Abstract
A critical component of ecosystem restoration projects involves using genetic data to select source material that will enhance success under current and future climates. However, the complexity and expense of applying genetic data is a barrier to its use outside of specialised scientific contexts. To help overcome this barrier, we developed Reef Adapt (www.reefadapt.org), an innovative, globally applicable and expandable web platform that incorporates genetic, biophysical and environmental prediction data into marine restoration and assisted gene flow planning. The Reef Adapt tool provides maps that identify areas with populations suited to user-specified restoration/recipient sites under current and future climate scenarios. We demonstrate its versatility and practicality with four case studies of ecologically and evolutionarily diverse taxa: the habitat-forming corals Pocillopora damicornis and Acropora kenti, and macroalgae Phyllospora comosa and Ecklonia radiata. Reef Adapt is a management-ready tool to aid restoration and conservation efforts amidst ongoing habitat degradation and climate change.
Similar content being viewed by others
Introduction
Our world is changing now more rapidly than ever before due to human activities1. Approximately one-third of the Earth’s arable land surface is degraded2 and between 50% and 90% of marine ecosystems are in an altered state3. The coral reef and kelp forest ecosystems that represent more than 50% of the world’s coastal ecosystems4 are now at very high risk of widespread, climate-mediated collapse5,6. Amidst these threats, global efforts to conserve and restore biodiversity and ecosystem resilience are intensifying (CBD/WG2020/REC/3/1, UNEP-WCMC 2022). This is reflected in strong growth in the scope and scale of marine restoration initiatives7,8. Central to the success of these efforts is the development of strategic, science-based approaches to restore lost and degraded ecosystems, particularly in the face of climate change9.
Provenance (i.e. the geographic source of material to be used in restoration re-introductions) and genetic diversity are critical issues to the survival and adaptability of restored populations10,11,12. Restoration practitioners have traditionally been advised to source material from local populations, to retain local genetic diversity13,14. However, current rates of environmental change—largely due to the effects of climate change—are outpacing natural rates of migration and adaptation15. There is an increasing need to consider more transformative interventions aimed at enhancing the resilience and adaptive capacity of both restored and natural populations to climate change16. One such emerging strategy is ‘assisted gene flow'17, which involves the translocation of individuals adapted to future climate conditions at the receiving site. This can be achieved using naturally occurring genotypes that are experimentally proven to be suited to projected conditions (‘predictive provenancing18’) or by sourcing seeds with a bias towards the direction of predicted climatic changes, though not exclusively, to account for climate prediction uncertainties (‘climate-adjusted provenancing19’). While more than 200 studies have experimentally assessed the potential outcomes of assisted gene flow in a range of species, very few have explicitly implemented this intervention as part of an official management effort or conservation tactic, and only a handful have done so in aquatic and marine ecosystems20. Nevertheless, using assisted gene flow in a conservation setting is currently being proposed or considered for the restoration and management of a wide range of terrestrial and marine habitats globally21,22.
An understanding of within-species patterns of genetic diversity is crucial in the design of both restoration and assisted gene flow strategies23,24,25. Genetic information can generally be broken down into two components: neutral genetic diversity, which provides information on historical demographic patterns, gene flow and adaptive potential, and adaptive genetic diversity, which is directly associated with organismal functional traits and fitness. Both types of genetic information are important for practitioners to consider when selecting suitable distances for translocation, or to identify populations that harbour genotypes to be used in assisted gene flow. Generation of genetic data is now relatively cost-effective26, and empirical inferences about whether genetic diversity is neutral or putatively adaptive can be gained through expert analysis of specific datasets27. However, existing data remain relatively inaccessible to end users and exist largely in the scientific literature28. This hinders the uptake of these techniques by restoration managers and practitioners, who may lack the necessary expertise to effectively harness genetic data29,30,31. The limited use of genetic data is particularly pronounced in marine compared to terrestrial systems32, even though widespread habitat declines have been documented for both realms33,34 and their ecological, social and economic importance is of a similar magnitude35,36,37.
In terrestrial ecosystems, many countries have already developed national guidelines for provenancing in restoration (e.g., National Seed Strategy in the US, Florabank Guidelines in Australia), and there are more than 20 resources that provide guidance to improve genetic management outcomes for fragmented or vulnerable populations38. However, in marine systems, there is a notable absence of global standards and local policies for delineating appropriate provenance and a lack of incorporation of genetic information into restoration activities39,40. Terrestrial systems have also seen recent innovations that incorporate and translate complex genetic information into user-friendly formats for non-specialist audiences to make informed provenance choices, such as the Seedlot Selection Tool41, Climate-Smart Restoration Tool (https://adaptwest.databasin.org/pages/adaptwest-climatena/), Provenancing Using Climate Analogues42, and the Restore and Renew framework43. These are broadly based on the development of modelling approaches that associate genetic information with current and future predicted environmental variables and generate predictions of genetic turnover (the change in allele frequencies) across the landscape44. Adapting similar approaches to marine contexts is pertinent, however, marine systems present unique challenges to generating genetic predictions and effective guidelines across the seascape. For example, in addition to environmental drivers of genetic turnover, there is a need to account for the influence of local and regional oceanographic currents on population connectivity, as these represent significant drivers of genetic structure in marine environments45. Tools that enable marine restoration practitioners, managers and policymakers to rapidly and effectively design and assess climate-smart restoration and management interventions are thus urgently needed.
Here, we leverage marine genetic, environmental and biophysical data to develop Reef Adapt—an accessible tool that generates species-specific models and predictions of genetic turnover and then provides instant, dynamic guidelines on where to source material for use in marine restoration and assisted gene flow activities. Below, we provide details on how Reef Adapt works and present case studies for four ecologically significant marine species to demonstrate its practical application. Designed to be continuously expandable and applicable to any marine taxa, Reef Adapt has the potential to substantially improve the way in which marine restoration and assisted gene flow strategies are designed and assessed.
Results
We developed a dynamic and user-friendly tool that accommodates data from multiple sources and is applicable to a wide range of geographical areas. This was achieved by employing a predictive modelling approach, which is accessible via an intuitive platform interface (Fig. 1). The platform, available on our website (www.reefadapt.org), is comprised of the following components:
-
I.
Dedicated webtool: An R shiny app that disseminates provenancing guidelines in a simple, easy-to-use manner. Users interact with drop-down menus to view species with available genetic data. An interactive map allows users to identify their target site, and visualise bespoke guidelines for either local provenancing or assisted gene flow activities that anticipate future (2050) conditions. The webtool has a variety of additional functions, including optional generation of a full report that outlines user inputs, background model details and source of the baseline data, and further explains the chosen provenancing approach. ‘Advanced’ features allow control over technical details. For example: users can divide the available genetic datasets based on their interest, into either neutral or adaptive genetic regions of their target species’ genome27,46. Users can also alter the threshold of genetic differentiation and overlay a survey gap analysis to display prediction confidence.
The webtool is underpinned by
-
II.
Database and data upload portal: Population genetic differentiation data (FST) are added to the Reef Adapt database along with associated metadata, including site coordinates, taxonomic and species life history information. Users can upload their own data to the database, enabling the tool to grow and include more species/taxa as genetic data becomes more available.
-
III.
Automated model pipeline: R software47 is used to extract environmental (e.g. sea surface temperature, SST) and biophysical data for each of the sampled sites and generates generalised dissimilarity models (GDM) and predictions of genetic turnover across the seascape for each species and molecular marker combination.
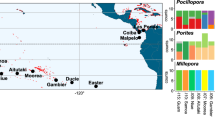
As a starting point to build the tool, we included data from existing literature for 25 species of habitat-forming seaweeds and two coral species found across temperate, polar and tropical marine environments globally (Fig. 2). To illustrate how the tool can be used for different species and management scenarios, we present an overview of the resulting guidelines for local provenancing for two corals (Pocillopora damicornis and Acropora kenti) and local and assisted gene flow provenancing for two macroalgae (Phyllospora comosa and Ecklonia radiata). These species were chosen as examples because they all currently have populations in decline and have available high-resolution genetic data across a broad geographic range. Technical details on the underlying sources of genetic data, environmental covariates and model-fitting are provided in the Methods and in the example downloadable Reef Adapt reports in Supplementary Materials 1.
Blue points represent 420 sampling locations for 27 species of macroalgae and coral. Species highlighted in our case studies are shown clockwise from left (image credits in brackets): Pocillopora damicornis (Karen Filbee-Dexter), Acropora kenti (John Edmondson), Phyllospora comosa (Leah Wood) and Ecklonia radiata (John Turnbull).
Local provenancing
To illustrate how Reef Adapt could help guide local provenancing for a brooding coral species, we chose a hypothetical restoration site for P. damicornis near Coral Bay (Ningaloo Reef, Australia). This is a common species of reef-building coral found throughout tropical and subtropical Indian and Pacific oceans. The GDM used genome-wide single nucleotide polymorphism (SNP) data from 397 samples at six sites48. This model revealed that P. damicornis’ genetic turnover was largely associated with maximum SST and oceanographic connectivity between populations (Fig. 3A). This model was used to generate predictions of genetic turnover across Ningaloo Reef with the default level of allowable genetic differentiation from the restoration site (FST = 0.05). The Reef Adapt output identified that the genetically ‘local’ area corresponded to ~10 km to the north and 50 km south of the target area (Fig. 3B). Practitioners aiming to use this species in restoration projects in this location are recommended to source material from as many populations within this area as possible.
A Turnover of genetic (neutral and adaptive) variation as a function of environmental and geographic variables. The shape of each function indicates how the rate of change in allele frequencies varies along the gradient. Points are site pairs, the line is the predicted relationship between predicted and observed genetic and ecological (or climatic) distance. Panels on the left represent model-fitted I-splines for each GDM model, showing predicted genetic distance/change against each of the biophysical variables included in the final model dataset. The distribution of raw data points from each covariate is indicated via rugplot on the x-axis, with the exception of the oceanographic distance unit, which is calculated internally from the NMDS coordinates during model construction. ±Standard error (in green) generated from 999 bootstrap iterations with 10% of the populations removed. B Reef Adapt webtool predictions showing suitable areas to source seedstock for restoration for P. damicornis. Species distribution highlighted in transparent white; ‘local’ areas with predicted genetic differentiation of up to FST 0.05 from the restoration site (red) are highlighted in green.
In contrast, to illustrate local provenancing for a broadcast spawning coral species example, we chose a hypothetical restoration site for A. kenti (formerly A. tenuis) at Opal Reef (Great Barrier Reef, Australia). This is a common branching coral that is widely distributed across inshore and offshore reefs of the Great Barrier Reef (GBR) and is the subject of active reef restoration initiatives across the GBR (https://www.coralnurtureprogram.org). The GDM used genome-wide SNP data from 141 samples across 13 sites from Matias et al. 49. This revealed that A. kenti’s genetic turnover was largely associated with SST, with oceanographic connectivity having a weak influence (Fig. 4A). Using the default setting of FST = 0.05, the Reef Adapt output identified that the local provenance area for A. kenti extended throughout the region where predictions were available, reaching >1100 km south and >800 km north from the restoration site. Given this large geographic scale, we also used an optional ‘advanced’ feature of Reef Adapt to trial lowering of the FST threshold to 0.01, such that only populations predicted to have genetic differentiation/FST values of 0.01 when compared to the restoration site were identified as suitable source material. Even with this change, the predictions identified areas >300 km south and >600 km north of the restoration site as suitable for provenancing (Fig. 4B).
A Turnover of genetic (neutral and adaptive) variation as a function of environmental and geographic variables. B Reef Adapt webtool predictions showing suitable areas to source seedstock for restoration for A. kenti using a genetic differentiation threshold value of up to FST 0.01 (green). Further details are presented in the Fig. 3 caption for brevity.
To illustrate an example of local provenancing in a dominant seaweed, we chose a restoration site for P. comosa (crayweed) at Manly (Sydney, Australia), where this species is currently under active restoration50,51. This is a large habitat-forming fucoid seaweed from south-eastern Australia. It relies on sexual reproduction, is dioecious and reproductive throughout the year. P. comosa is characterised by gas-filled floats that are thought to facilitate connectivity between populations. GDM used genome-wide SNP data from 331 samples across 13 sites by Wood et al. 52. This revealed that P. comosa genetic turnover was associated with maximum SST, oceanographic connectivity between populations and SST range (Fig. 5A). Using the default settings, practitioners aiming to restore this species to this location are recommended to source material from populations up to 40 km north of this restoration site (Fig. 5B).
A Turnover of genetic (neutral and adaptive) variation as a function of environmental and geographic variables. B Reef Adapt webtool predictions showing suitable areas to source seedstock for restoration for P. comosa (green). Further details are presented in the Fig. 3 caption for brevity.
Assisted gene flow
To illustrate how Reef Adapt could help identify donor populations for assisted gene flow strategies, we chose a hypothetical recipient site for E. radiata outside Port Phillip Bay (Melbourne, Australia). E. radiata is a globally significant kelp that has been severely affected by marine heatwaves and ocean warming53,54,55 and is the subject of active restoration efforts across the southern states of Australia (www.greengravel.org). Both genomic data56 and quantitative experiments57 suggest that E. radiata is locally adapted to temperature, making it an ideal candidate for assisted gene flow research. GDM models were based on genome-wide SNP data collected across 165 samples across nine sites from Minne et al.58. E. radiata genetic turnover was associated with maximum SST (Fig. 6A). Using the default settings, Reef Adapt identified populations containing genetic variation likely favourable to the restoration site under 2050 conditions (Fig. 6B). The closest suitable sites identified were >300 km away, indicating material collections would need to be conducted close to the eastern border or outside of the state of Victoria if they are to include genotypes putatively adapted to conditions expected under 2050 climate projections.
A Turnover of genome-wide genetic variation as a function of environmental variables, as predicted by a generalised dissimilarity model. B Reef Adapt webtool predictions showing suitable areas to source material for assisted gene flow strategies for E. radiata. Species distribution is highlighted in transparent white; areas with predicted genetic composition suitable for 2050 conditions in the focal site (red) are highlighted in red. Further details are presented in the Fig. 3 caption for brevity.
Survey gaps and model confidence
Genetic sampling is logistically challenging and expensive in marine systems. Reef Adapt users can view model predictions for areas where genetic data are unavailable and overlay survey gap data to assess model confidence. In the case of P. damicornis, this analysis identified several gaps in genetic information, with the lowest prediction confidence occurring in offshore, low-latitude remote environments (Fig. 7). These ‘dark’ locations may be candidates for further genetic data collection. If data-poor areas are considered as restoration targets and Reef Adapt is used to provide provenance recommendations, the model uncertainty must be carefully considered.
Discussion
Here, we show that Reef Adapt can be used to rapidly determine where a representative ‘local’ stock is for any given area within the distribution of a reef species based on the best available genetic data. This knowledge is critical for restoration because the size of any genetically ‘local’ area can vary significantly depending on the species and scenario. We also show how Reef Adapt can help practitioners identify areas to source material that are likely to be better adapted to near-future (2050) conditions via the E. radiata case study. Such information is critical for marine management organisations currently undertaking research and risk assessments for implementing climate-smart strategies such as assisted gene flow59.
Informing local provenancing
Local provenancing is currently the default strategy for most interventions in marine systems and is often required by permitting bodies for restoration60. Yet, restoration practitioners rarely have access to the genomic information and skills needed to inform provenance decision-making. Instead, proponents often need to make ad hoc generalisations based on limited information, which can include using genetic data from the same or similar species and/or locations, replicating what might have been done elsewhere, or simply sourcing from the ‘closest available’ reef. The Reef Adapt tool thus has immediate and direct practical value to restoration projects by: (i) enabling practitioners and managers to efficiently progress planning and activities whilst adhering to permitting requirements, (ii) opening up the marine restoration space to a broader diversity of practitioners and (iii) providing a basis for which further research on provenancing can be based, collectively leading to the increased robustness and speed at which restoration activities can be achieved.
Ultimately, provenance guidelines delivered via Reef Adapt are dependent on the FST values used to delineate or define appropriate translocation limits. Because the tool has been developed to aid a non-specialist audience, we provide default FST threshold values as a recommended default value. In addition to these ‘generic’ local seed-sourcing zones, however, Reef Adapt outputs can be tailored to suit specific management needs. For example, in the case of the default output identifying a very large area to cover for collections which can be undesirable (e.g., crossing jurisdictional boundaries), it may be preferable to decrease FST (e.g. the A. kenti example). The converse may be suitable if the tool identifies an area too restrictive where, for example, only minimal available source populations occur and there is a known minimal risk of genetic pollution from introducing more distantly related populations (e.g. via field and/or laboratory provenance trials to quantify the transfer risks61,62). Optimal FST thresholds could also vary between species, markers or populations of a particular species, and future research should focus on defining these. Research trials could be directly informed by the Reef Adapt tool by identifying populations to be used within experiments. To avoid users changing the FST threshold without a thorough understanding of the potential consequences, we have added a popup warning box when this function is activated. We recommend that users monitor the health, survival and recruitment of restored donors and neighbouring populations (controls) to assess the appropriateness of different sourcing distances and provide feedback to Reef Adapt on their project’s success so that we can continue to validate or refine the guidelines for use as the tool expands.
Informing assisted gene flow
Restoration with the aim of maintaining populations via local provenancing can no longer be considered a ‘safe’ or risk-free position18 given the rate of ongoing environmental change. However, assisted gene flow does come with some inherent risks, such as outbreeding depression or introduced locally maladapted genetic variation, which can decrease the fitness of the receiving population after several generations63 (although see ref. 61). Reef Adapt, while not eliminating such risks, provides a science-backed framework to make informed provenancing decisions for assisted adaptation strategies. Moreover, the tool has the capability for users to provide feedback on the empirical success of assisted adaptation in action, which can be used to robustly adjust guidance decisions over time.
Given that both neutral and adaptive genetic variation are important to consider when conducting translocations, we have set the default dataset to include both neutral and adaptive genetic data where these distinctions are available. Critically, however, the efficacy of the Reef Adapt tool for this ‘climate-proofing’ application depends on the degree of local adaptation present, and the importance of predictive environmental variables in the Reef Adapt GDM models. The tool has been developed with this use in mind, such that if climate-proofing is a key objective for a project, users can select model outputs based on peer-reviewed ‘adaptive’ SNP data only via the advanced settings. Going forward, it will be important to ground-truth Reef Adapt predictions through in situ experiments64, both to quantify the adaptive landscape and to test how translocated genotypes that might be better adapted to 2050 conditions perform under current conditions at a recipient site. Updates are planned that will include new prediction ranges (e.g. 2080, 2100) as new scenarios (such as Shared Socioeconomic Pathway scenarios of CMIP665,66 and performance against them are released.
Reef Adapt’s assisted gene flow provenancing tool is just one of a suite of climate-smart strategies that may be used to facilitate climate adaptation. The most effective strategy to slow climate impacts is undoubtedly to immediately reduce greenhouse gas emissions67. Further, it is critical to ensure that restored or managed populations have sufficient genetic diversity to respond to a variety of environments and stressors. Where the level of genetic diversity that can be conferred by translocating individuals sourced from the wild is not considered enough to confer climate resilience other strategies such as selective breeding, the introduction of non-traditional species, or even gene editing may be considered, but genetic homogenisation of populations should be avoided68.
Versatility across different taxa and inclusion of new species
Reef Adapt currently includes data for 27 species of macroalgae and corals, but this versatile platform can be readily applied to diverse marine taxa such as seagrasses, sponges, oysters, and fishes. Reef Adapt is now available to rapidly and cost-effectively put the best available genetic data in the hands of users at a critical time when projects are initiating and scaling up. We have provided a mechanism for expansion via upload of new genetic data by users on the website, however, it is worthwhile noting that the Reef Adapt tool is currently limited to the spatial extent and resolution of the environmental data available (currently BioOracle-derived data, which excludes estuaries69,70). As global scale products become available at increasing spatial and temporal resolutions (e.g., Himawari-9 for SST), the environmental conditions around coastal areas and complex and dynamic areas such as estuaries will be better resolved.
Corals and some other taxa present common taxonomic challenges, whereby cryptic, but genetically distinct species are discovered within previously morphologically defined species49,71. Failing to identify these species-level genetic divisions may unintentionally bias conservation and restoration plans informed by genetic data. In such cases, FST values may either be spuriously inflated if cryptic species are geographically restricted within the broader sampling range, or underestimated if co-distributed species are grouped together in the analysis72. While the coral genetic datasets used in our case studies were pre-filtered to ensure that population genetic statistics are derived from a single taxon, detecting and delineating hidden genetic biodiversity is a significant challenge. We will continue to update species distribution ranges and environmental and genetic data in Reef Adapt to follow taxonomic iterations to help alleviate this problem.
Genetic information and interpreting different molecular markers
Where there is more than one type of molecular marker available for a species in Reef Adapt, the selection of markers is discretionary and can impact results. In such circumstances, we recommend prioritising SNP markers derived from genomic datasets, which yield more precise estimates of population-level diversity and higher power to identify genetic differentiation between populations by considering both neutral and adaptive genomic diversity73. Within SNP datasets, there are also likely to be minor differences in predictions generated using either neutral, adaptive or all (default) regions of the genome74. While microsatellite markers have been used to generate most genetic datasets in the past, inferences based on such low-density genetic nuclear markers will lack the resolution to appropriately resolve cryptic species diversity and subtle genetic differentiation and should be considered a secondary choice for projects that are aiming to make climate-proof provenancing decisions. Further research is needed to determine optimum FST thresholds, and we plan to establish an adaptive management feedback loop to update Reef adapt guidelines as/when new information on species/marker-specific thresholds is made available.
Future work
There are many other potentially useful metrics for management and conservation decision-making available from the GDM approach that could be incorporated into Reef Adapt. For example, Reef Adapt GDM models can be used to rapidly produce maps of genomic bioregions, genomic vulnerability, genomic uniqueness or genetic diversity within MPA networks75. These metrics could help identify areas of high management value for biobanking, genetic reinforcement or spatial protection. The inclusion of additional factors that are likely to influence restoration/assisted gene flow success, e.g. ecological interactions, microhabitat data, or insights gained from other “omic”-based research (disease resistance, how long target genes take to spread throughout an existing population, etc.) may also be integral in coping with environmental changes, and are exciting areas for future expansion. Further avenues for research include population and ecosystem-wide implications of mixing genetically distinct individuals (i.e. conducting assisted gene flow) at evolutionarily relevant timescales (i.e., across multiple generations) and determining whether genetic turnover is likely to change in the future (e.g., via the strengthening of boundary currents) in order to identify priority areas for genetic management interventions. In both cases, combining current knowledge of underlying genetics with in silico simulations76,77,78 would be pertinent.
Finally, the application of Reef Adapt in marine restoration must be sensitive to governance arrangements such as permitting regulations and jurisdictions. Incorporating existing knowledge of target species population density, abundance and health is important to reduce impacts on wild populations during collection of source material. Consideration of issues of cultural and indigenous significance and boundaries is also important. In future iterations of Reef Adapt we plan to integrate features such as sea country boundaries (in Australia), to facilitate Indigenous group participation in the provenance decision-making process.
Methods
Population genetic information
To build the initial database for the Reef Adapt platform, we developed a semi-automated analysis pipeline based on genetic input data gathered from the existing literature, or directly input by users via a web tool. FST is a widely reported metric of genetic differentiation between populations79. The standard data consists of (i) a metadata file for each genetic dataset (number of sites, number of sampled individuals, water depth, molecular markers used and whether the genetic data (FST) were derived from all, neutral or putatively adaptive genome regions, any additional taxonomic and trait information, (ii) coordinates of sampled sites and (iii) pairwise FST values for all sites sampled. Additional metrics such as F’ST, GST and RST can be incorporated into future versions of the tool. To be accepted, studies must provide evidence of peer-review or submit a QAQC form and include >5 sampling sites, with at least 10 km separation. Templates for users to upload their own data to the platform are supplied on the website under the “Submit Data” tab and in Supplementary Data 1.
Predictor variables
Environmental predictors
The geophysical and environmental covariates were obtained from the BioOracle v2.20 database74,75, which has a spatial resolution of 5 arcmin (~9.2 km at the equator) for both present (2000–2014) and predicted future (2040–2050) conditions. The 2040–2050 timeframe was selected as it represents a realistic mid-term goal for climate resilient restoration efforts, while RCP 8.5 most closely tracks current and expected future carbon emissions80. A diverse range of variables were extracted (e.g., sea surface temperature (SST) and photosynthetically active radiation; full list in Supplementary Data 1). Redundant covariates were removed during the model fitting process, allowing each species’ model to fit with the most appropriate set of covariates.
Biophysical predictors
The Marine Ecoregions of the World database was used to represent historical barriers to gene dispersal in the model81. A shapefile of the ecoregions was converted to a spatial raster with cell values indicating the ecoregion identification number for model fitting (see the section “Generalised dissimilarity modelling”). Linear geographic distance of each sample site from the nearest mainland was also estimated using the distance function in the raster package in R, with maps of continental mainlands from the ‘natural earth’ database82.
Species-specific distribution raster layers were used to determine every population’s relative position in their respective current distributional range (i.e. centre vs. range edge populations). The Reef Adapt database uses distribution layers for macroalgal species from Fragkopoulou et al.83 and coral species distribution layers from the IUCN spatial data portal (https://www.iucnredlist.org/resources/spatial-data-download). Range position is estimated using the median latitudinal value of each species range (i.e. ‘range centre’) calculated in R, with cell values transformed to represent the percentile distance from the range centre to the latitudinal extremes in each hemisphere. To account for temperate (seaweed) species occupying multiple continents (with several range centres and edges), range positions were calculated separately for each quadrant of the globe, using the −30° longitudinal meridian (which falls between most continents) to separate the quadrants. For tropical species (corals), range position was calculated across global distribution.
Pairwise connectivity distance variables were computed for every grid cell (representative of populations) to incorporate oceanographic connectivity. The pairwise distances were calculated as the shortest path in a global ocean transport network (Supplementary Note 1), where each node in the network represented an ocean grid cell of 0.1° resolution and each directional link represented the transport between nodes by ocean surface currents within 1 day. Links with a probability of transport of <0.05 were removed from the network before calculating the distances. Long-distance dispersal events were given an upper limit of 33 days84,85 to constrain the influence of extreme dispersal events.
To transform the matrices into coordinates on a two-dimensional space suitable for modelling86, the resulting distance matrices were then inputted to a non-metric multidimensional scaling (nMDS) analysis using the metaMDS function in the vegan package in R87. The Euclidean distances of the resulting coordinates were then used in the generalised dissimilarity models (see below). In the case of a model not converging using this pairwise connectivity distance term, the standard Euclidean distance term generated in the GDM was used.
Seascape genetic modelling
Variable selection
For each unique genetic dataset (i.e., each dataset of species-specific FST values calculated from a set of loci), environmental and biophysical data were extracted for each grid cell with corresponding genetic samples. If the location of a site fell on a cell with no data in a particular layer, data were extracted from the nearest cell using the move function in the gecko R-package88. To exclude redundant variables, one of any highly correlated variables (correlation > 0.8) was included in the final set of covariates.
Generalised dissimilarity modelling
Generalised dissimilarity modelling (GDM) was used to model and predict genetic turnover. GDM is a powerful extension of generalised linear modelling89,90,91, and has been used extensively over the past decade to model and predict non-linear associations between environmental variables and pairwise biological measures such as assemblage composition, trait variation and genetic distances.
Genetic matrices and predictor data were formatted using the formatsitepair function in the gdm92 package. This was done separately for each FST-based dataset, as genetic data from different methods or genomic regions cannot be directly compared. In some cases, model fit was improved by normalising FST values using the scale function, so two models were fit; one with raw FST data and one with normalised FST data, with the final model chosen based on the percentage of deviance explained. Significance of fit was tested using matrix permutation with the gdm.varimp function, using 999 permutations for each step and the default three-i-spline basis function. Genetic turnover was assessed using the output plots (e.g. Fig. 3A) using the maximum height of each curve, whereby the height of the curve reflects the relative importance of the environmental variable in influencing genetic distance. The predictive power of the model was cross-validated using a permutational approach over 1000 iterations, excluding a random 20% subset of sites for validation.
Spatial predictions and provenance guidelines
Models with ≥20% of null deviance explained were selected as suitable for use in the webtool. Spatial prediction maps of genetic composition were produced by predicting from the model splines using the covariate rasters with the gdm.transform and predict.gdm functions. The predictions were confined to the spatial extent of each species’ distribution and the bioregions for which genetic data was present, in order to avoid over-extrapolation and generation of spurious provenancing guidelines93,94,95. Future additions to the Reef Adapt database will aim to improve the coverage of sampled areas.
To delineate provenances, we compared the predicted genetic composition of a target site to the genetic composition expected at all other sites across the seascape96. Assisted gene flow provenancing decisions were generated by replacing the environmental covariates with predicted future values for 2050 (where available, or current values were used for specific covariates where future predictions were not available), then comparing the predicted future genetic composition of the target site to the current genetic composition at all other sites. Details on the covariates used in the model predictions are highlighted in the downloadable report. A default FST value threshold of 0.05 was used to mask and highlight predictions as suitable for local provenancing (green on Reef Adapt outputs) or for assisted gene flow (red on Reef Adapt outputs). This threshold value was based on a preliminary examination of a variety of thresholds across species and is a conservative value that should remain within the realm of “within populations” for most species across markers. There is also an option for users to adjust this threshold in the advanced settings (thereby adjusting the permissible level of genetic differentiation a population may have from that predicted at a restoration site) if management preferences (e.g., to avoid crossing jurisdictional boundaries) or additional data (e.g., FST-fitness threshold data) are available. The FST threshold is currently the same for both SNPs and microsatellites by default, as there was not a consistent statistical difference in FST estimates between markers in our datasets where both marker types were available. We recommend that users source material from as many populations as possible within the suitable range highlighted by Reef Adapt to maximise genetic diversity.
Survey gap analysis
For each GDM model generated in Reef Adapt, a survey gap analysis was run using the locations of the data used to fit the GDM, following the methods described in Mokany et al. 75. This analysis identifies ‘dark’ regions that would benefit from further genetic data collection. The Reef Adapt tool includes the option to overlay this information on outputs.
Code availability
All analyses reported in this study used the statistical software R. R script files have been deposited publicly on the corresponding authors’ Github account at https://github.com/georgina-wood/REEF-ADAPT.
References
Steffen, W. et al. Planetary boundaries: guiding human development on a changing planet. Science 347, 1259855 (2015).
Gibbs, H. K. & Salmon, J. M. Mapping the world’s degraded lands. Appl. Geogr. 57, 12–21 (2015).
Lotze, H. K. et al. Depletion, degradation, and recovery potential of estuaries and coastal seas. Science 312, 1806–1809 (2006).
Wernberg, T., Krumhansl, K., Filbee-Dexter, K. & Pedersen, M. F. Status and trends for the world’s Kelp forests. In World Seas: An Environmental Evaluation 2nd edn (ed. Sheppard, C.) Ch. 3, 57–78 (Academic Press, 2019).
Cornwall, W. Florida coral restoration in hot water. Science 383, 576–577 (2024).
Wernberg, T. et al. Impacts of climate change on marine foundation species. Annu. Rev. Mar. Sci. 16, 247–282 (2024).
Eger, A. M. et al. Global kelp forest restoration: past lessons, present status, and future directions. Biol. Rev. 97, 1449–1475 (2022).
Bay, L. K., Gilmour, J., Muir, B. & Hardisty, P. E. Management approaches to conserve Australia’s marine ecosystem under climate change. Science 381, 631–636 (2023).
Burdett, H. L. et al. Including environmental and climatic considerations for sustainable coral reef restoration. PLoS Biol. 22, e3002542 (2024).
Reynolds, L. K., McGlathery, K. J. & Waycott, M. Genetic diversity enhances restoration success by augmenting ecosystem services. PLoS ONE 7, e38397 (2012).
SERA. National Standards for the Practice of Ecological Restoration in Australia 2nd edn (SERA, 2017).
Whiteley, A. R., Fitzpatrick, S. W., Funk, W. C. & Tallmon, D. A. Genetic rescue to the rescue. Trends Ecol. Evol. 30, 42–49 (2015).
McKay, J. K., Christian, C. E., Harrison, S. & Rice, K. J. “How local is local?”—A review of practical and conceptual issues in the genetics of restoration. Restor. Ecol. 13, 432–440 (2005).
Mortlock, W. Demand and Supply of Native Seed and Seedlings in Community Revegetation: A Survey (FloraBank, Commonwealth Natural Heritage Trust, 1999).
Radchuk, V. et al. Adaptive responses of animals to climate change are most likely insufficient. Nat. Commun. 10, 3109 (2019).
Jordan, R., Harrison, P. A. & Breed, M. The eco‐evolutionary risks of not changing seed provenancing practices in changing environments. Ecol. Lett. 27, e14348 (2024).
Aitken, S. N. & Whitlock, M. C. Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 44, 367–388 (2013).
Sgrò, C. M., Lowe, A. J. & Hoffmann, A. A. Building evolutionary resilience for conserving biodiversity under climate change. Evol. Appl. 4, 326–337 (2011).
Prober, S. M. et al. Climate-adjusted provenancing: a strategy for climate-resilient ecological restoration. Front. Ecol. Evol. 3, 65 (2015).
Twardek, W. M. et al. The application of assisted migration as a climate change adaptation tactic: an evidence map and synthesis. Biol. Conserv. 280, 109932 (2023).
Gann, G. D. et al. International principles and standards for the practice of ecological restoration. Restor. Ecol. 27, S1–S46 (2019).
Stanturf, J. A., Ivetić, V. & Kasten Dumroese, R. Framing recent advances in assisted migration of trees: a special issue. Ecol. Manag. 551, 121552 (2024).
Breed, M. F. et al. The potential of genomics for restoring ecosystems and biodiversity. Nat. Rev. Genet. 20, 615–628 (2019).
Chen, Z. et al. Applying genomics in assisted migration under climate change: Framework, empirical applications, and case studies. Evol. Appl. 15, 3–21 (2022).
Frankham, R. et al. A Practical Guide for Genetic Management of Fragmented Animal and Plant Populations (Oxford University Press, 2019).
Kardos, M. et al. The crucial role of genome-wide genetic variation in conservation. Proc. Natl Acad. Sci. USA 118, e2104642118 (2021).
Xuereb, A. et al. Incorporating putatively neutral and adaptive genomic data into marine conservation planning. Cons. Biol. 35, 909–920 (2020).
van Oppen, M. J. H. & Coleman, M. A. Advancing the protection of marine life through genomics. PLoS Biol. 20, e3001801 (2022).
Mohr, J. J., Harrison, P. A., Stanhope, J. & Breed, M. F. Is the genomics ‘cart’ before the restoration ecology ‘horse’? Insights from qualitative interviews and trends from the literature. Philos. Trans. R. Soc. B Biol. Sci. 377, 20210381 (2022).
Taft, H. R. et al. Research–management partnerships: an opportunity to integrate genetics in conservation actions. Conserv. Sci. Pract. 2, e218 (2020).
Taylor, H. R., Dussex, N. & van Heezik, Y. Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Glob. Ecol. Conserv. 10, 231–242 (2017).
Mijangos, J. L., Pacioni, C., Spencer, P. B. S. & Craig, M. D. Contribution of genetics to ecological restoration. Mol. Ecol. 24, 22–37 (2015).
Halpern, B. S. et al. Spatial and temporal changes in cumulative human impacts on the world’s ocean. Nat. Commun. 6, 1–7 (2015).
IPBES. Global Assessment Report on Biodiversity and Ecosystem Services (IPBES Secretariat, 2019).
Costanza, R. et al. Changes in the global value of ecosystem services. Glob. Environ. Change 26, 152–158 (2014).
Eger, A. M. et al. The value of ecosystem services in global marine kelp forests. Nat. Commun. 14, 1894 (2023).
Teh, L. S. L., Teh, L. C. L. & Sumaila, U. R. A global estimate of the number of coral reef fishers. PLoS ONE 8, e65397 (2013).
Broadhurst, L. et al. Restoration genetics—a consideration of lessons and opportunities. In Ecological Restoration (eds Florentine, S., Gibson-Roy, P., Dixon, K. W. & Broadhurst, L.) 473–519 (Springer International Publishing, Cham, 2023).
Wood, G. V. et al. Upscaling marine forest restoration: challenges, solutions and recommendations from the Green Gravel Action Group. Front. Mar. Sci. 11, 1–15 (2024).
Baums, I. B. et al. Considerations for maximizing the adaptive potential of restored coral populations in the western Atlantic. Ecol. Appl. 29, e01978 (2019).
St.Clair, J. B. et al. Seedlot Selection Tool and Climate‐Smart Restoration Tool: web‐based tools for sourcing seed adapted to future climates. Ecosphere 13, e4089 (2022).
Harrison, P. A., Vaillancourt, R. E., Harris, R. M. B. & Potts, B. M. Integrating climate change and habitat fragmentation to identify candidate seed sources for ecological restoration. Restor. Ecol. 25, 524–531 (2017).
Rossetto, M. et al. Restore and Renew: a genomics‐era framework for species provenance delimitation. Restor. Ecol. 27, 538–548 (2019).
Fitzpatrick, M. C. & Keller, S. R. Ecological genomics meets community‐level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 18, 1–16 (2015).
Selkoe, K. et al. A decade of seascape genetics: contributions to basic and applied marine connectivity. Mar. Ecol. Prog. Ser. 554, 1–19 (2016).
Bernatchez, L. et al. Genomics for monitoring and understanding species responses to global climate change. Nat. Rev. Genet 25, 165–183 (2024).
R Core Team. A Language and Environment for Statistical Computing (R Core Team, Vienna, Austria, 2023).
Thomas, L., Kennington, W. J., Evans, R. D., Kendrick, G. A. & Stat, M. Restricted gene flow and local adaptation highlight the vulnerability of high‐latitude reefs to rapid environmental change. Glob. Change Biol. 23, 2197–2205 (2017).
Matias, A. M. A. et al. Cryptic diversity and spatial genetic variation in the coral Acropora tenuis and its endosymbionts across the Great Barrier Reef. Evol. Appl. 16, 293–310 (2023).
Wood, G. et al. Using genomics to design and evaluate the performance of underwater forest restoration. J. Appl. Ecol. 57, 1988–1998 (2020).
Vergés, A. et al. Operation Crayweed: ecological and sociocultural aspects of restoring Sydney’s underwater forests. Ecol. Manag. Restor. 21, 74–85 (2020).
Wood, G. et al. Genomic vulnerability of a dominant seaweed points to future‐proofing pathways for Australia’s underwater forests. Glob. Change Biol. 27, 2200–2212 (2021).
Wernberg, T. et al. Climate-driven regime shift of a temperate marine ecosystem. Science 353, 169–172 (2016).
Young, M. A. et al. Mapping the impacts of multiple stressors on the decline in kelps along the coast of Victoria, Australia. Divers. Distrib. 29, 199–220 (2023).
Vergés, A. et al. Long-term empirical evidence of ocean warming leading to tropicalization of fish communities, increased herbivory, and loss of kelp. Proc. Natl Acad. Sci. USA 113, 13791–13796 (2016).
Vranken, S. et al. Genotype–environment mismatch of kelp forests under climate change. Mol. Ecol. 30, 3730–3746 (2021).
Veenhof, R. J. et al. Projecting kelp (Ecklonia radiata) gametophyte thermal adaptation and persistence under climate change. Ann. Bot. 133, 153–168 (2024).
Minne, A. J. P. Incorporating Genomic Data to Assess The Adaptive Potential of Australian Kelp Forests to Climate Change. Doctoral dissertation, The University of Western Australia (2024).
Bay, L. K. et al. Reef restoration and adaptation program: intervention technical summary. A report provided to the australian government by the reef restoration and adaptation program. (2019).
Commander, L. E. An update of the florabank guidelines: national guidelines for best practice native seed collection and use. Australas. Plant Conserv. J. Aust. Netw. Plant Conserv. 31, 21–23 (2022).
Frankham, R. et al. Predicting the probability of outbreeding depression: predicting outbreeding depression. Conserv. Biol. 25, 465–475 (2011).
Gellie, N. J., Breed, M. F., Thurgate, N., Kennedy, S. A. & Lowe, A. J. Local maladaptation in a foundation tree species: Implications for restoration. Biol. Conserv. 203, 226–232 (2016).
Grummer, J. A. et al. The immediate costs and long‐term benefits of assisted gene flow in large populations. Conserv. Biol. 36, e13911 (2022).
Layton, K. K. S. et al. Predicting the future of our oceans—evaluating genomic forecasting approaches in marine species. Glob. Change Biol. 30, e17236 (2024).
Séférian, R. et al. Tracking improvement in simulated marine biogeochemistry between CMIP5 and CMIP6. Curr. Clim. Change Rep. 6, 95–119 (2020).
Riahi, K. et al. The shared socioeconomic pathways and their energy, land use, and greenhouse gas emissions implications: an overview. Glob. Environ. Change 42, 153–168 (2017).
IPCC. in Climate Change 2022: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (eds Pörtner, H.-O. et al.) 3–33 (Cambridge University Press, 2022).
Coleman, M. A. et al. Restore or redefine: future trajectories for restoration. Front. Mar. Sci. 7, 237 (2020).
Tyberghein, L. et al. Bio‐ORACLE: a global environmental dataset for marine species distribution modelling. Glob. Ecol. Biogeogr. 21, 272–281 (2012).
Assis, J. et al. Bio‐ORACLE v2.0: extending marine data layers for bioclimatic modelling. Glob. Ecol. Biogeogr. 27, 277–284 (2018).
Grupstra, C. G. et al. Integrating cryptic diversity into coral evolution, symbiosis and conservation. Nat. Ecol. Evol. 8, 1–15 (2024).
Riginos, C. et al. Cryptic Species and Hybridisation in Corals: Challenges and Opportunities for Conservation and Restoration https://doi.org/10.32942/X2502X (2024).
Zimmerman, S. J., Aldridge, C. L. & Oyler-McCance, S. J. An empirical comparison of population genetic analyses using microsatellite and SNP data for a species of conservation concern. BMC Genom. 21, 382 (2020).
Lind, B. M. & Lotterhos, K. E. The accuracy of predicting maladaptation to new environments with genomic data. Mol. Ecol. Resourc. e14008. https://doi.org/10.1111/1755-0998.14008 (2024).
Mokany, K., Ware, C., Woolley, S. N. C., Ferrier, S. & Fitzpatrick, M. C. A working guide to harnessing generalized dissimilarity modelling for biodiversity analysis and conservation assessment. Glob. Ecol. Biogeogr. 31, 802–821 (2022).
Matz, M. V., Treml, E. A., Aglyamova, G. V. & Bay, L. K. Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral. PLoS Genet. 14, e1007220 (2018).
Matz, M. V., Treml, E. A. & Haller, B. C. Estimating the potential for coral adaptation to global warming across the Indo‐West Pacific. Glob. Change Biol. 26, 3473–3481 (2020).
Quigley, K. M., Bay, L. K. & Van Oppen, M. J. H. The active spread of adaptive variation for reef resilience. Ecol. Evol. 9, 11122–11135 (2019).
Wright, S. The genetical structure of populations. Ann. Eugen. 15, 323–354 (1949).
Schwalm, C. R., Glendon, S. & Duffy, P. B. RCP8.5 tracks cumulative CO2 emissions. Proc. Natl Acad. Sci. USA 117, 19656–19657 (2020).
Spalding, M. D. et al. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience 57, 573–583 (2007).
South, A., Michael S. & Massicotte, P. rnaturalearthdata: world vector map data from natural earth used in ‘rnaturalearth’. R package version 1.0.0.9000. https://github.com/ropensci/rnaturalearthdata (2024).
Fragkopoulou, E. et al. Global biodiversity patterns of marine forests of brown macroalgae. Glob. Ecol. Biogeogr. 31, 636–648 (2022).
Deysher, L. & Norton, T. A. Dispersal and colonization in Sargassum muticum (Yendo) Fensholt. J. Exp. Mar. Biol. Ecol. 56, 179–195 (1981).
Wu, K., Chen, C.-N. N. & Soong, K. Long distance dispersal potential of two seagrasses Thalassia hemprichii and Halophila ovalis. PLoS ONE 11, e0156585 (2016).
Coutant, O. et al. Environmental DNA reveals a mismatch between diversity facets of Amazonian fishes in response to contrasting geographical, environmental and anthropogenic effects. Glob. Change Biol. 29, 1741–1758 (2023).
Oksanen, J. et al. The vegan package. Community Ecol. Package 10, 719 (2007).
Branco, V. V., Correia, L. & Cardoso, P. SPECTRE: standardized global spatial data on terrestrial SPECies ThREats. Preprint at bioRxiv https://doi.org/10.1101/2023.11.26.568516 (2023).
Ferrier, S. Mapping spatial pattern in biodiversity for regional conservation planning: where to from here? Syst. Biol. 51, 331–363 (2002).
Ferrier, S. et al. Mapping more of terrestrial biodiversity for global conservation assessment. BioScience 54, 1101–1109 (2004).
Ferrier, S., Manion, G., Elith, J. & Richardson, K. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Divers. Distrib. 13, 252–264 (2007).
Fitzpatrick, M. C., Mokany, K., Manion, G., Nieto-Lugilde, D. & Ferrier, S. gdm: Generalized dissimilarity modeling. R package version 1.5. https://github.com/fitzLab-AL/gdm/ (2021).
Elith, J. & Leathwick, J. R. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40, 677–697 (2009).
Miller, J. R., Turner, M. G., Smithwick, E. A., Dent, C. L. & Stanley, E. H. Spatial extrapolation: the science of predicting ecological patterns and processes. BioScience 54, 310–320 (2004).
Araújo, M. B. & New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 22, 42–47 (2007).
Supple, M. A. et al. Landscape genomic prediction for restoration of a Eucalyptus foundation species under climate change. eLife 7, e31835 (2018).
Acknowledgements
G.W. was supported by an Australian Research Council (ARC) Linkage Grant (LP190100346) and an ARC Industry Fellowship (IE230100464). T.W., M.A.C. and K.F.D. were supported by ARC grants (DP190100058 and LP190100346 to T.W., M.A.C., K.F.D. and DP20010020119 to T.W. and M.A.C.). T.W., K.F.D., G.W. and M.A.C. were also funded by the Norwegian Research Council (GecoKelp Project no. 335371). MFB was supported by ARC grants (LP190100051, LP190100484, DP210101932). M.J.H.v.O. was supported by an ARC Laureate Fellowship (FL180100036). The authors would like to thank Quinten van der Leest and Eratos (https://www.eratos.com/) for application co-development, Karel Mokany for statistical advice, Luke Thomas and Cynthia Riginos for data provision, Mauricio van den Berg for converting the oceanographic connectivity calculation to an R function and Emma Camp for photos.
Author information
Authors and Affiliations
Contributions
G.W.: Conceptualisation, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, visualisation, writing—original draft preparation, writing—review and editing; K.G.: Formal analysis, investigation, methodology, software, writing—review and editing. MVDM: Data curation, methodology, software, writing—review and editing; M.F.B.: Methodology, writing—review and editing; J.E.: Data curation, writing—review and editing; C.G.: Methodology, writing—review and editing; A.M.: Data curation, visualisation, writing—review and editing; I.P.: Data curation, writing—review and editing; K.F.D.: Funding acquisition, resources, methodology, writing—review and editing; M.J.H.v.O.: Data curation, methodology, writing—review and editing; T.W.: Funding acquisition, resources, writing—review and editing; M.A.C.: Conceptualisation, funding acquisition, resources, writing—original draft preparation, writing—review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Anastazia T. Banaszak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Linn Hoffmann and Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wood, G.V., Griffin, K.J., van der Mheen, M. et al. Reef Adapt: A tool to inform climate-smart marine restoration and management decisions. Commun Biol 7, 1368 (2024). https://doi.org/10.1038/s42003-024-06970-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-06970-4
This article is cited by
-
Charting a course for incorporating restoration in marine planning
Discover Conservation (2025)
-
Coral reef thermal microclimates mapped from the International Space Station
Coral Reefs (2025)