Abstract
Excessive alarmins S100A8/A9 escalate the inflammation and even exacerbate immune-driven thrombosis and multi-organ damage. However, the regulatory mechanisms of S100A8/A9 expression in infectious diseases remain unclear. In this study, high-dimensional transcriptomic data analyses revealed a high proportion of CD14+FCN1hi macrophages within the pulmonary niche post-severe SARS-CoV-2 infection. By constructing the S100-coexpression gene list and supervised module scoring, we found that CD14+FCN1hi macrophages presented the highest scores of alarmin S100, and possibly served as the trigger and amplifier of inflammation in severe COVID-19. These CD14+FCN1hi cells lacked the positive regulatory activity of transcription factor PPARγ, and lost their differentiation ability towards mature macrophages. Ex vivo experiments further validated that the epithelial cells with high ORF-3a expression promoted the expression and secretion of S100A8/A9 through ANXA1/SAA1-FPR1 signaling. S100A8/A9 heterodimers, as well as the co-localization of S100A8/A9 with microtubules, were both diminished by the FPR1 inhibitor. Phospho-kinase protein array indicated that STAT3 promoted transcription, and PLC-γ and ERK1/2 pathways were involved in the hetero-dimerization and unconventional secretion of S100A8/A9. Our study highlights the pivotal role of FPR1 signaling in the excessive production of S100A8/A9 and provides a promising target for the prevention and control of severe COVID-19 and post-acute COVID-19 sequelae.
Similar content being viewed by others
Introduction
Since SARS-CoV-2 emerged in late 2019 in Wuhan, various subvariants have led to breakthrough infections and superinfections, causing multiple waves of epidemics worldwide. In May 2023, the WHO declared that COVID-19 may persist as a long-term endemic, and emphasized the need for continued vigilance against surges in cases and deaths caused by new SARS-CoV-2 variants1,2. In this context, high-risk individuals remain vulnerable to severe and potentially life-threatening COVID-19. Additionally, the increase in both infection times and disease severity raises the risk of long-COVID3,4. Therefore, exploring the mechanisms by which SARS-CoV-2 infection progresses to severe COVID-19 remains crucial and relevant.
The SARS-CoV-2 infection can trigger dysregulated host immune responses, leading to inflammation and disease progress. Severe COVID-19 impacts innate immune phenotypes and even reshapes the epigenetics of hematopoietic stem and progenitor cells (HSPCs), with effects persisting for more than one year5,6. The mononuclear phagocyte system (MPC) is not only an important component of innate immunity but also plays a crucial role in COVID-19 pathology. Previous studies indicated that monocyte-derived macrophage infiltration is a typical histological feature of the lungs in COVID-19 patients. In severe COVID-19, monocytes/macrophages were abnormally activated and led to a harmful loop of proinflammatory cytokine release and recruitment of inflammatory immune cells, thereby exacerbating damage to host tissues7,8,9,10.
Alarmins are essentially endogenous immunostimulatory DAMP molecules released in response to infection, as a result of degranulation or cell death11,12. The S100 family is recognized as a major group of alarmins, with prominent members S100A8 and S100A9. S100A8 and S100A9 typically exist as heterodimers (also known as calprotectin), and are widely reported to play proinflammatory roles in host infections, sterile inflammation, and autoimmune diseases13,14,15,16. Previous studies reported that the high level of S100 calprotectin correlated with disease severity, making them valuable biomarkers for distinguishing severe from mild/moderate cases17,18,19,20. Mechanistically, excessive S100A8/A9 promoted the emergency myelopoiesis and release of immature myeloid cells, further escalating the inflammatory response and exacerbating tissue damage, and also triggering immune-driven thrombosis by supporting the formation of procoagulant platelets and fibrin19,20. Although previous studies emphasized the crucial role of S100 alarmins in COVID-19 pathology, there is very limited research on their source and regulatory mechanisms of their expression and release.
In this study, high-dimensional transcriptomic data analyses revealed a high proportion of CD14+FCN1hi macrophages within the pulmonary niche post-severe SARS-CoV-2 infection. CD14+FCN1hi cluster presented the highest scores of calprotectin S100, held an ‘Off’ state of PPARg regulon, and lost the differentiation ability towards mature macrophages. Ex vivo experiments further validated that lung epithelial cells induced S100A8 and S100A9 expression through ANXA1/SAA1-FPR1 signaling. S100A8/A9 heterodimers, as well as the co-localization levels of S100A8/A9 with microtubes, were both diminished by the FPR1 inhibitor treatment. Phospho-kinase protein assay further suggested that STAT3 promoted the transcription of S100A8/A9, and PLC-γ and ERK1/2 pathways were involved in the hetero-dimerization and unconventional secretion of S100A8/A9. Our findings underscore the pivotal role of FPR1 signaling in inducing the excessive production and release of S100 proteins by immature macrophages, and provide a promising target for the prevention and control of severe COVID-19 and post-acute sequelae of COVID-19.
Results
scRNA-seq identifies a large proportion of monocyte-derived macrophages (MDMs) within the pulmonary niche of COVID-19 patients
To explore cell compartment changes in the lungs of COVID-19 patients, we reanalyzed the single-cell transcriptome sequencing data from bronchoalveolar lavage fluid (BALF) of healthy controls, and moderate and severe COVID-19 patients. After quality control, a total of 66,452 high-quality cells were used for downstream analyses. The single-cell transcriptomes of individuals were integrated using the Seurat package. Unsupervised dimension reduction and clustering analyses revealed 33 distinct clusters (Fig. 1a). Based on the expression of known markers, ten cell types were identified, including macrophage, neutrophil, epithelial cell, mDC, pDC, mast cell, T cell, NK cell, B cell, and plasma cell. These cell types exhibited distinct distribution patterns across three groups (Fig. 1b). The expression of select signatures used for annotation in each cluster is shown in Fig. 1c and Supplementary Fig. S1A.
a, b Uniform manifold approximation and projection (UMAP) of all quality-controlled cells. BALF cells were categorized into 33 clusters (a), and the distribution of annotated cell types across different groups is presented (b). c Dot plot of marker genes used for annotations within each cell type. d Stacked bar plot displaying the proportions of each cell type in healthy controls, moderate COVID-19 patients, and severe COVID-19 patients. e Bubble plot showing the extent of global transcriptional alternations in BALF cells after COVID-19 infection, evaluated by the total number of differentially expressed genes (DEGs) and cumulative fold change (FC) within each cell type shown in (b). Left panel, upregulated genes in COVID-19 patients; right panel, downregulated genes. The size of bubbles represents the number of cells for each type. f Expression of origin-associated features in macrophages. Left panel, the dimplot shows the two-dimensional distribution of macrophages from different groups (health and COVID-19); right panel, split-violin plots depict the expression of MDM/TRM features in macrophages from 14 clusters. Blue represents healthy people and red for COVID-19.
We quantified the proportions of different cell types in BALF of three groups (Fig. 1d). In COVID-19 moderate patients, there were 58.5% macrophages and 29.3% T lymphocytes. Compared to moderate patients, severe patients had much more macrophages (68.6%), along with decreased T cells (11.2%). We further calculated a ‘perturbation score’ for specific cell types based on the number of differentially expressed genes (DEGs) and the cumulative fold change (FC) to characterize the extent of transcriptional changes. The extent of gene expression was most pronounced in macrophages compared to other types of cells. Also, macrophages in COVID-19 showed higher gene expression compared to healthy donors (Fig. 1e). These data highlighted significant alterations in macrophages among COVID-19 pneumonia.
Alveolar pulmonary macrophages can be classified into tissue-resident macrophages (TRMs) and circulating MDMs according to their source21. Transcriptional analysis of origin-specific signatures showed that lung macrophages were mostly TRMs under homeostasis in healthy people, which could be maintained by local self-renewal. In contrast, a large proportion of lung macrophage compartments were filled by MDMs in moderate and severe COVID-19 patients (Fig. 1f and Supplementary Figs. S1B and S2), which was in line with the apparent differences in their transcriptional profiles.
The CD14+FCN1hiS100hi MDM subpopulation exhibits the greatest distribution divergence between moderate and severe patients
Macrophages from BALF of COVID-19 patients were subset and divided into 13 clusters (Fig. 2a), and the expression of characteristic genes for each cluster was shown in Fig. 2b and Supplementary Fig. S3. Clusters 1, 6, and 10; clusters 3, 4, 5, and 11; clusters 2 and 7; and clusters 8 and 9 respectively exhibit strong correlations, representing likewise transcriptional profiles (Supplementary Fig. S3A). Cluster 1-associated clusters expressed high levels of chemokines such as CCL2, and CCL7 and pathways enriched in inflammatory responses, cell mobility, NF-Kappa B signaling, and cytokine production, which resembled M1 macrophage. Cluster 2 showed a correlation with cluster 7 and upregulation of FCN1 and VCAN, suggesting monocyte-like characteristics. Clusters 3, 4, 5, 8, 9, and 11 exhibited high expression of HLA-DQA1, C1QC, and APOE. Among these, clusters 3 and 5 showed high expression of TREM2, and cluster 4 showed high FABP4, MARCO, and MRC1 levels. DEGs of C3-like clusters were related to antigen processing and presentation, mitochondrial and lysosomal organization, and peptide metabolic process, potentially resembling mature alveolar macrophages. Clusters 8 and 9 enriched pathways included oxidative phosphorylation, ribosomal protein complexes, and protein-RNA complex assembly, suggesting active translation processes (Supplementary Figs. S3B and S3C). Differences in macrophages between moderate patients and healthy people were discussed in Supplementary Fig. S4.
a Macrophages from COVID-19 patients were subset and re-clustered. Left panel, 13 unsupervised clusters; right panel, the distribution of macrophages among different groups. M, moderate; S, severe. b Expression of marker genes for the 13 clusters in (A), depicted as stacked violin plots. c Venn diagram illustrating the overlap among co-expressed genes of S100A8, S100A9, and S100A12. The intersection contained 28 genes. These S100 co-expressed genes were used to conduct the S100 scoring. d Dimplot of macrophages colored based on their S100 scores. e Violin plots of the S100 scores for 13 macrophage clusters from COVID-19. Dashed lines represent the threshold for assigning ‘S100 level’ identity, with cells above the upper threshold (0.5) classified as S100hi, below the lower threshold (0) as S100lo, and those in between as S100mid. f Projection of cells on dimplot after assigning them S100hi/mid/lo identities according to the threshold shown in (e). g Proportion of S100hi cells in macrophages from moderate or severe COVID-19 patients. Each dot represents a patient.
Co-expression genes for S100A8/A9/A12 were retrieved separately from the COXPRESdb database, and the intersection of these genes yielded 28 genes highly correlated with S100A8/A9/A12, including FPR1, FCN1, CSTA, etc. (Fig. 2c). Subsequently, we calculated the ‘S100 score’ for each cell based on the expression level of the gene module containing S100A8/A9/A12 and 28 co-expressing genes. Using the AddModuleScore function, each cell was scored, ranging from −0.56 to 1.69. These macrophages exhibited a stepwise distribution pattern in low-dimensional projections, and Cluster 2 (CD14+FCN1hi macrophage) showed the highest S100 scores among 13 clusters (Fig. 2d, e).
Based on their scores, these macrophages were assigned the identity of S100hi, S100mid, and S100lo (Fig. 2f). Further quantification of cell proportions revealed that the S100hi macrophages were enriched only in severe patients (14%), while they were nearly absent in moderate patients (only 0.3%) (Fig. 2g). These findings suggest that the CD14+FCN1hiS100hi cells are closely associated with the severity of COVID-19 and may play a critical role in disease pathogenesis.
The CD14+FCN1hiS100hi cells are characterized by low maturity, blocked differentiation, and downregulated phagocytosis and lipid metabolism capabilities
To better characterize the severe-specific CD14+FCN1hiS100hi cells, we performed ROC testing on the scRNA-seq data to evaluate DEGs that distinguish cluster 2 from all other clusters, and then conducted GO/KEGG enrichment analysis. Signatures were enriched in pathways related to monocyte chemotaxis, cell migration, and pro-inflammatory responses, accompanied by upregulation of secretion granules, cytoplasmic vesicles, and cellular skeleton entries (Fig. 3a). Conversely, pathways involved in phagocytosis, lysosomal acidification, antigen processing and presentation, and lipid metabolism were downregulated in the CD14+FCN1hiS100hi cluster (Fig. 3b). These downregulated genes and pathways are typically associated with the classical functions of differentiated tissue macrophages, which prompted us to consider whether the S100hi cluster represents a population of immature macrophages.
a, b Enrichment analysis for DEGs in CD14+FCN1hi cluster: a upregulated pathways and b downregulated pathways. c Differentiation trajectory of randomly sampled 10% of macrophages, evaluated by Monocle 2. From left to right, cells in each panel were colored by developmental states (state), pseudotime values, S100 levels, and different sample groups, respectively. d Expression trend of the top ten DEGs determining the order of cells along the pseudotime trajectory. e Heatmap of DEGs associated with pseudotime (clustered into six groups based on their expression patterns). Enrichment terms of Gene Ontology (GO) for each cluster of genes are listed on the right. f The relative activity of PPARg(+) and PPARg(−) regulon within macrophages of the four developmental states along the trajectory trunk identified in (c), assessed by SCENIC analysis.
Next, we performed a pseudotime analysis of macrophages to assess the positions of each subpopulation in the lineage inference. The position of cells along the pseudotime sequence was mapped onto a trajectory tree using the monocle DDRTree algorithm, defining seven differential states (Fig. 3c). Notably, the S100hi cluster had the lowest pseudotime values, and was placed at the beginning of the developmental trajectory (Fig. 3c). It is worth mentioning that, opposite to the S100hi cluster, macrophages from moderate COVID-19 patients were predominantly distributed at the other end, possibly indicating a mature state of terminally differentiated macrophages (Fig. 3c).
Figure 3d illustrates the top ten specific genes determining the order of cells along the pseudotime axis. As the pseudotime state progressed, S100A8, S100A9, and S100A12 were all downregulated, while HLA-DQA1 and HLA-DRB1 gradually increased. IL1RN and the chemokine CXCL11 also exhibited high levels of expression at the initial developmental stage. In contrast, genes encoding complement components C1Q and the scavenger receptor MARCO reached their highest expression at the end of development.
Using regression algorithms, we identified 4832 DEGs that changed with cell progression along the pseudotime. These genes were clustered based on their expression patterns into six distinct clusters (Fig. 3e). In our scRNA-seq data, different biological processes were enriched at the starting, middle, and ending stages of macrophage development. The initial stage retained monocyte characteristics and exhibited a tendency towards proinflammatory and degranulation activities. Features of the intermediate transition state were associated with ribosome biogenesis and synthesis and transport of active proteins, reflecting the transition from resting to cell differentiation, which was consistent with previous reports22. The terminal end, representing mature macrophages, was enriched with genes related to homeostasis, and emphasized the prominent roles of redox, mitochondria, and lipid metabolism (Fig. 3e).
Spatial-temporal heterogeneity of gene expression exists in response to transcription factor activity. SCENIC gene regulatory analysis revealed significant differences in regulon activity among macrophage subsets in different developmental states. PPAR gamma (PPARg) is a transcription factor crucial for alveolar macrophage maturation23. PPARg positively regulated genes formed regulon ‘PPARg(+)’ and its negatively correlated target genes composed regulon ‘PPARg(−)’. Remarkably, ‘PPARg(+)’ displayed much lower activity in the early stage of the developmental trajectory (state 1), while ‘PPARg(−)’ was highly active. In contrast, terminally differentiated state 6 exhibited the highest PPARg(+) activity and the lowest PPARg(−) activity (Fig. 3f). Gene expression initiated by PPARg is essential for cell differentiation and maturation23, thus, the dysfunction of this transcription factor in state 1 may indicate a stunted developmental progression of CD14+FCN1hiS100hi cells.
FPR1 signaling of macrophages dysregulated by epithelial cells is associated with the S100 levels
The microenvironment and cellular interactions can modulate the metabolism and differentiation of immune cells, and consequently influence their functions24. We used the R package CellChat to investigate whether the overexpression of S100 alarmins was associated with the communication between epithelial cells and macrophages. Macrophage clusters 2, 1, and 3, predominantly originated from severe cases, and presented high, medium, and low levels of S100 respectively (named S100hiM, S100midM, and S100loM). The epithelial cells from severe COVID-19 had four clusters which were assigned as E1–E4 (Fig. 4a). These epithelial clusters were then integrated with the three representative macrophage clusters to construct the CellChat object.
a Extracted epithelial cells from severe patients were assigned new identity labels (E1–E4) for usage in cell communication analysis. b Overview of epithelial-macrophage interactions. Cellchat analysis predicted the number of interactions (left) and interaction strength/weights (right) between epithelial cell clusters and macrophage subpopulations. c Hierarchy plot depicting the ANNEXIN and SAA signaling pathway for epithelial-macrophage communications. Line width represents communication strength. d Dot plot of the ligand-receptor pairs within the ANNEXIN and SAA signaling between different epithelial/macrophage clusters. Colors indicate interaction probability, while dot size represents statistical significance. e Scatter plot of the correlation between FPR1 expression levels and S100 scores of macrophages, evaluated by Pearson correlation coefficient.
The ‘human’ database served as the reference, and ‘secreted signaling’ was utilized for the cell communication analysis. We analyzed the communication between the epithelial cells and the macrophage clusters with high (S100hi), medium (S100mid), and low (S100lo) S100 expression levels. Results showed each of the epithelial clusters had the highest communication strength with the S100hi cluster, compared to that with the S100mid and S100lo macrophage clusters (Fig. 4b).
We identified six signaling pathways involved in the interactions between epithelial cells and macrophages (S100hiM, S100midM, and S100loM), including MIF, MK, COMPLEMENT, SEMA3, ANNEXIN, and SAA signaling. Epithelial cells exerted the most significant influence on the S100hi macrophage population via ANNEXIN and SAA signaling, followed by the S100mid cluster (Fig. 4c). The specific receptor-ligand interactions analyses indicated that communication probabilities of ANXA1-FPR1 and ANXA1-FPR2 were significantly upregulated between epithelial cells and the S100hi macrophage cluster, surpassing that communications with S100mid and S100lo clusters. Similarly, the communications mediated by SAA1-FPR1 and SAA1-FPR2 displayed a likewise trend. ANNEXIN and SAA1 both showed a higher chance of binding to the FPR1 receptor (Fig. 4d). Additionally, Pearson correlation analysis revealed a strong positive correlation (R = 0.52) between the FPR1 receptor expression and S100 scores of macrophages (Fig. 4e). This suggests that epithelial cells may induce the expression of S100 alarmins through FPR1-related signaling.
Ex vivo assays indicate viral protein-expressed epithelial cells produce ANXA1 and SAA1, and are involved in S100A8/A9 production of CD14+ monocytes
SARS-CoV-2 ORF3a (referred to as ORF3a hereafter), a major accessory protein, affects multiple physiological processes in host cells25. Human lung epithelial cells (A549) were transfected with SARS-CoV-2 ORF3a, and stable cell lines expressed ORF3a (referred to as A549-3a) were generated after puromycin selection. The stable expression of ORF3a in A549-3a cells was subsequently confirmed by western blotting (Fig. 5a). ANXA1 and SAA1 expression increased in A549-3a cells to varying degrees (Fig. 5b).
a The expression of SARS-CoV-2 ORF3a protein in A549-3a cells (A549 cells transfected with ORF3a) was detected by western blotting. b, c mRNA levels of certain genes were analyzed by real-time quantitative PCR, and the relative expression was normalized against GAPDH (n = 6). FPR1 ligands ANXA1 and SAA mRNAs in A549/A549-3a cells were detected (b) and mRNAs of S100A8, A100A9, S100A12, and other inflammatory cytokines like IL-6 within monocytes after co-culture with A549/A549-3a for 24 h were assessed (c). d After 24 h coculture, the supernatants in the lower chamber were collected and the amounts of secretory S100A8 were determined via ELISA (n = 4). Results are expressed as the mean ± SD. b, d Two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns stands for not significant.
To verify the interaction between epithelial cells and CD14+ monocytes, we designed a non-contact co-culture system using Transwell inserts. A549 or A549-3a cells were seeded in upper inserts in advance, and CD14+ monocytes from the peripheral blood of healthy donors were placed in the lower chambers. The system avoided direct cell contact while allowing the exchange of soluble substances. After 24 h of co-culture, CD14+ monocytes from the lower chamber were collected for subsequent experiments.
A higher proportion of FCN1hi monocytes in the A549-3a co-cultured group was revealed by flow cytometry, along with increased S100A8-high cells (Supplementary Fig. S5A). RT-qPCR results showed significant induction of S100A8, S100A9, S100A12, and other inflammatory molecules in CD14+ monocytes co-cultured with A549-3a cells as opposed to those with A549 cells (Fig. 5c). Additionally, RT-qPCR results confirmed the expression of the FPR1 receptor in circulating CD14+ monocytes. The levels of S100A8 proteins in the supernatant were examined by ELISA and significantly increased in the A549-3a co-culture group, mostly derived from monocytes (Fig. 5d). We also conducted stimulation of recombinant protein ANXA1 or SAA1, and both elevated the S100 level in monocytes (Supplementary Fig. S5B). This suggests that, under COVID-19 pathological conditions, lung epithelial cells may induce the expression and secretion of alarmin S100 in CD14+ monocytes through ANXA1/SAA-FPR1 signaling.
FPR1 inhibition facilitates the differentiation of circulating CD14+ monocytes and decreases S100A8/A9/A12 production and unconventional secretion
An FPR1 inhibitor, HCH6-1, was applied in the co-culture system to verify whether FPR1 signaling played a pivotal role in the crosstalk between COVID-19 epithelial cells and CD14+ monocytes, and regulated the S100 alarmins expression and secretion of CD14+ monocytes. Interestingly, we observed significant differences in monocyte differentiation progress between groups with or without HCH6-1 treatment. Morphologically, the CD14+ monocytes without the treatment of HCH6-1 maintained a round or elliptical shape, resembling their original form in circulating blood. In contrast, CD14+ monocytes with HCH6-1 treatment exhibited extending pseudopods or filopodia, presenting the characteristics when cells respond to tissue chemotaxis and differentiate into macrophages (Fig. 6a and Supplementary Fig. S6A, S6B). The PPARg level also increased after HCH6-1 treatment (Supplementary Fig. S6C).
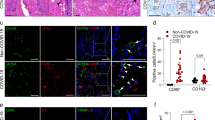
CD14+ Monocytes were co-cultured with A549-3a cells with or without FPR1 inhibitor HCH6-1 (10 μM or 20 μM) for 36 h (morphological observations) or 24 h (RT-qPCR, ELISA and confocal microscopy). a Cell morphology was observed using differential interference contrast (DIC) imaging. Scale bar = 10 μm. bThe mRNA expression levels of S100A8, S100A9, and S100A12 in monocytes were determined by RT-qPCR and normalized to GAPDH (n = 6). c S100A8 contents in the supernatants were measured by ELISA (n = 4). d, e Monocytes were examined by confocal microscopy for the localization and expression of S100A9 (red) and S100A8 (green) (d) or S100A9 (red) and β-tubulin (green) (e). Nuclei were visualized using DAPI counterstain (blue). Scale bar = 10 μm. Pearson’s correlation was calculated for merged images as colocalization indicators. b, c Results are expressed as the mean ± SD, and statistical analysis is performed using a two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns stands for not significant.
After HCH6-1 treatment, the transcription levels of S100A8, S100A9, and S100A12 in monocytes decreased (Fig. 6b). S100A8 and S100A9 exist predominantly as heterodimers in the extracellular space, known as calprotectin, and have been identified as key players in inflammation signaling and immune regulation13. While co-culture with HCH6-1 treatment, the concentration of extracellular S100A8 was reduced in the supernatant, and immunofluorescence and laser confocal imaging demonstrated the reduction of fluorescence intensity of S100A8 and S100A9 in CD14+ monocytes, as well as a decrease in their intracellular co-localization (Fig. 6c, d and Supplementary Fig. S6D).
Furthermore, laser confocal imaging also showed that CD14+ monocytes co-cultured with A549-3a exhibited clearer fibers and a network-like distribution of microtubules. FPR1 blockade attenuated these features, accompanied by a reduction of S100 co-localization with microtubules (Fig. 6e and Supplementary Fig. S6E). These results suggest that FPR1 blockade not only inhibits the transcription of S100 alarmin, but is also associated with the formation of the S100A8/A9 heterodimers, and their unconventional secretion in CD14+ monocytes.
Phospho-kinase protein arrays show that STAT3, PLC-γ and ERK1/2 pathways promote the transcription, hetero-dimerization and unconventional secretion of S100A8/A9
To determine by which FPR1 induced S100 upregulation in CD14+ monocytes, we employed a human phospho-kinase antibody array to assess the phosphorylation levels of kinases and their substrate. Following co-culturing with or without HCH6-1 treatment, CD14+ monocytes from the lower chambers were harvested, and protein extracts were subsequently detected and quantified (Fig. 7a and Supplementary Fig. S7A).
After co-culture with A549-3a in the presence or absence of the FPR1 inhibitor HCH6-1 (20 μM), CD14+ monocytes were harvested, and protein extracts were subsequently detected and quantified. a Immunoassay image of the protein chip. Table listing the types and amounts of target proteins spotted on the membrane. b Heatmap presenting quantitative results of phosphorylation levels of kinase and transcription factors. The color bar represents the relative expression levels. c Bar diagram of the pathways involved in FPR1 signaling, determined by enrichment analysis of differentially expressed proteins. The horizontal axis represents-log10(p).
In comparison to co-culture without HCH6-1, we observed decreases in phosphorylation levels of transcription factors, notably STAT3, which has been previously reported to enhance the transcription of S100A8 and S100A926. Functional annotation and enrichment analyses also indicated that altered kinases and substrates were associated with STAT signal transduction (Fig. 7b, c and Supplementary Fig. S7B).
GPCR-mediated ligand-independent transactivation of RTKs is an important factor in the diversity of intracellular signal regulation. In line with that, co-culture with HCH6-1 downregulated Src and receptor tyrosine kinase EGFR activity. Downstream, phosphor-PLC-γ1 was also reduced (Fig. 7b, c and Supplementary Fig. S7B), possibly leading to PKC suppression that decreased intracellular S100A8/A9 dimers27,28.
Functional annotation and enrichment analyses also showed that altered kinases and substrates were involved in MAPK activity regulation, intracellular protein transport, and regulation of cytoskeletal organization. Moreover, ERK1/2, which modulates microtubule function29 and is associated with calprotectin interaction with microtubules, was downregulated in co-culture with HCH6-1 (Fig. 7b, c and Supplementary Fig. S7B).
Based on the above results, FPR1 was involved in multiple downstream signal transduction pathways to regulate transcription, intracellular active heterodimers, and microtubule-dependent release of S100A8 and S100A9. Thus, FPR1 signaling could be developed as a promising target for the treatment of severe COVID-19 and the prevention of post-acute COVID-19 sequelae.
Discussion
Immunopathogenesis studies have demonstrated that dysregulated responses of monocytes/macrophages in blood and lungs exacerbated inflammation and disease progression in severe COVID-19 patients8,30,31. Here we identified a distinct cluster of MDMs (CD14+FCN1hiS100hi) retained low maturity, which were aberrantly regulated by epithelial cells through ANXA1/SAA1-FPR1 signaling in the lungs of severe COVID-19, exacerbating inflammation and pulmonary pathology.
Alveolar macrophages are classified into tissue-resident macrophages (TRMs) derived from the fetal liver (yolk sac as well in early gestational stages) and circulating MDMs according to their source32,33. TRMs have distinct developmental trajectories and physiological functions to the circulating MDMs21,34. In our study, the analysis of origin-specific signatures shows that macrophages in BALF are primarily TRMs under homeostasis in healthy people, which could be maintained by local self-renewal. However, in moderate and severe COVID-19 patients, a large proportion of macrophage compartments in BALF are filled by CD14+FCN1hi MDMs.
Liao’s study classified macrophages into 4 groups based on FCN1, SPP1, and FABP4 expression patterns, and the ‘Group1’ (FCN1hi macrophages) exhibited upregulated expression of S100 genes compared to other groups35. Through unsupervised clustering, S100 co-expression gene scoring, and developmental trajectory analyses, we established a multi-layer map of COVID-19 pulmonary macrophages and especially revealed the aberrant CD14+FCN1hi cluster2 MDMs from severe COVID-19 patients presented the highest S100 scores. We provided a more refined and specific definition for S100hi clusters, excluding healthy cells under homeostasis. This further narrowed down the range of “harmful” cells which may act as an amplifier of the local or even systemic inflammation cascade. In mouse models, MDMs were rapidly recruited and replaced the resident alveolar macrophage population, and promoted acute inflammation, lung injury, and even pulmonary fibrosis36,37,38. BMO-AMs (Bone marrow monocyte-derived alveolar macrophages) produced more IFN-γ and IL-6, and led to high inflammatory responses and severe pathological damages during recurrent IAV infection in different-age mouse models39. Our findings, in line with these reports, further demonstrate the roles of aberrant MDM in inflammatory responses and lung damage under infection conditions.
CD14+FCN1hiS100hi cells were at the beginning of the developmental trajectory inferred from pseudo-time analyses, and presented inactivity of PPARγ, a transcription factor indispensable to alveolar macrophages23. CD14+FCN1hiS100hi cells lacked the robust phagocytic and lipid metabolism capabilities of mature macrophages, with MHC-II expression and antigen-presenting functions significantly diminished. These aberrant MDM cells were dysfunctional and incomplete in the differentiation process in the lungs, thus unprofitable to the pathogen clearance and effective activation of adaptive immunity, resulting in a prolonged duration of virus infection and inflammation40,41.
Tissue-specific niches involve extensive signals and crosstalk, regulating the dynamic immune responses42,43. FPR1 on CD14+FCN1hi MDMs served as the receptor for both ANXA1 and SAA1, and the communication intensity of ANXA1/SAA1-FPR1 between pulmonary epithelial cells and the CD14+FCN1hiS100hi cells was significantly enhanced in severe COVID-19. FPR1 encodes formyl peptide receptor, a member of the G protein-coupled receptor superfamily44. Interestingly, FPR1 is co-expressed with S100A8, S100A9, and S100A12, further linking this receptor to the production of S100 alarmins as its potential pathogenic mechanism. FPR1 inhibitor (HCH6-1) promoted differentiated characteristics of MDMs, and reduced S100A8/A9/A12 production.
The roles of S100A8/A9 and S100A12 in immune cascades and inflammation amplification have been extensively described in several independent studies13,14,15,16. However, there is limited research on the regulatory mechanisms of their expression and release. The promoters of S100A8 and S100A9 contain STAT3-binding sites26, and the significant down-regulation of p-STAT3 was detected by phosphokinase protein chip after FPR1 inhibitor treatment, suggesting that FPR1-mediated intracellular signaling may regulate the transcriptional activation of multiple S100 genes concurrently.
S100A8 and S100A9 present as a heterodimer, the main active form of pro-inflammatory calprotectin. Existing evidence indicates that S100A8 and S100A9 lack of signal peptides and might be secreted through unconventional pathways28. Laser confocal imaging indicated the decreases of S100A8/A9 heterodimers, and the subcellular co-localization of S100A8/A9 and microtubules after FPR1 inhibitor treatment. Phospho-kinase protein chip arrays subsequently suggest that PLC-γ and ERK1/2 pathways were involved in the hetero-dimerization and unconventional secretion of calprotectin S100A8/A9. The competitive FPR1 inhibitor, HCH6-1, has also demonstrated a protective effect against acute lung injury and liver cancer progression by inhibiting myeloid cell activation and migration45,46. Our research provides a supplement to the underlying mechanisms of FPR1 signaling in pulmonary infection and other inflammatory diseases.
In conclusion, our findings enhance the understanding of the molecular determinants that regulate the inflammatory response and organ damage in severe COVID-19. Immature CD14+FCN1hi S100hi MDMs were specifically enriched in the lungs of severe COVID-19 patients, which did not undergo the normal differentiation process towards mature alveolar macrophages. Ex vivo experiments demonstrated that ORF3a-expressing epithelial cells regulated S100A8/A9 production and secretion in CD14+FCN1hi cells, which can be effectively reduced by FPR1 inhibitors.
Our findings suggest that FPR1 plays a crucial role in inflammatory responses and lung injury in severe COVID-19. Severe immunopathology during infection is linked to pulmonary fibrosis and other long-term symptoms3,47, thus timely blockade of FPR1 and inhibition of S100 expression could aid in preventing severe COVID-19 and post-acute sequelae of COVID-19 (PASC). The limitation of this study is that ORF3a-expressing cells are not equivalent to infected lung epithelial cells under COVID-19 pathological conditions, and future studies on genetic evidence could fully validate FPR1 signaling and investigate the clinical utility of FPR1 inhibitors.
Methods
Cell lines and plasmids
HEK-293T and A549 cell lines were obtained from the American Type Culture Collection (ATCC, USA). Plasmids (pLVX-EF1α-SARS-CoV-2-orf3a-2xStrep-IRES-Puro) were generated by Nevan Krogan’s lab and supplied by Addgene, USA. Lentivirals were packaged using the GAG packaging plasmid and VSV-G envelope plasmid from Addgene, USA.
Data source
Single-cell RNA sequencing data of human BALF from 4 healthy individuals, 3 moderate and 6 severe/critical COVID-19 patients were collected from the GEO database, GSE145926, and GSM3660650 from GSE128033 datasets35,48.
scRNA-seq data processing and analysis
scRNA-seq data was processed using Seurat (4.3.0) package. Quality control was conducted on the single-cell RNA sequencing data to filter the cells with gene counts between 200 and 6000, UMI counts under 1000, and mitochondrial RNA percentages less than 10%. Data were normalized and scaled using the NormalizeData and ScaleData functions, and 2000 highly variable genes were identified with the FindVariableFeatures function.
Principal component analysis (PCA) was conducted based on those high-variable genes. Based on the top 50 principal components, cell clustering is conducted by the FindNeighbors and FindClusters functions with a resolution of 1.2. Cell clusters were visualized using the UMAP dimensional reduction method. FindAllMarkers was used to identify marker genes for each cell cluster. Cell type annotation was then performed based on the expression of signature genes, referred to the published lung scRNA sequencing studies49. The FindMarkers function in Seurat (4.3.0) is used to identify DEGs of different groups among various cell types. In this study, differential gene expression was determined based on the criteria of P.adj < 0. 01. In addition, the SCP package (0.5.6) was used for single-cell analysis visualization.
Barcodes of macrophages were obtained to subset the corresponding cells from the original data. The extracted cells are then re-integrated after being split according to the sample source. The macrophage population is normalized and scaled. After that, linear and non-linear dimensional reduction and clustering were performed based on the top 30 principal components with a resolution of 0.5.
S100 score
The co-expressed genes of S100A8, S100A9, and S100A12 were acquired in the COXPRESdb database (https://coxpresdb.jp/), respectively. The intersection of genes co-expressed with all three S100 molecules was determined. Based on the transcription levels of S100 molecules and their co-expressed genes, the AddModuleScore function is used to score the macrophages from COVID-19 patients.
Gene enrichment analysis
Differential genes for cluster 2 are identified using the Seurat FindMarkers function. Marker genes for cluster 2 were used to perform GO and KEGG enrichment analysis by ClusterProfiler (4.2.2). GO terms and KEGG pathways with significant enrichment are selected based on adj. p-values and enrichment scores, and the results are visualized in bar plots.
Pseudotime analysis
The pseudotime algorithm Monocle2 (2.22.0) is utilized for reconstructing cell differentiation trajectories. 10% of the macrophages from COVID-19 patients were randomly sampled to construct the monocle object. Highly variable genes are selected for cell clustering, and the DDRTree method is employed for trajectory inference. The differentialGeneTest function is used to identify genes associated with the pseudotime axis, and a clustered heatmap is plotted based on gene expression patterns.
Regulatory network inference
SCENIC (1.3.1) was used to infer the gene regulatory networks based on co-expression and DNA motif analysis. TFs were identified using GENIE3 and compiled into regulons. Subsequently, RcisTarget was employed for cis-regulatory motif analysis on these regulons, and performed gene-motif ranking of two categories: around transcription start sites (TSS) within 10 kb and upstream 500 bp. The activity of regulons in each cell was scored using AUCell.
Cell-cell communications
CellChat (1.4.0) was utilized to analyze intercellular communication between epithelial and macrophage clusters. The secreted signaling section in the CellChatDB.human database was selected for reference to uncover the communication patterns of source cells and target cells. The netVisual_circle and netVisual_bubble functions were employed for visualizing the strength of signaling pathways, as well as important ligand-receptor interactions.
Stable cell line construction
HEK-293T cells were used for lentivirus packaging and amplification. The 293 T cells were seeded in a 6-well plate at 60-70% confluency. Plasmids were pre-mixed with PEI (MW 25000 Da, Sigma, USA) and transfected into 293 T cells, followed by a 48-h incubation. Lentiviral particles were collected and filtered through a 0.22 μm filter. Polybrene (Sigma, USA) was added to the filtered supernatant (8 μg/mL), then A549 cells were infected for 24 h. A549-3a cells were selected using puromycin and the stable expression of ORF3a was verified by immunoblotting.
RNA extraction and RT-qPCR
Total cellular RNA was extracted using the RNAsimple Kit (Tiangen, CHN) following the manufacturer’s instructions. The purity and concentration of the extracted RNA were assessed, and cDNA was synthesized by the ABS Script II RT Mix (ABclonal, CHN). Real-time quantitative PCR was performed using the 2× Universal SYBR Green Fast qPCR Mix (ABclonal, CHN), and the fluorescence signals were collected by the CFX96 Touch PCR system (Bio-Rad, USA). Primer sets used in experiments are listed in Supplementary Table 1. The relative expression levels of genes in each sample were calculated using the 2−ΔΔCt method.
Western blotting and ELISA
For detection of stable expression of viral protein ORF3a, A549, and A549-3a cells were harvested and lysed using RIPA lysis buffer on ice. The supernatant was collected after centrifugation, and the total protein concentration was determined by BCA assay (Biosharp, CHN). The protein samples were separated by SDS-PAGE and transferred to a PVDF membrane. Membranes were blocked with 5% non-fat milk at room temperature for 1 h and then incubated overnight at 4 °C with specific primary antibodies against GAPDH (1:10000, 10494-1-AP, Proteintech), Strep II-Tag (1:2500, AE066, ABclonal). After washing, the membrane was incubated with an HRP-conjugated goat anti-rabbit IgG secondary antibody (1:5000, AS014, ABclonal) at room temperature for 1 h. The chemiluminescence signal was developed using an ECL reagent and captured using the ChemiDoc XRS+ imaging system (Bio-rad, USA).
For the validation of several phosphorylated kinases, protein samples were extracted from different groups of monocytes (no treatment/co-cultured with A549-3a/co-cultured with A549-3a and HCH6-1 treatment). The methods for detecting are similar as above, with the antibodies used being: PPARg (1:2000, 16643-1-AP, Proteintech), p-PLCG1 (1:500, 29566-1-AP, Proteintech), p-ERK1/2 (1:2000, 28733-1-AP, Proteintech), and p-STAT3 (1:2000, D3A7, 9145S, CST).
The supernatant from monocytes was collected after being co-cultured for 24 h and was used to measure the concentration of secreted S100A8 using the S100A8 ELISA kit (ABclonal, CHN) according to the manufacturer’s instructions.
Isolation of CD14+ monocytes and co-culture
PBMCs from healthy volunteers were isolated using density gradient centrifugation. Briefly, PBS buffer diluted whole blood samples were carefully layered onto PBMC separation solution (TBDscience, CHN) in centrifuge tubes, and centrifuged at room temperature for 30–40 min. The white cloudy layer containing PBMCs was collected, washed, and counted. Anti-Human CD14 Magnetic Particles (BD Biosciences, USA) were added to the cells at a ratio of 1 × 107 cells/50 μL. CD14+ monocytes were obtained by placing the magnetic bead-cell suspension in a magnetic field following the manufacturer’s instructions.
For co-culture experiments, A549/A549-3a epithelial cells were seeded in the upper inserts (6.5 mm Transwell® with 0.4 µm pore polyester membrane, #3470, Corning, USA) 24 h in advance. CD14+ monocytes obtained from blood were placed in the lower chambers. Ensure that the culture medium contacts the insert bottom. The system was then incubated at 37 °C, 5% CO2 for 24–36 h for different experiments.
Differential interference contrast (DIC) imaging
After co-culture, the insert was removed and the monocytes in the lower chamber were observed under an inverted DIC microscope. The polarizer, analyzer, aperture, and condenser height were adjusted to appropriate positions to obtain gray-scale images with embossed appearance and moderate contrast. Representative pictures were taken on the Olympus IX73 inverted microscope (Olympus, JPN) at the 40× objective.
Immunofluorescence and confocal imaging
Round climbing slides were placed in advance in the lower chambers, and a centrifuge was performed after coculture to make the monocytes adhere to the slides. Cells were fixed and permeabilized by absolute ethanol and blocked for 1 h with TBST containing 3% BSA. For the co-localization detection of S100A8 and S100A9, cells were incubated with S100A8 (66853-1-Ig, Proteintech) and S100A9 primary antibodies (26992-1-AP, Proteintech) overnight at 4 °C. The next day, cells were washed and incubated with FITC- and TRITC-conjugated secondary antibodies corresponding to the species of the primary antibodies, respectively. For the co-localization detection of S100A9 and microtubules, cells were incubated with S100A9 primary antibody and CoraLite®488 labeled β-tubulin antibody (CL488-66240, Proteintech) overnight at 4 °C. Nuclei were counterstained with DAPI. Apply FV3000 confocal laser scanning microscope (Olympus, JPN) for scanning and imaging in the 405 nm, 488 nm, and 561 nm excitation wavelengths under a 100× silicon oil objective.
Images procession, quantitative estimation of immunofluorescence, and co-localization levels calculation were using Fiji ImageJ (https://fiji.sc/). Relative fluorescence intensity was calculated using the mean fluorescence intensity and normalized to the group of monocytes co-cultured with A549-3a (no HCH6-1). For quantitative analysis of fluorescence co-localization, Pearson’s correlation coefficients were calculated using the Colocalization Finder plugin after splitting the red and green channels of the fluorescence images. Values closer to 1 indicate stronger co-localization between the two. Three or more nonoverlapping images were analyzed per group.
Human phospho-kinase array
The Proteome Profiler Human Phospho-Kinase Array Kit (R&D, USA) was used to examine the expression levels of phosphorylated kinases in monocytes after co-culture, according to the manufacturer’s instructions. In brief, monocytes treated with or without inhibitors were collected and protein was extracted on ice. Equal amounts of protein from both groups were incubated overnight with the blocked array membranes. Captured proteins were detected with a biotinylated detection antibody cocktail and then visualized using chemiluminescent detection reagents with the UVP Chemstudio touch imaging system (Analytik Jena, DEU).
Quantitative analysis of scanned images was performed using ImageJ software. Firstly, the imported images were converted to 8-bit grayscale and inverted. Choose the measurements: check “Centroid” “Integrated Density” and “Min and Max Gray Values”. The Circle Tool (an ImageJ plugin) was used, set appropriate radius to ensure complete coverage of individual immunoblot dot. This radius remained constant. After setup, each spot on the image was selected sequentially for integrated pixel density measurement. The average of the two groups was calculated, followed by determining the fold change relative to the mean value for each spot. Heatmap was generated using TBtools.
Flow cytometry
Monocytes were co-cultured with A549 or A549-3a epithelial cells for 24 h, then were harvested for flow cytometry. After fixation and permeabilization, monocytes were incubated with S100A8 antibody (PA5-86063, Invitrogen) and FCN1 antibody (ABS 036-05-02, Invitrogen) for 60 min, and followed by incubation with respective fluorescence-conjugated secondary antibodies in the dark. Finally, all samples were examined with BD FACSCelesta™ (BD Biosciences, USA) and data was analyzed by FlowJo (v10, https://www.flowjo.com/).
Statistics and reproducibility
R (4.1.3) was used for scRNA-seq data analyses and graphics processing. Ex vivo experiments were independently repeated at least three times. Statistical analyses between two groups were performed using unpaired two-tailed Student’s t-test, and multiple groups were statistically compared with one-way ANOVA in GraphPad Prism 10. The following criteria were used to determine the statistical significance: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, and ns for no significant difference.
Institutional Review Board statement
The investigations involving human participants have complied with all relevant ethical regulations and obtained informed consent from all participants. This study was reviewed and approved by the Ethics Committee of Wuhan University of Science and Technology (#216).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data used in this study is publicly available in the GEO database under the accession numbers GSE145926 and GSE128033. The source data for graphs in figures are provided in the Supplementary Data file, and uncropped blots are shown in Supplementary Fig. S8. All relative data are available from the corresponding author upon reasonable request.
References
World Health Organization. WHO COVID-19 Dashboard. https://covid19.who.int (2023).
World Health Organization. WHO Director-General’s Opening Remarks at The Media Briefing, 5 May 2023. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing---5-may-2023 (2023).
Davis, H. E., McCorkell, L., Vogel, J. M. & Topol, E. J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21, 133–146 (2023).
Su, Y. et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 185, 881–895.e820 (2022).
Wong, L. R. & Perlman, S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses—Are we our own worst enemy? Nat. Rev. Immunol. 22, 47–56 (2022).
Cheong, J. G. et al. Epigenetic memory of coronavirus infection in innate immune cells and their progenitors. Cell 186, 3882–3902.e3824 (2023).
Rendeiro, A. F. et al. The spatial landscape of lung pathology during COVID-19 progression. Nature 593, 564–569 (2021).
Merad, M. & Martin, J. C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 20, 355–362 (2020).
Sefik, E. et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 606, 585–593 (2022).
Wendisch, D. et al. SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell 184, 6243–6261.e6227 (2021).
Yang, D., Han, Z. & Oppenheim, J. J. Alarmins and immunity. Immunol. Rev. 280, 41–56 (2017).
Danieli, M. G. et al. Alarmins in autoimmune diseases. Autoimmun. Rev. 21, 103142 (2022).
Jukic, A., Bakiri, L., Wagner, E. F., Tilg, H. & Adolph, T. E. Calprotectin: from biomarker to biological function. Gut 70, 1978–1988 (2021).
Scott, N. R. et al. S100A8/A9 regulates CD11b expression and neutrophil recruitment during chronic tuberculosis. J. Clin. Invest. 130, 3098–3112 (2020).
Vogl, T. et al. Autoinhibitory regulation of S100A8/S100A9 alarmin activity locally restricts sterile inflammation. J. Clin. Invest. 128, 1852–1866 (2018).
Manfredi, M. et al. Circulating calprotectin (cCLP) in autoimmune diseases. Autoimmun. Rev. 22, 103295 (2023).
Silvin, A. et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 182, 1401–1418.e1418 (2020).
Chapuis, N. et al. Dynamics of circulating calprotectin accurately predict the outcome of moderate COVID-19 patients. EBioMedicine 80, 104077 (2022).
Guo, Q. et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe 29, 222–235.e224 (2021).
Colicchia, M. et al. S100A8/A9 drives the formation of procoagulant platelets through GPIbα. Blood 140, 2626–2643 (2022).
Liu, Z. et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525.e1519 (2019).
Evren, E. et al. Distinct developmental pathways from blood monocytes generate human lung macrophage diversity. Immunity 54, 259–275.e257 (2021).
Schneider, C. et al. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15, 1026–1037 (2014).
Rivera, A., Siracusa, M. C., Yap, G. S. & Gause, W. C. Innate cell communication kick-starts pathogen-specific immunity. Nat. Immunol. 17, 356–363 (2016).
Bianchi, M., Borsetti, A., Ciccozzi, M. & Pascarella, S. SARS-Cov-2 ORF3a: mutability and function. Int J. Biol. Macromol. 170, 820–826 (2021).
Cheng, P. et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 205, 2235–2249 (2008).
Cattaneo, F. et al. Cell-surface receptors transactivation mediated by g protein-coupled receptors. Int J. Mol. Sci. 15, 19700–19728 (2014).
Rammes, A. et al. Myeloid-related protein (MRP) 8 and MRP14, calcium-binding proteins of the S100 family, are secreted by activated monocytes via a novel, tubulin-dependent pathway. J. Biol. Chem. 272, 9496–9502 (1997).
Laks, D. R. et al. A molecular cascade modulates MAP1B and confers resistance to mTOR inhibition in human glioblastoma. Neuro Oncol. 20, 764–775 (2018).
He, B. et al. The metabolic changes and immune profiles in patients with COVID-19. Front Immunol. 11, 2075 (2020).
Zhou, J. et al. The metabolic and immunological characteristics of pregnant women with COVID-19 and their neonates. Eur. J. Clin. Microbiol. Infect. Dis. 40, 565–574 (2021).
Hoeffel, G. & Ginhoux, F. Ontogeny of tissue-resident macrophages. Front Immunol. 6, 486 (2015).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Lazarov, T., Juarez-Carreño, S., Cox, N. & Geissmann, F. Physiology and diseases of tissue-resident macrophages. Nature 618, 698–707 (2023).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Maus, U. A. et al. Resident alveolar macrophages are replaced by recruited monocytes in response to endotoxin-induced lung inflammation. Am. J. Respir. Cell Mol. Biol. 35, 227–235 (2006).
Misharin, A. V. et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med. 214, 2387–2404 (2017).
Pervizaj-Oruqaj, L. et al. Alveolar macrophage-expressed Plet1 is a driver of lung epithelial repair after viral pneumonia. Nat. Commun. 15, 87 (2024).
Li, F. et al. Monocyte-derived alveolar macrophages autonomously determine severe outcome of respiratory viral infection. Sci. Immunol. 7, eabj5761 (2022).
Xiao, F. et al. Fibrinogen-like protein 2 deficiency inhibits virus-induced fulminant hepatitis through abrogating inflammatory macrophage activation. World J. Gastroenterol. 28, 479–496 (2022).
Schneider, C. et al. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 10, e1004053 (2014).
Mothes, R. et al. Distinct tissue niches direct lung immunopathology via CCL18 and CCL21 in severe COVID-19. Nat. Commun. 14, 791 (2023).
Sutherland, T. E., Dyer, D. P. & Allen, J. E. The extracellular matrix and the immune system: a mutually dependent relationship. Science 379, eabp8964 (2023).
Chen, K. et al. Regulation of inflammation by members of the formyl-peptide receptor family. J. Autoimmun. 85, 64–77 (2017).
Yang, S. C. et al. Dipeptide HCH6-1 inhibits neutrophil activation and protects against acute lung injury by blocking FPR1. Free Radic. Biol. Med. 106, 254–269 (2017).
Wu, L. et al. An invasive zone in human liver cancer identified by Stereo-seq promotes hepatocyte-tumor cell crosstalk, local immunosuppression and tumor progression. Cell Res. 33, 585–603 (2023).
Cai, J. et al. A one-year follow-up study of systematic impact of long COVID symptoms among patients post SARS-CoV-2 omicron variants infection in Shanghai, China. Emerg. Microbes Infect. 12, 2220578 (2023).
Morse, C. et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. https://doi.org/10.1183/13993003.02441-2018 (2019).
Sikkema, L. et al. An integrated cell atlas of the lung in health and disease. Nat. Med. 29, 1563–1577 (2023).
Acknowledgements
We would like to thank Nevan Krogan’s lab and Addgene for their generosity in providing the plasmids used in this study. This study was funded by the Department of Science and Technology of Hubei Province (#2022EHB035) and the Hubei Chutian Program (#1180011).
Author information
Authors and Affiliations
Contributions
Conceptualization: J.Z. and Z.-H.L.; methodology: J.Z., Z.W., Y.W., Q.Y., J.H., and Z.-H.L.; data curation: Z.W., Y.W., and Q.Y.; formal analysis: Z.W., Y.W., Q.Y., Y.F., C.C., Q.H., T.L., S.Y., and J.H.; investigation: Z.W., Y.W., Q.Y. Y.F., C.C., Q.H., and J.H.; validation: Z.W., Y.W., Q.Y., Y.F., and T.L.; visualization: Z.W., Y.W., Q.Y., Y.F., C.C., and S.Y.; writing—original draft: J.Z., Z.W., J.H., and Z.-H.L.; writing—review and editing: J.Z. and Z.-H.L.; supervision and funding acquisition: J.Z. and Z.-H.L. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Learta Pervizaj-Oruqaj, Francisco Mesquita, and Jack Leslie for their contribution to the peer review of this work. Primary handling editors: Si Ming Man and David Favero.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Z., Wang, Y., Yan, Q. et al. FPR1 signaling aberrantly regulates S100A8/A9 production by CD14+FCN1hi macrophages and aggravates pulmonary pathology in severe COVID-19. Commun Biol 7, 1321 (2024). https://doi.org/10.1038/s42003-024-07025-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07025-4