with the patterns reported for outcrossing species52,53. Because dioecy reduces trade-offs between male and female functions, it may have been selected and accelerated species diversification in Juniperus. Givnish54 demonstrated a strong correlation between dioecy and the presence of fleshy or succulent tissues in the seeds or cones of conifers. The characteristic fleshy-dioecious combinations are stable reproductive strategies as well as the combination of monoecy with dry cones30,55, and these traits may have increased the speciation rates of Juniperus.
Several studies discovered polyploidy is associated with higher diversification rates in several plant lineages such as Allium36, Rosaceae56. Polyploidy or whole genome duplication is rare in conifers, however, at least 16 Juniperus species are polyploid, a trait that is higher in frequency compared to other conifers31. All the polyploid Juniperus species are belong to sect. Sabina and most distributed in the region of Himalaya-Hengduan Mountains. However, there was no diversification rates shift of the polyploid Juniperus species (Supplementary Fig. 9).
The genus Juniperus is one of the most diverse genera of conifers, and different clades have significant niche divergence (Fig. 2 and Supplementary Fig. 5). Our results are consistent with the hypothesis28 that the ancestor of Juniperus originated during the middle of the Paleocene in Europe and Asia (Fig. 4). The niche reconstruction indicated that the ancestor of Juniperus evolved in a warm and semi-arid environment (Fig. 2). In the early period of divergence, the Juniperus species underwent geographic expansion by multiple long-distance dispersal events that allowed them to occupy different environments and regions owing to the niche expansion. Juniperus exhibited accelerated rates of species diversification along with key innovations concurrent with geographic and niche expansion. The key innovations of berry-like seed cones and dioecy have a well-established adaptive basis in gymnosperms54. The berry-like seed cones and dispersal by birds could promote allopatric speciation.
In the genus Juniperus, the niche variation among different sections and clades underwent significant differentiation during the early divergence (Fig. 2). Those species in the high-altitude QTP, species that are tolerant of a colder environment, were derived in the middle Oligocene (Fig. 2 and Supplementary Fig. 3) and colonized new habitats, resulting in range expansion. The serrate leaf Juniperus species are endemic and are adapted to the arid environments of North America, and the ancestral niche has diverged with other clades (Fig. 2)57. In parallel with the niche and geographic expansion, Juniperus experienced a rapid divergence of phenotypes. At the section level, Juniperus exhibited significant morphological differences in reproductive traits, female cone size, texture and color of the cones, and leaf shape25,26. Furthermore, there are also significant morphological differences in the four clades in sect. Sabina25,28: the turbinate, single-seeded, entire leaf margin junipers; the multi-seeded, entire leaf margin junipers, and the serrate leaf margin junipers. These morphological traits such as the number of seeds, the type of the leaves, and the color of seed cones are adaptive traits30,57,58,59,60,61, and they have well-established adaptive bases in Juniperus.
The exact ecological processes and the key adaptions behind the Juniperus diversification remain insufficiently understood. One possible explanation is that the colonization of different habitats induced adaptive diversification in cone and seed traits (including number, size, and color)30,62 and hydraulic traits (such as SLA: leaf area/leaf mass)57. Additionally, in the island-distributed species, morphological divergence was influenced in part by novel ecological opportunities, and there are some ecotypes that exist on islands63. Consistent with this interpretation, Juniperus exhibits the highest number of species distributed on islands24,29. The species endemism pattern coincides with the level of species richness (Fig. 1), indicating the effect of local adaptation on species diversity.
Conclusion
Our results showed that rapid diversification of the widespread Juniperus was characterized by key innovations and expansions of ecological niches. The evolution of berry-like seed cones and dioecy as key innovations had driven the rapid diversification of Juniperus through increased dispersal ability, thereby promoting allopatric speciation. The geological and climatic changes were also relevant to the long-distance dispersal of Juniperus. Moreover, our findings suggest that Juniperus was able to inhabit arid and semi-arid landscapes throughout the Northern Hemisphere not only because it had the capacity for long-distance dispersal but also the ability to adapt to new environments. Our study suggests that species richness and widely distributed lineages can be better understood as processes in which a key innovation acts in synergy with niche variation. Combing ecological and evolutionary data together can extend our understanding about species richness and distribution range dynamics of extant species and improve our knowledge for planning conservation actions under future scenarios of climate change.
Methods
Taxon sampling
We sampled leaf material from 65 accessions of Juniperus, representing 55 species (Supplementary Table 2) that were newly contributed by the present study. The materials were obtained from the herbarium of Beijing Forestry University (BJFC), the Plant DNA Bank of China in the Institute of Botany, the Chinese Academy of Sciences, and field sampling. The other 32 accessions were downloaded from GenBank; these included 20 species only containing chloroplast gene fragments in order to obtain more comprehensive sampling coverage (Supplementary Table 3 and Supplementary Table 4). Five species are duplicated obtained from the two methods. Finally, a total of 97 Juniperus accessions were selected representing 70 Juniperus species, or 93.3% of living Juniperus diversity (from a total of 75 species according to Adams24).
In addition, in order to estimate accurate divergence times, examine character evolution, and compare the rates of diversification, we also sampled a total of 56 species with whole chloroplast genome sequences from GenBank representing all of the other genera in the Cupressaceae sensu lato (Supplementary Table 3).
Chloroplast genomes assembly and annotation
Genomic DNA was extracted from silica gel-dried leaves using a modified CTAB method64. Total DNA was sheared using an ultrasonicator into 350 bp fragments, and paired-end sequencing libraries were constructed on an Illumina HiSeq X-ten device platform according to the manufacturer’s instructions using a paired-end sequencing of 2 × 150 bp runs. Each sample yielded 5 Gb of raw data. The chloroplast genomes were assembled and annotated following the methods of Dong et al.65.
Phylogenetic analysis
We first constructed a phylogeny for the Cupressaceae based on a dataset of 107 species. Due to the short inversions ( < 300 bp) and chloroplast rearrangements among Cupressaceae groups, we manually adjusted the sequences according to the gene order of Juniperus. The chloroplast genomes were aligned using MAFFT version 7.49066, and trimAl version 1.3 was used to delete ambiguous regions based on the automated trimming method67. This dataset alignment was 82,203 bp in length, including 25,141 variable sites and 19,871 information sties.
For the Juniperus phylogenic analysis, we chose Cupressus sensu lato, Microbiota, Platycladus, Tetraclinis, Calocedrus, and Chamaecyparis as the outgroups. Complete chloroplast sequences were aligned using MAFFT. The 11 gene fragments from GenBank for 20 unsampled species (Supplementary Table 4) were aligned independently and were manually added to the sequence matrix. The final dataset contained 116 accessions covering 19 outgroups and all 97 Juniperus samples. The length of this dataset was 139,888 bp, including 24,735 variable sites and 15,631 information sites. We used the partition method to assess the best models for the complete chloroplast genome sequence dataset and the concatenation of the 11 gene fragments dataset to reduce the influence of the missing data.
Both maximum likelihood (ML) and Bayesian inference (BI) methods were performed to infer the phylogenetic relationships of the Cupressaceae and Juniperus. The best-fit nucleotide substitution model referred to the results of ModelFinder68. ML phylogenies were inferred using IQ-TREE69 in PhyloSuite software70. The BI analysis was performed in MrBayes v3.271 with two independent Markov Chain Monte Carlo runs. Each chain began with a random tree and was run for 2,000,000 generations with a sampling frequency of once every 100 generations. The first 25% of the sampled trees were discarded as a burn-in.
Molecular dating and macroevolutionary rate estimation
The divergence times of Juniperus and the Cupressaceae were calculated using BEAST 2 under the Bayesian method and the GTR model, respectively. According to the results of Mao et al.28, five fossils were used to estimate the divergence time of Juniperus (Supplementary Table 5). Cupressinocladus interruptus (Fossil A, ~99.6 Ma) was used for the root node, and Calocedrus suleticensis (Fossil B, ~28.4 Ma) was used to constrain the crown age of Tetraclinis articulata. Juniperus pauli (Fossil C, ~33.9 Ma) was placed at the crown group divergence time of Juniperus. J. creedensis and J. desatoyana (Fossil D and E, ~23.0 and 16.0 Ma) were positioned on the inner branch nodes of the serrate leaves clade in Juniperus.
For the estimation of the divergence times of the Cupressaceae, we added four additional calibration priors: the crown time of Cupressoideae ( ~ 123.0 Ma) and the stem ages of Cunninghamioideae, Sequoiodeae, and Tacodioideae ( ~ 211.0 Ma, 174.0 Ma, and 159.0 Ma), referring to Mao et al.72. Moreover, the earliest known fossil record (with seed cones) of Chamaecyparis (Fossil J, <121 Ma) from the Lower Cretaceous Guyang Formation of the Guyang Basin, Inner Mongolia Autonomous Region, North China73 was used to constrain the stem of Chamaecyparis.
The details for the calibration priors of six fossils and four secondary calibrations are shown in Supplementary Table 5. For fossil calibrations, a uniform prior was used to begin, while the secondary calibrations were obtained using a standard normal distribution. Convergence was assessed using effective sample size (ESS > 200) values of the runs in Tracer v1.6 after eliminating the first 25% of the samples. The maximum clade credibility tree, including credibility intervals for time and posterior probabilities (PPs) for nodes, was then assembled using TreeAnnotator74.
Bayesian Analysis of Macroevolutionary Mixtures (BAMM)75, which uses a reversible-jump MCMC to sample a large number of possible diversification regimes from a time-calibrated phylogeny, was used to estimate speciation and extinction rates through time and to identify shifts of diversification rate in both Cupressaceae and Juniperus. The MCMC was run for two million generations and sampled every 1000 generations. Prior values were selected using the “setBAMMpriors” function. Visualization used the R package BAMMtools v.2.1776. The initial 25% of the samples of the MCMC run were discarded as a burn-in. In order to avoid the impacts on estimation caused by sampling bias of different clades, we set the proportions of sampling in different clades. The sampling proportion of Cupressaceae and Juniperus are shown in the Supplementary Tables 6 and 7.
Juniperus distributions
To explore the global distribution patterns of Juniperus, data were downloaded from online databases (NSII, http://www.nsii.org.cn/; GBIF, https://www.gbif.org/), herbaria (PE, K, BJFC) and the literature24,29,37. The species distribution points were carefully checked according to the descriptions by Adams24, and duplicate data were removed.
A set of 1° latitude × 1° longitude grids was drawn using ArcGIS v10.8 on the global scale to avoid the influence of administrative divisions on estimates of the species richness (SR) distribution patterns. The number of species in each grid was counted and visualized using a heat map. A phylogenetic approach (phylogenetic diversity, PD) was used to avoid the influence of artificial classification and to infer the patterns of species richness based on evolutionary relationships between species. This represented one of the components of species diversity77. The sum of the branch lengths within each grid was counted78, and the heat map for PD was drawn based on the phylogenetic tree using the R package Picante79. Endemic species were defined as those restricted entirely to a specified region; the spatial distribution of endemism can provide crucial information on the origin and driving factors of biodiversity patterns at the landscape scale. These can be used to explain the underlying patterns of speciation or range restriction as well as their importance for conservation80,81. Species endemism was measured using weighted endemism (WE) calculated as the reciprocal of the number of species distribution grids to the common degree of all species in each grid. The common degree of all species in each grid was calculated as the sum of species endemic diversity. All of the above visualizations were done with ArcGIS v10.8.
Environmental variables
In order to explore how environmental variables have driven the current global distribution pattern of Juniperus, we collected 19 bioclimatic and two topographical variables (Elevation range and Mean elevation) (Supplementary Table 8). All bioclimatic and topographical variables were collected from WorldClim (https://www.worldclim.org/data/worldclim21.html/) at a resolution of 2.5 arc minutes. The values of 21 environmental variables within each grid were calculated by taking the mean values of each grid for subsequent analyses. Pearson correlation based on the results of stepwise regression with SR as response variable using the Akaike Information Criterion (AIC) was employed to avoid internal correlations between factors that may affect the stability of the model.
Species niche variables and reconstructing ancestral niches
According to the results of the field investigation and previous studies, different distribution regions of Juniperus appeared to show niche variation, and this differentiation also seemed to be related to the phylogenetic relationships within the genus28. Based on the phylogenetic results, we constructed ecological niches of different clades based on the averages of ecological factors. Five environmental variables (bio5: Max temperature of the warmest month; bio11: Mean temperature of the coldest quarter; bio14: Precipitation of the driest month; bio16: Precipitation of the wettest quarter; ME: Mean elevation) were selected based on the above correlation analysis and their potential contributions to the species distribution to reconstruct historical dynamics of the niches occupied by species of Juniperus. The existing niches of Juniperus species were obtained based on the averages of the environmental variables of the grids (Supplementary Table 9). A significance difference according to Duncan’s test was used and was implemented in the agricolae package for R. The ancestral niche was calculated by RevBayes82 using 10 million iterations and was visualized using the ggplot2 package for R.
Biogeographic analysis
Biogeographic analysis was used to explore whether dispersal or isolation have affected the Juniperus global distribution pattern in a geological context. We divided the world into seven geographical regions based on the results of Mao et al.28 and Uckele et al.32: Western North America (A), Eastern North America (B), Central America (C), Europe plus the Mediterranean coast (D), Africa and southern Arabia (E), Asia (F), and Japan and Taiwan Islands (G). The distribution areas of each Juniperus species are shown in Supplementary Table 10. The estimation of ancestral geographical ranges was conducted using RASP83. We tested the six models provided by BioGeoBEARS based on the Akaike information criterion. The model of DEC + J was used to infer the historical biogeography of Juniperus. Owing to the communication ability of species between geographical regions not being the same under the local geological background, the dispersal probabilities among regions (Supplementary Table 11) were set to three categories following Dong et al.84: 0.01 for well-separated areas, 0.1 for moderately separated areas, and 0.5 for well-connected areas.
Trait-dependent diversification
The species of Juniperus have many unique character states that may have promoted the diversification of the genus. For example, the appearance of berry-like seed cones is unique in the entire class of conifers, and this attribute facilitates the dispersal of Juniperus seeds over greater distances by birds; the appearance of shrubby forms implies better adaptation to harsh environments such as drought and arctic-alpine biomes, and the evolution of dioecy (although a few species have remained monoecious) avoids the possibility of self-fertilization, traits that have allowed junipers to rapidly evolve and adapt to changing environments.
We tested the impact of traits on the diversification of Juniperus by concurrently estimating their impacts on speciation, extinction, and transition rates using BiSSE85. Four binary traits of the breeding system (monoecious vs. dioecious), cone type (normal seed cones vs. berry-like seed cones), life form (tree vs. shrub), and leaf type (scale-like leaves vs. acicular leaves) were selected for the analysis (Supplementary Table 12). Character scorings were obtained from our own field and herbarium observations, and in several cases from the species descriptions in Adams24. The Diversitree package for R86 was used to evaluate whether a change in a species trait from “state 0” to “state 1” influenced the BiSSE model parameters. The MCMC sampler was obtained by running 10,000 steps using the range of observed samples as a measure of speciation rates. We then plotted the marginal distribution and 95% credibility intervals for the diversification rate.
Analyses were performed using the R package Diversitree 0.7-686. Once the best-fitting model was selected, CIs for each parameter were estimated for the tree. We used an exponential prior following Fitzjohn86 and began the MCMC with the ML estimates. We ran the MCMC for 20,000 generations and applied a burn-in of 2000 steps. We then computed the net diversification rate for each trait.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The newly sequenced raw reads of this study are deposited the GenBank database under the BioProject PRJNA1132768. Details can be found in Supplementary Table 2. All supplementary files can be found in Supplementary Information.
References
Sanderson, M. J. & Donoghue, M. J. Reconstructing shifts in diversification rates on phylogenetic trees. Trends Ecol. Evol. 11, 15–20 (1996).
Wiens, J. J. & Donoghue, M. J. Historical biogeography, ecology and species richness. Trends Ecol. Evolution 19, 639–644 (2004).
Jablonski, D., Roy, K. & Valentine, J. W. Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314, 102 (2006).
Li, E. et al. Historical climate change and vicariance events contributed to the intercontinental disjunct distribution pattern of ash species (Fraxinus, Oleaceae). Commun. Biol. 7, 603 (2024).
Mittelbach, G. G. et al. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 10, 315–331 (2007).
Jansson, R. & Davies, T. J. Global variation in diversification rates of flowering plants: energy vs. climate change. Ecol. Lett. 11, 173–183 (2008).
Ding, W.-N., Ree, R. H., Spicer, R. A. & Xing, Y.-W. Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369, 578 (2020).
Ronco, F. et al. Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature 589, 76–81 (2021).
Blaimer, B. B. et al. Key innovations and the diversification of hymenoptera. Nat. Commun. 14, 1212 (2023).
Vamosi, J. C. & Vamosi, S. M. Key innovations within a geographical context in flowering plants: towards resolving Darwin’s abominable mystery. Ecol. Lett. 13, 1270–1279 (2010).
Meier, J. I. et al. The coincidence of ecological opportunity with hybridization explains rapid adaptive radiation in Lake Mweru cichlid fishes. Nat. Commun. 10, 5391 (2019).
Wang, Y., Li, E., Sun, J., Zhang, Z. & Dong, W. Phylogenetic diversity and interspecies competition shaped species diversity in adaptive radiated Ligustrum (Oleaceae). J. Syst. Evol. https://doi.org/10.1111/jse.13117 (2024).
Zhou, B.-F. et al. Phylogenomic analyses highlight innovation and introgression in the continental radiations of Fagaceae across the Northern Hemisphere. Nat. Commun. 13 https://doi.org/10.1038/s41467-022-28917-1 (2022).
Bolmgren, K. & Eriksson, O. Fleshy fruits – origins, niche shifts, and diversification. Oikos 109, 255–272 (2005).
Svenning, J.-C., Eiserhardt, W. L., Normand, S., Ordonez, A. & Sandel, B. The influence of paleoclimate on present-day patterns in biodiversity and ecosystems. Annu. Rev. Ecol. Evolution, Syst. 46, 551–572 (2015).
Wang, Q., Guo, Q., Chi, X., Zhu, S. & Tang, Z. Evolutionary history and climate conditions constrain the flower colours of woody plants in China. J. Plant Ecol. 15, 196–207 (2022).
Jin, W. T. et al. Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines. Proc. Natl Acad. Sci. USA 118, e2022302118 (2021).
Lapoint, R. T., Magnacca, K. N. & O’Grady, P. M. Phylogenetics of the antopocerus-modified tarsus clade of Hawaiian Drosophila: diversification across the Hawaiian Islands. PLoS ONE 9, e113227 (2014).
Fragniere, Y. et al. Biogeographic overview of Ulmaceae: diversity, distribution, ecological preferences, and conservation status. Plants (Basel) 10, 1111 (2021).
Wang, H.-F. et al. Spatial patterns and determinants of Moraceae richness in China. J. Plant Ecol. 15, 1142–1153 (2022).
Yoder, J. B. et al. Ecological opportunity and the origin of adaptive radiations. J. Evolut. Biol. 23, 1581–1596 (2010).
Matuszak, S., Favre, A., Schnitzler, J. & Muellner-Riehl, A. N. Key innovations and climatic niche divergence as drivers of diversification in subtropical Gentianinae in southeastern and eastern Asia. Am. J. Bot. 103, 899–911 (2016).
Yang, Y., Bian, Z., Ren, G., Liu, J. & Shrestha, N. Niche conservatism limits the distribution of Medicago in the tropics. Ecography 2022, e06085 (2022).
Adams, R. P. Junipers of the world: the genus Juniperus. (Trafford Publishing, 2014).
Adams, R. P. & Schwarzbach, A. E. Phylogeny of Juniperus using nrDNA and four cpDNA regions. Phytologia 95, 179–187 (2013).
Jagel, A. & Dörken, V. Morphology and morphogenesis of the seed cones of the Cupressaceae-part II Cupressoideae. Bull. CCP 4, 51–78 (2015).
Yang, Y. et al. Recent advances on phylogenomics of gymnosperms and a new classification. Plant Diversity 44, 340–350 (2022).
Mao, K., Hao, G., Liu, J., Adams, R. P. & Milne, R. I. Diversification and biogeography of Juniperus (Cupressaceae): variable diversification rates and multiple intercontinental dispersals. N. Phytol. 188, 254–272 (2010).
Adams, R. P. Juniperus of Canada and the United States: taxonomy, key and distribution. Lundellia 21, 1–34 (2008).
Leslie, A. B., Beaulieu, J. M., Crane, P. R. & Donoghue, M. J. Explaining the distribution of breeding and dispersal syndromes in conifers. Proc. R. Soc. B: Biol. Sci. 280, 20131812 (2013).
Farhat, P. et al. Polyploidy in the conifer genus Juniperus: An unexpectedly high rate. Front. Plant Sc. 10 https://doi.org/10.3389/fpls.2019.00676 (2019).
Uckele, K. A., Adams, R. P., Schwarzbach, A. E. & Parchman, T. L. Genome-wide RAD sequencing resolves the evolutionary history of serrate leaf Juniperus and reveals discordance with chloroplast phylogeny. Mol. Phylogenet. Evol. 156, 107022 (2021).
Cabral, F. N. et al. Phylogeny, divergence times, and diversification in Calophyllaceae: linking key characters and habitat changes to the evolution of Neotropical Calophylleae. Mol. Phylogenet. Evol. 157, 107041 (2021).
Lagomarsino, L. P., Condamine, F. L., Antonelli, A., Mulch, A. & Davis, C. C. The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). N. Phytol. 210, 1430–1442 (2016).
Peng, H.-W. et al. The synergy of abiotic and biotic factors correlated with diversification of Fumarioideae (Papaveraceae) in the Cenozoic. Mol. Phylogenet. Evol. 186, 107868 (2023).
Han, T.-S. et al. Polyploidy promotes species diversification of Allium through ecological shifts. N. Phytol. 225, 571–583 (2020).
Yang, Y. Diversity and distribution of gymnosperms in China. Biodivers. Sci. 23, 243–246 (2015).
Beaulieu, J. M. & Donoghue, M. J. Fruit evolution and diversification in campanulid angiosperms. Evolution 67, 3132–3144 (2013).
Liu, X. et al. Why is the beautyberry so colourful? Evolution, biogeography, and diversification of fruit colours in Callicarpa (Lamiaceae). Plant Diversity 45, 6–19 (2023).
Igor, M. K. et al. Drivers of dispersal and diversification in bromeliads. https://doi.org/10.1101/2022.11.04.515068 (2022).
Adams, R. P. & Thornburg, D. Seed dispersal in Juniperus: a review. Phytologia 92, 424–434 (2010).
Bello, C. & Barreto, E. The footprint of evolution in seed dispersal interactions. Science 372, 682–683 (2021).
de Queiroz, A. Contingent predictability in evolution: key traits and diversification. Syst. Biol. 51, 917–929 (2002).
Renner, S. S. & Ricklefs, R. E. Dioecy and its correlates in the flowering plants. Am. J. Bot. 82, 596–606 (1995).
Vamosi, J. C., Otto, S. P. & Barrett, S. C. H. Phylogenetic analysis of the ecological correlates of dioecy in angiosperms. J. Evolut. Biol. 16, 1006–1018 (2003).
Vamosi, J. C. & Vamosi, S. M. The role of diversification in causing the correlates of dioecy. Evolution 58, 723–731 (2004).
Käfer, J. et al. Dioecy is associated with higher diversification rates in flowering plants. J. Evolut. Biol. 27, 1478–1490 (2014).
Käfer, J., Marais, G. A. B. & Pannell, J. R. On the rarity of dioecy in flowering plants. Mol. Ecol. 26, 1225–1241 (2017).
Heilbuth, J. C. Lower species richness in dioecious clades. Am. Naturalist 156, 221–241 (2000).
Sabath, N. et al. Dioecy does not consistently accelerate or slow lineage diversification across multiple genera of angiosperms. N. Phytol. 209, 1290–1300 (2016).
Thomson, J. D. & Brunet, J. Hypotheses for the evolution of dioecy in seed plants. Trends Ecol. Evol. 5, 11–16 (1990).
Sertse, D., Gailing, O., Eliades, N.-G. & Finkeldey, R. Anthropogenic and natural causes influencing population genetic structure of Juniperus procera Hochst. ex Endl. in the Ethiopian highlands. Genet. Resour. Crop Evol. 58, 849–859 (2011).
Vanden-Broeck, A. et al. Genetic structure and seed-mediated dispersal rates of an endangered shrub in a fragmented landscape: a case study for Juniperus communis in northwestern Europe. BMC Genet. 12, 73 (2011).
Givnish, T. J. Ecological constraints on the evolution of breeding systems in seed plants: dioecy and dispersal in gymnosperms. Evolution 34, 959–972 (1980).
Wilson, W. G. & Harder, L. D. Reproductive uncertainty and the relative competitiveness of simultaneous hermaphroditism versus dioecy. Am. Naturalist 162, 220–241 (2003).
Vamosi, J. C. & Dickinson, T. A. Polyploidy and Diversification: A Phylogenetic Investigation in Rosaceae. Int. J. Plant Sci. 167, 349–358 (2006).
Willson, C. J., Manos, P. S. & Jackson, R. B. Hydraulic traits are influenced by phylogenetic history in the drought-resistant, invasive genus Juniperus (Cupressaceae). Am. J. Bot. 95, 299–314 (2008).
Vamosi, JanaC., Zhang, Y. & Wilson, WilliamG. Animal dispersal dynamics promoting dioecy over hermaphroditism. Am. Naturalist 170, 485–491 (2007).
Stetter, M. G., Vidal-Villarejo, M. & Schmid, K. J. Parallel seed color adaptation during multiple domestication attempts of an ancient new world grain. Mol. Biol. Evol. 37, 1407–1419 (2020).
Lev-Yadun, S. & Ne’eman, G. Bimodal colour pattern of individual Pinus halepensis Mill. seeds: a new type of crypsis. Biol. J. Linn. Soc. 109, 271–278 (2013).
Pierce, S., Brusa, G., Sartori, M. & Cerabolini, B. E. L. Combined use of leaf size and economics traits allows direct comparison of hydrophyte and terrestrial herbaceous adaptive strategies. Ann. Bot. 109, 1047–1053 (2012).
Dimitri, L. A., Longland, W. S. & Vander Wall, S. B. Cone and seed traits of two Juniperus species influence roles of frugivores and scatter-hoarding rodents as seed dispersal agents. Acta Oecologica 85, 93–103 (2017).
Elias, R. B. & Dias, E. The recognition of infraspecific taxa in Juniperus brevifolia (Cupressaceae). Phytotaxa 188, 241–250 (2014).
Li, J., Wang, S., Jing, Y., Wang, L. & Zhou, S. A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 48, 72–78 (2013).
Dong, W. et al. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 20, 92 (2022).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Zhang, D. et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355 (2019).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Mao, K. et al. Distribution of living Cupressaceae reflects the breakup of Pangea. Proc. Natl Acad. Sci. USA 109, 7793–7798 (2012).
Xu, X.-H. et al. A new discovery of Chamaecyparis from the Lower Cretaceous of Inner Mongolia, North China and its significance. Rev. Palaeobot. Palynol. 257, 64–76 (2018).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comp. Biol. 10, e1003537 (2014).
Rabosky, D. L. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543 (2014).
Rabosky, D. L. et al. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707 (2014).
Winter, M., Devictor, V. & Schweiger, O. Phylogenetic diversity and nature conservation: where are we? Trends Ecol. Evol. 28, 199–204 (2013).
Faith, D. P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 (1992).
Kembel, S. W. et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010).
Harold, A. S. & Mooi, R. D. Areas of endemism: definition and recognition criteria. Syst. Biol. 43, 261–266 (1994).
Bonn, A., Rodrigues, A. S. L. & Gaston, K. J. Threatened and endemic species: are they good indicators of patterns of biodiversity on a national scale? Ecol. Lett. 5, 733–741 (2002).
Hohna, S. et al. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst. Biol. 65, 726–736 (2016).
Yu, Y., Harris, A. J., Blair, C. & He, X. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49 (2015).
Dong, W. et al. Phylogenomics and biogeography of Catalpa (Bignoniaceae) reveal incomplete lineage sorting and three dispersal events. Mol. Phylogenet. Evol. 166, 107330 (2022).
Maddison, W. P., Midford, P. E. & Otto, S. P. Estimating a Binary Character’s Effect on Speciation and Extinction. Syst. Biol. 56, 701–710 (2007).
Fitzjohn, R. G. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol. Evolution 3, 1084–1092 (2012).
Acknowledgements
This work was supported by Science and Technology Basic Resources Investigation Program of China (Grant No. 2021FY100200), the Survey of Herbaceous Germplasm Resources in Shandong Province (Grant No. Lu Financial [2021]1) and the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK050202). We would like to thank the Plant DNA Bank of China in the Institute of Botany, Chinese Academy of Sciences for providing some materials.
Author information
Authors and Affiliations
Contributions
W.D. conceived and designed the research. K.L., X.C., Y.W., C.X., Z.S., and Z.Z. collected samples, performed the experiments and collected the data. K.L., and E.L. analyzed the data. K.L. produced the figures and tables. K.L. and W.D. wrote the manuscript. All co-authors edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Baosheng Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Matteo Dell’Acqua, Luke Grinham and Benjamin Bessieres. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, K., Li, E., Cui, X. et al. Key innovations and niche variation promoted rapid diversification of the widespread Juniperus (Cupressaceae). Commun Biol 7, 1002 (2024). https://doi.org/10.1038/s42003-024-06687-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-06687-4