Abstract
Loss-of-function mutations in CLN3 cause juvenile Batten disease, featuring neurodegeneration and early-stage neuroinflammation. How loss of CLN3 function leads to early neuroinflammation is not yet understood. Here, we have comprehensively studied microglia from Cln3∆ex7/8 mice, a genetically accurate disease model. Loss of CLN3 function in microglia leads to lysosomal storage material accumulation and abnormal morphology of subcellular organelles. Moreover, pathological proteomic signatures are indicative of defects in lysosomal function and abnormal lipid metabolism. Consistent with these findings, CLN3-deficient microglia are unable to efficiently turnover myelin and metabolize the associated lipids, showing defects in lipid droplet formation and cholesterol accumulation. Accordingly, we also observe impaired myelin integrity in aged Cln3∆ex7/8 mouse brain. Autophagy inducers and cholesterol-lowering drugs correct the observed microglial phenotypes. Taken together, these data implicate a cell-autonomous defect in CLN3-deficient microglia that impacts their ability to support neuronal cell health, suggesting microglial targeted therapies should be considered for CLN3 disease.
Similar content being viewed by others
Introduction
The neuronal ceroid lipofuscinoses (NCLs) are a group of rare inherited lysosomal disorders with overlapping clinical and pathological features, including seizures, progressive motor and cognitive decline and accumulation of autofluorescent material in the lysosome1. The NCLs are caused by mutations in one of 13 distinct genes (CLN1–CLN8, CLN10–CLN14), whose proteins have different characteristics, including subcellular localization and function1,2. Despite the diversity of NCL proteins, their loss of function similarly causes aberrant lysosomal function, lysosomal accumulation of electron-dense autofluorescent storage material (primarily comprised of subunit c of the mitochondrial ATP synthase, saposins, and phospholipids), and cell death3,4. NCL affects all ages and ethnicities, and the NCL subtypes are categorized with respect to the gene that causes the disorder5. Autosomal recessive CLN3 mutations cause CLN3 disease (also known as juvenile NCL or Batten disease), which is the most common form of neurodegeneration in children worldwide1. Patients first develop vision loss, typically between 4 and 10 years of age, which is followed by onset of seizures, loss of motor and cognitive function, and premature death, typically by the mid-twenties3,4. Unfortunately, there are no effective treatments for this devastating disorder.
Although the primary function of CLN3 is still unknown, numerous studies revealed that this endo/lysosomal transmembrane protein has roles associated with endocytic trafficking, autophagy, cytoskeletal organization, lysosomal pH homeostasis, lipid metabolism, and calcium signaling6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. While CLN3 disease is primarily a neurodegenerative disease, recent studies have demonstrated that peripheral organs are also clinically impacted at later disease stages21. Despite the ubiquitous expression of CLN322,23, expression levels vary in different tissues and cell populations, suggesting the biological role of CLN3 may differ in different cell types22,24.
Neuroinflammation has been documented at early disease stages in CLN3 disease mouse models. For example, astrocyte activation was observed already in 1–3-month-old homozygous Cln3∆ex7/8 mice, whereas neuronal loss was not obvious until later ages (12–18 months)25,26. Moreover, in vitro studies revealed the neurotoxic effects of CLN3-deficient primary microglia on neuronal growth in neuron-glia co-culture models27,28,29. Constitutive activity of caspase-1 and concomitant inflammasome activation were also demonstrated in CLN3-deficient microglia, with increased levels of pro-inflammatory cytokines, including TNFα, IL-1β or IL-1α, following exposure to CLN3 disease-specific pathophysiological stimuli, such as ceramide or neuronal lysate28. Taken together, these studies suggest neuroinflammation likely plays an important role early in the CLN3 disease process. Indeed, neuroinflammation has emerged as a candidate therapeutic target, with several recent studies investigating the impact of dampening glial activation or T cell infiltration on CLN3 neuropathogenesis with promising results27,29,30,31,32. However, the mechanism by which neuroinflammation impacts disease onset and progression, especially how microglia play a role, is poorly understood. Establishing a clear role for loss of function mechanisms in CLN3-dependent neurodegeneration and a mechanistic connection to chronic inflammation has high potential to aid in developing novel therapeutic strategies in CLN3 disease. Here, we have established a model system to study CLN3-deficient microglia using acutely isolated microglia from homozygous Cln3∆ex7/8 mice, which were engineered to recapitulate the most common CLN3 disease-associated mutation33,34. Herein, we confirm a pro-inflammatory state and describe newly discovered molecular and functional changes in CLN3-deficient microglia that collectively point towards cell-autonomous defects in microglial function that likely impact their ability to support neuronal cell health in CLN3 disease.
Results
Cln3 ∆ex7/8/∆ex7/8 mouse microglia exhibit characteristic pathological features of CLN3 disease
To study CLN3-deficient microglia, we used a MACS approach to establish highly enriched microglial fractions from adult homozygous Cln3∆ex7/8 mouse brain for downstream primary culture and morphological and biochemical analysis33 (Supplementary Fig. 1a). The hallmark pathology of CLN3 disease is electron-dense autofluorescent ceroid lipopigment accumulation inside lysosomes1. Thus, we first isolated microglia from homozygous Cln3∆ex7/8 mice and wild-type littermates at timepoints between 3 and 20 months of age, which were then evaluated for the relative burden of storage material. As early as 3 months of age, isolated microglia from homozygous Cln3∆ex7/8 mice had significantly more autofluorescent storage material than that observed in wild-type microglia (Fig. 1a; yellow signal). With increasing age, the amount of autofluorescent material increased in both wild-type and homozygous Cln3∆ex7/8 microglia, but this was more dramatic in the homozygous Cln3∆ex7/8 microglia (Fig. 1a). Subunit c of the mitochondrial ATP synthase (hereafter referred to as “subunit c”) is the main accumulated protein inside CLN3-deficient lysosomes3,4, and indeed, the storage material in homozygous Cln3∆ex7/8 mouse microglia was immunopositive for subunit c. As shown in Fig. 1b, the relative burden of subunit c-positive storage material substantially increased with age, such that by 18 months of age, large subunit c-positive deposits were detected in swollen Lamp1-positive vesicles. These findings were consistent with a previous study in which a subpopulation of microglia was shown to accumulate autofluorescent lysosomal storage material in aging wild-type mice, and this population of microglia from homozygous Cln3∆ex7/8 mice accumulated dramatically more autofluorescent material compared to that observed in age-matched wild-type littermate microglia35.
a Representative fluorescent images of cultured microglia from 3- and 20-month-old (mo) Cln3∆ex7/8/∆ex7/8 mice and wild-type (Cln3+/+) littermates. Microglia from aged mice show broad-spectrum autofluorescence (obtained using excitation wavelengths of 488 and 555), which is more abundant in the Cln3∆ex7/8/∆ex7/8 microglia. Scale bar = 25 μm. b Representative micrographs of cultured primary isolated microglia from 3-, 12-, and 18-month-old homozygous Cln3∆ex7/8 mice and from 18-month-old wild-type littermate mice for comparison. Microglia were immunostained with Lamp1 (red) and subunit c antibodies (green). Scale bar = 20 μm. c Transmission electron microscopy (TEM) images of acutely isolated microglia from Cln3∆ex7/8/∆ex7/8 mice and littermate wild-type controls at the age of 12 months. Scale bars = 800 nm. d, e Bar graphs showing the quantification of storage material (asterisks) and lipid storage bodies (arrowheads) from TEM images. N = 114 cells/genotype, from 3 mice per genotype (38 cells/mouse). Student’s t-test, ****P ≤ 0.0001.
To examine the ultrastructure of the lysosomal storage material in the homozygous Cln3∆ex7/8 mouse microglia, we also performed TEM on isolated microglia from 12-month-old mice. As shown in Fig. 1, the cytoplasm of the homozygous Cln3∆ex7/8 microglia was filled with electron-dense, membranous storage bodies, which at higher power magnification exhibited a multilamellar fingerprint-like profile, consistent with the classical NCL-type storage material that is seen in CLN3 disease (Fig. 1c, d)1. By comparison, only occasional membranous structures were observed in the cytoplasm of wild-type microglia, which also frequently contained electron-lucent lipid storage bodies similar to those previously described in aged autofluorescent-positive microglia35. However, interestingly, these lipid storage bodies were virtually absent in the microglia from homozygous Cln3∆ex7/8 mice (Fig. 1e).
It has been shown in neurons and in lower organism models that, in addition to endolysosomal abnormalities, other organelle systems are also impacted by the loss of CLN3 function6,15,36,37. However, to the best of our knowledge, this has not been examined in microglia. Therefore, we next wanted to examine late endosomes/lysosomes more carefully and survey the membrane organelles in microglia isolated from homozygous Cln3∆ex7/8 mice. To accomplish this, we probed the isolated microglia from homozygous Cln3∆ex7/8 and wild-type littermate mice with additional markers for endolysosomes, endoplasmic reticulum, Golgi apparatus, and mitochondria (lysosome-associated membrane protein 2a ‘Lamp 2a’, LysoTracker™, PDI, GM130, Grp75, respectively). Consistent with our observations using Lamp 1 antibody (Fig. 1b), Lamp 2a immunostaining highlighted a significantly expanded lysosomal network in homozygous Cln3∆ex7/8 microglia compared to wild-type controls. The overall cellular Lamp 2a signal intensity was higher and the diameter of the Lamp 2a structures was larger in the homozygous Cln3∆ex7/8 microglia compared to wild-type controls (Fig. 2a, e, f). Further staining using LysoTracker™, a pH-sensitive lysosomotropic dye, supported these observations, where LysoTracker™ stained vesicles were reduced in number in the homozygous Cln3∆ex7/8 microglia compared to wild-type (Fig. 2b, g). Notably, the relative intensity of LysoTracker™ signal in the stained vesicles was also reduced in the homozygous Cln3∆ex7/8 microglia compared to wild-type (Fig. 2h). Although we did not directly measure the lysosomal pH of CLN3-deficient microglia in the present study, we note that altered lysosomal pH has been reported in several other cell models of CLN3 disease14,15,16. However, LysoTracker™ staining differences may alternatively reflect differences in the permeability of the lysosomal membrane or in dye uptake/diffusion.
Cultured microglia from 6-month-old mice were stained with lysosomal markers, Lamp2a (red) (a) or LysotrackerTM (b), mitochondrial marker, Grp75 (white) (c), or Golgi marker, GM130 (white) (d). Quantification of morphological changes is shown in the bar graphs (e–k). GM130 compactness determination using CellProfiler (k) assigns circular objects a compactness value of 1 and irregular objects, or objects with holes, compactness values greater than 1, such that a higher value indicates reduced compactness. N = 300 cells/genotype (3 mice/genotype from 3 independent experiments and 100 cells/mouse). Unpaired Student’s t-test with a two-tailed distribution was used to compare the two groups. Scale bars = 25 μm. Data are shown as mean ± s.d.; **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 Student’s t-test.
In addition to the lysosomal abnormalities observed in homozygous Cln3∆ex7/8 microglia, we also observed altered morphology of the mitochondrial network and the Golgi apparatus (Fig. 2c, d, i–k). More specifically, the mitochondrial network appeared more fragmented in homozygous Cln3∆ex7/8 microglia compared to wild-type (Fig. 2i, j), and the Golgi was less compact and more spread away from the nucleus in the homozygous Cln3∆ex7/8 microglia compared to wild-type (Fig. 2k). No obvious morphological differences were observed in the ER, as highlighted by PDI immunostaining (Supplementary Fig. 2).
Cln3 ∆ex7/8/∆ex7/8 mouse microglia harbor a significantly altered lysosomal proteome
To gain further unbiased insights into the impact of CLN3 deficiency on microglia, we prepared acutely isolated microglia samples from 3-month-old and 12-month-old homozygous Cln3∆ex7/8/∆ex7/8 mice and littermate controls (wild-type or heterozygous littermates), which were then subjected to proteomics analysis by liquid chromatography/mass spectrometry (LC/MS). Publicly available RNA sequencing data indicated that the Cln3 expression level in mouse brain is relatively higher in microglia compared to other cell types, including neurons, oligodendrocytes, and astrocytes38. Notably, CLN3-specific peptides were detected by LC/MS in both 3-month-old and 12-month-old control microglial samples, and CLN3 was also detected by immunoblot analysis in microglia-enriched samples from wild-type mice, more than in the microglia-depleted fractions from wild-type mice containing primarily neurons and astrocytes, consistent with the RNAseq data (Supplementary Data 1; Supplementary Fig. 1). To the contrary, no CLN3 peptides were detected by LC/MS in the homozygous Cln3∆ex7/8/∆ex7/8 microglial samples (Supplementary Data 1).
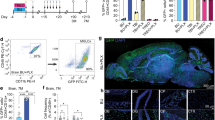
As depicted by the volcano plots in Fig. 3, our proteomics analysis revealed prominent upregulation of lysosomal proteins already at 3 months of age (Fig. 3a), and this was exacerbated further with age, with both a greater degree of change and a greater number of lysosomal proteins with increased abundance at 12 months of age in homozygous Cln3∆ex7/8 microglia compared to control microglia (Fig. 3b). Moreover, multiple components of the mTOR pathway were also upregulated at both 3 and 12 months of age (Fig. 3a, b), including the mTORC1 signaling complex (mTOR, Rptor, Lamtor1, 2, 3, and 5, and Mlst8); mTORC1 is a major signaling hub associated with the lysosome and metabolic regulation39. The two most dramatically changed proteins, which were both increased ~10-fold or more in homozygous Cln3∆ex7/8 microglia compared to control microglia at both the 3- and 12-month timepoints, were likely novel lysosomal membrane transporters, PTTG1IP and TTYH3 (Fig. 3a, Supplementary Data 1)40. Among the lysosomal proteins upregulated were Lamp 1 and Lamp 2, consistent with our immunostaining studies and prior reports (Fig. 2,35), and further validated by immunoblot (Fig. 3c, Supplementary Fig. 3a).
Relative levels of detected proteins for homozygous Cln3Δex7/8 microglia compared to littermate heterozygous and wild-type control microglia (CON) are depicted in the volcano plots from 3-month (a) and 12-month-old mice (b). Open red dots indicate proteins with a significant change in abundance [greater than false discovery rate (FDR, hyperbolic curve) threshold], while open blue dots represent proteins that were detected but did not significantly differ between Cln3Δex7/8/Δex7/8 and controls. Lysosomal proteins are depicted by solid orange dots, with significant lysosomal protein dots being bounded by a red border and non-significant lysosomal proteins being bounded by a blue border c.) Representative immunoblots are shown for the validation of the indicated protein changes in the microglial (CD11b + ) fractions from wild-type (+/+) and homozygous Cln3Δex7/8 (∆ex7/8/∆ex7/8) microglia and relevant molecular weight markers are shown. Bar graphs show the quantification of immunoblots using ImageJ (N = 3 biological replicate samples/genotype). Data are shown as mean ± s.d.; *P ≤ 0.05, **P ≤ 0.01 Student’s t-test. d GO analysis results for ‘Cellular component’ (top) and ‘Biological process’ (bottom). e Graphs represent the results of a representative migration assay on microglia isolated from homozygous Cln3Δex7/8 and wild-type control mice. Real-time imaging was performed every 15 min for 5 h to generate the migration plots, with each track from an individual cell aligned at the starting point. Axis units in μm from the starting point. Quantification of migration velocity (μm/min) over 5 h was performed using ImageJ and Ibidi’s Chemotaxis and Migration Tool plugin. N = 37 cells for wild-type and N = 45 cells for homozygous Cln3Δex7/8 microglia. Data are shown as mean ± s.d.; ****P ≤ 0.0001 Student’s t-test. f Cultured microglia from 9-month-old wild-type (+/+) or homozygous Cln3Δex7/8 (∆ex7/8/∆ex7/8) mice were stained with ganglioside markers, GM1 (red, left panels) or GM3 (red, right panels). Quantification of mean GM1 intensity per cell (left) and mean GM3 intensity per cell (right) are shown in the bar graphs. 250 cells/genotype were analyzed. Data are shown as mean ± s.d.; **P ≤ 0.01, ***P ≤ 0.001, Two-way ANOVA followed by Sidac’s multiple comparison test. Scale bar = 25 μm. Heatmap represents relative levels of sphingolipid degradation pathway proteins where the color scale depicts relative abundance in 3 month (3 m) or 6 month (6 m) homozygous Cln3Δex7/8 microglia samples compared to control. “X” = not detected.
We next performed a Gene Ontology (GO) analysis on the 12 months of age proteomic datasets to gain more comprehensive insights into the pathways affected by loss of CLN3 function in microglia. Not surprisingly, the lysosome/lytic vacuole was the most prominently changed cellular component, with the intermediate filament cytoskeleton next most prominent (Fig. 3d). GO biological processes that were significantly altered in homozygous Cln3∆ex7/8 microglia compared to wild-type microglia were mRNA processing, WNT Signaling, Transcription, Cytoskeleton Organization, Chromatin Organization, Cell Motility/Migration, mRNA Cleavage, Tissue Morphogenesis, and Sphingolipid, Glycolipid Metabolism, revealing numerous interesting pathways for further study (Fig. 3d).
In addition to the GO biological process analysis revealing enrichment in the “Sphingolipid, Glycolipid Metabolism” pathway, we also noted a number of individual lipid-related proteins with significantly altered levels in the early and later time points (3 and 12 months of age, respectively). Examples include proteins linked to glutathione metabolism [glutathione S-transferase omega 1 (GSTO1)], myelin-associated oligodendrocyte basic protein (MOBP), myelin proteolipid protein 1 (PLP), apolipoprotein E (APOE), prosaposin (PSAP), and low-density lipoprotein receptor-related protein 1 (LRP1), which is important in lipoprotein metabolism and cell motility41. Finally, consistent with prior reports, homozygous Cln3∆ex7/8 microglia showed elevated levels of multiple inflammatory markers, including apolipoprotein E (APOE), integrin alpha-X (ITGAX), CD68, vimentin, galectin 3 (LGALS3) (Supplementary Data 1, Supplementary Fig. 3a, b, Fig. 3c). However, these were less prominent changes than what was seen for lysosomal proteins, and mostly these were only significantly upregulated at the 12-month timepoint. Validation of changes for a selected set of proteins identified in the proteomics studies (LAMP2A, APOE, LRP1, PLP, LAMP1, galectin 3) was performed on additional microglial samples (Fig. 3c and Supplementary Fig. 3a, b).
To follow up on the GO analysis results, we tested the motility of CLN3-deficient microglia (GO Biological Process: Cell Motility/Migration) and examined glycosphingolipids in CLN3-deficient microglia (GO Biological Process: Sphingolipid, Glycolipid Metabolism). Notably, prior studies reported on cytoskeleton and motility defects and alterations in the sphingolipid degradation pathway in other cell types with CLN3-deficiency42,43,44,45,46. Consistent with these observations, motility of the cultured microglia isolated from homozygous Cln3∆ex7/8 mice, measured using live cell time-lapse imaging, was significantly reduced over a 5 h period, as compared to wild-type microglia (Fig. 3e). Glycosphingolipid alterations were also detected, with reduced GM1 ganglioside and elevated GM3 ganglioside immunostaining seen in the cultured homozygous Cln3∆ex7/8 microglia, as compared to wild-type microglia (Fig. 3f). These changes were consistent with the prior published reports detecting glycosphingolipid changes in CLN3-deficient cells using LC/MS44,45.
Abnormal myelin turnover and cholesterol accumulation in Cln3 ∆ex7/8/∆ex7/8 microglia
As reported above, levels of several myelin-component and myelin-related proteins were altered in the Cln3∆ex7/8/∆ex7/8 mouse microglial proteomes, including PLP, MOBP, and LRP1 (an essential receptor for microglial myelin phagocytosis47) (Supplementary Data 1). We therefore next sought to evaluate myelin phagocytosis and turnover in CLN3-deficient microglia to test whether the proteomic signatures observed were accompanied by functional changes.
Myelin is an important insulating component for neurons that facilitates the transduction of electrical impulses. During developmental remodeling and throughout aging, myelin is taken up by microglia via phagocytosis, where it is trafficked to the lysosome for degradation by proteases and lysosomal lipases. The cholesterol component of myelin cannot be degraded by lysosomal enzymes and instead is exported from the lysosome for extracellular release, or it is transported to the ER where it is esterified and stored in lipid droplets (LD) as cholesterol esters. Cholesterol esters can then be used by the cell as energy fuel48. These pathways for handling free cholesterol are critical for cellular health since accumulated cholesterol is toxic to the cell49.
To first study myelin uptake in homozygous Cln3∆ex7/8 microglia, we incubated green fluorescent-labeled myelin with isolated microglia and analyzed its uptake by fluorescent microscopy. As shown in Fig. 4a, the labeled myelin was readily taken up by both the wild-type and homozygous Cln3∆ex7/8 microglia, although a comparison of the relative green fluorescent intensities revealed reduced signal in the homozygous Cln3∆ex7/8 microglia compared to wild-type on both the per vesicle and per cell level, suggesting a slight reduction in the phagocytosis of the labeled myelin (Fig. 4a).
a Representative micrographs of green-fluorescent myelin uptake (2-hour incubation) by wild-type and homozygous Cln3∆ex7/8 microglia. Myelin (green), Lamp2 (red), DAPI= nuclei (blue). See Supplementary Fig. 4a for low-power images. Quantification of green fluorescent-myelin intensity coming from each Lamp2a positive lysosomal vesicle (N = 870 lysosomes, wild-type; N = 790 lysosomes, Cln3∆ex7/8/∆ex7/8 microglia, from 3 independent experiments). Each dot represents one lysosome. Quantification of green fluorescent-myelin intensity coming from each cell (N = 119 cells, wild-type; N = 110 cells, Cln3∆ex7/8/∆ex7/8 microglia, from 3 independent experiments). Each dot represents one cell. Data are shown as mean ± s.d.; ****P ≤ 0.0001, Student’s t-test. b Representative micrographs of green-fluorescent myelin degradation at 48 and 72 h for wild-type and homozygous Cln3∆ex7/8 microglia. Myelin (green), Iba-1 (red), DAPI= nuclei (blue). See Supplementary Fig. 4b for low-power images. Quantification of green fluorescent-myelin at 72 h (N = 203 cells, wild-type; N = 248 cells, Cln3∆ex7/8/∆ex7/8 microglia, from 3 independent experiments). Each dot represents one cell. Data are shown as mean ± s.d.; ****P ≤ 0.0001, Student’s t-test. See Supplementary Fig. 4b for the quantification of green fluorescent-myelin at 48 h. c Representative micrographs of lipid droplets labeled with LipiDyeII following unlabeled myelin uptake and 48 h post myelin washout. Bar graph shows quantification of lipid droplet number per cell (N = 297 cells, wild-type; N = 316 cells, Cln3∆ex7/8/∆ex7/8). d Representative micrographs following Filipin staining during myelin uptake and turnover. Basal panels show Filipin staining prior to myelin uptake. PDI (green), Iba-1 (red), and Filipin (blue). e Bar graph shows quantification of Filipin intensity per cell. (N = 134 cells, wild-type; N = 150 cells, Cln3∆ex7/8/∆ex7/8, from 3 independent experiments). Data are shown as mean ± s.d.; ns: non-significant, **P ≤ 0.01, ****P ≤ 0.0001, Two-way ANOVA followed by Sidac’s multiple comparison test. f Zoomed in representative images from unchallenged (i.e. no myelin phagocytosis) 6-month-old wild-type and Cln3∆ex7/8 mice microglia co-stained with Filipin (blue) and lysosomal marker Lamp2a (red). See Supplementary Fig. 5a for low-power images. Scale bars = 25 μm.
Next, to examine the turnover of the labelled myelin, we washed the cultured microglia post-myelin uptake and subsequently incubated the cells in regular media for another 48 or 72 h. Green-fluorescent signal was similar in the wild-type and Cln3∆ex7/8/∆ex7/8 microglia at the 48 h timepoint, although the green fluorescence took on a more punctate appearance versus the more diffuse signal seen immediately following the myelin uptake, likely reflecting the degradation process (Fig. 4b, Supplementary Fig. 4b). By 72 h post-green-fluorescent-myelin washout, very little green signal was observed in the wild-type cells, indicating these cells had almost completely degraded the myelin. To the contrary, significant green punctate signal was still observed in the Cln3∆ex7/8/∆ex7/8 microglia at 72 hours post-green-fluorescent-myelin washout, indicating these cells were significantly less efficient in the degradation of the phagocytosed green fluorescent-myelin compared to wild-type microglia (Fig. 4b).
We next studied LD formation following myelin phagocytosis. To do so, we fed the cells unlabeled myelin for 6 h, which was followed by a washout and regular media incubation for an additional 48 h. LDs were then labeled with LipiDyeII. At 48 h post-myelin washout, wild-type cells displayed nicely rounded spherical LDs that were strongly LipiDyeII positive (Fig. 4c). To the contrary, Cln3∆ex7/8/∆ex7/8 microglia displayed very few LDs labeled with LipiDyeII, with only a few cells harboring small LipiDyeII-positive LDs (Fig. 4c).
The recycling of cholesterol is an important process following myelin uptake, and given the observation that APOE, a key cholesterol handling protein, was significantly elevated in the homozygous Cln3∆ex7/8 microglial proteome, we followed cholesterol handling by Filipin staining throughout the myelin uptake assay. Even prior to challenge by myelin uptake, we identified significant accumulation of cholesterol in the cultured homozygous Cln3∆ex7/8 microglia, as revealed by Filipin staining, which localized to vesicles surrounded by Lamp2a, suggesting the accumulated cholesterol localized to lysosomes (Fig. 4d, f, Supplementary Fig. 5a). Following the 6-hour myelin incubation, Filipin staining was robustly increased in both wild-type and Cln3∆ex7/8/∆ex7/8 microglia (Fig. 4d, e). At 48 h post-myelin washout we observed a major reduction in the level of Filipin stain in wild-type cells and nearly complete loss of Filipin stain by 72 h (Fig. 4d, e). By contrast, Cln3∆ex7/8/∆ex7/8 microglia still showed significantly elevated Filipin at 48 h and 72 h post-myelin washout, even over the basal level observed, (Fig. 4d, e). Notably, ganglioside levels also significantly differed following myelin washout (Supplementary Fig. 5b). Altogether, these data suggest that CLN3-deficient microglia are defective in the efficient turnover of myelin and the handling of its associated lipids.
Autophagy inducers and cholesterol-lowering drugs rescue the myelin turnover defects in Cln3 ∆ex7/8/∆ex7/8 microglia
We have previously identified compounds that promote autophagy-lysosomal function in CLN3-deficient cells, which notably also included several FDA-approved cholesterol-lowering drugs, including statins, which are inhibitors of isoprenoid (e.g. cholesterol) biosynthesis50. We next tested whether the compounds promoting autophagy-lysosomal function and/or the cholesterol-lowering drugs had any significant impact on the ability of Cln3∆ex7/8/∆ex7/8 microglia to efficiently degrade phagocytosed myelin and handle the myelin-associated cholesterol. For this, Lovastatin, the most effective cholesterol-lowering drug from prior studies50, Verapamil (an mTOR-independent autophagy inducer), and ML-SA5 [a TRPML-1 agonist that promotes TFEB activation and autophagy-lysosomal function in CLN3 disease models51, Supplementary Fig. 6] were tested. Microglia from wild-type and Cln3∆ex7/8/∆ex7/8 mice were fed unlabeled myelin for 6 h, followed by a thorough washout and subsequent incubation in regular media for an additional 48 h. In the final 24 h of the 48 h incubation period, we introduced ML-SA5, Verapamil, or Lovastatin (or DMSO vehicle control) to assess their potential in correcting the LD formation phenotype, using LipiDyeII to visualize LD formation. As seen in our prior experiments, CLN3-deficient microglia (Fig. 5b) produced fewer LDs than wild-type microglia (Fig. 5a, p). Interestingly, all three drugs tested partially rescued the reduced LD formation (Fig. 5c, d, e, p), with ML-SA5 being the most efficacious. We next performed the same assay but, this time, stained with Filipin to see the effect of drug treatments on cholesterol levels. The cholesterol-reducing drug, Lovastatin, was the most effective in reducing cholesterol in Cln3∆ex7/8/∆ex7/8 mutant microglia (Fig. 5j) to a level seen in wild-type cells (Fig. 5f, r), but ML-SA5 also showed a significant reduction in accumulated cholesterol in the Cln3∆ex7/8/∆ex7/8 mutant microglia post myelin uptake/washout (Fig. 5h, r). We also tested these drugs for their ability to clear the autofluorescent storage material (Fig. 5k–o). Following a 24-hour incubation using doses previously determined to be effective in a neuronal model system50, all three drugs significantly reduced the relative abundance of autofluorescent storage material, with ML-SA5 again being the most efficacious (Fig. 5s).
a–j Representative micrographs of wild-type and Cln3∆ex7/8/∆ex7/8 microglia, treated with small molecules or DMSO vehicle, as indicated, and subsequently labeled with LipiDyeII or Filipin, 48 h post-myelin washout. k–o For autofluorescence, microglia were unchallenged (no myelin) and unlabeled. Scale bars = 25 μm. p Bar graph shows quantification of lipid droplet number (N = 200 cells/genotype or treatment, from 3 independent experiments). r Bar graph shows quantification of average Filipin intensity (Mean intensities of 240 cells/genotype or treatment, from 3 independent experiments). s Bar graphs show quantification of average autofluorescence intensity (Mean intensities of 180 cells/genotype or treatment, from 3 independent experiments). Data are shown as mean ± s.d.; ns: non-significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, One-way ANOVA followed by Tukey’s multiple comparisons test was used.
Impaired myelin integrity in aged Cln3 ∆ex7/8/∆ex7/8 mouse brain
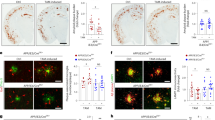
The critical role of microglia in clearing myelin debris after acute demyelinating injury to facilitate remyelination has been well-established49. Removal of aberrant myelin in development and aging is also mediated by microglial uptake and lysosomal degradation52,53,54. Notably, thinning of the corpus callosum and degeneration of optic nerves, major white matter tracts that primarily consist of myelinated nerve fibers, was previously reported in CLN3 disease patients and Cln3 mice55,56,57. Given these prior reports and the intriguing observations from our proteomics and myelin turnover studies, we hypothesized that CLN3 deficiency may impact myelin formation or maintenance, which would lead to a reduction in myelin integrity in the aging CLN3-deficient brain. We therefore immunostained brain sections from aged wild-type and homozygous Cln3∆ex7/8/∆ex7/8 mice with the compact myelin markers MBP and PLP. Both markers showed significantly reduced signal in 20-month-old Cln3∆ex7/8/∆ex7/8 mouse brain sections compared to wild-type mice (Fig. 6a-e, Supplementary Fig. 7). Moreover, TEM analysis of optic nerves from 18-month-old Cln3∆ex7/8/∆ex7/8 mice revealed an increased frequency of several aberrations in myelinated fiber integrity compared to wild-type mice (Fig. 6f-h). In addition to previously described hallmarks of axonal damage and degeneration31, we detected an accumulation of fibers with myelin infoldings, outfoldings, redundant or fragmented myelin, as well as myelin vacuoles (Fig. 6g). Quantification of fibers with these myelin aberrations showed a significant increase in Cln3∆ex7/8/∆ex7/8 compared to wild-type mice, while amyelinated or thinly myelinated fibers were not changed in frequency (Fig. 6h). Occasionally, we detected microglia with an accumulation of lysosomal storage material in optic nerves of Cln3∆ex7/8/∆ex7/8 mice, indicating decreased clearance function related to myelin degradation54. These observations lend support to our functional data that loss of CLN3 in microglia leads to altered myelin turnover in a manner that likely impacts neuronal cell health and function, particularly in the aging brain.
Representative brain sections from 20-month-old wild-type (a, b) and Cln3∆ex7/8/∆ex7/8 (c, d) mice immunostained for myelin using anti-MBP (red) antibody. Hoechst nuclear counterstaining is shown in blue. Scale bar = 500 μm. The bar graph represents the average of mean MBP intensities (e). N = 9 brain sections (3 mice per genotype, 3 brain sections per mouse, matched to represent comparable brain regions for control and Cln3∆ex7/8/∆ex7/8). Data are represented as mean ± standard error of the mean (SEM); ****P ≤ 0.0001, Student’s t-test. f Representative electron micrographs of transverse sections through the optic nerves from 18-months-old wild-type and Cln3-deficient mice. Scale bar= 2 µm. g Representative examples of distinct structural aberrations in myelin sheaths of Cln3-deficient mice. Myelin infoldings or outfoldings (arrowheads), redundant myelin (arrows), myelin fragments (hashtags), and myelin vacuoles (circles) are frequent. Occasionally, microglial cells (MG, green pseudocolor) containing lysosomal storage material (asterisks) can be detected. h Quantification of fibers with cumulative myelin aberrations (top) and amyelinated or thinly myelinated fibers (bottom) in the optic nerves from 18-months-old wild-type and Cln3-deficient mice (n = 5 mice per group, two-sided unpaired t test. top: t = 10.35, d.f. = 8, bottom: t = 10.35, d.f. = 8). Data are presented as the mean ± s.d.
Discussion
Microglia are the primary immune cells of the central nervous system (CNS) and have fundamental neuroprotective and tissue homeostatic functions through sensing the neuronal microenvironment. They protect the brain via phagocytosis and cytokine release to remove debris/pathogens and protect against infection. In response to an imbalance of microglial functions, they may become reactive and drive neuroinflammatory processes58. Microglia also play a critical role in synaptic development and remodeling and in the response to neuronal injury through their ability to turnover synapse and myelin components59. Notably, neuroinflammation and synaptic alterations have been reported in models of CLN3 disease and other forms of NCL8,28,30,32,60,61,62,63. Consistent with findings in those prior studies, our research on microglia from a genetically accurate mouse model of CLN3 disease, Cln3∆ex7/8 mice, presented in this study, also highlights evidence of elevated neuroinflammation. In addition, we have now significantly expanded upon the cell biological processes affected by the loss of CLN3 function in microglia, which likely have a significant impact on the degenerative disease process observed in CLN3 disease.
Changes in the lysosomal, Golgi and mitochondria subcellular organelles have previously been documented in neurons and other cell types upon loss of CLN3 function6,15,36,37. In the current study, we have similarly observed significant changes in these membrane organelles in microglia isolated from homozygous Cln3∆ex7/8 mice. Moreover, consistent with the primary localization of the CLN3 protein to the endolysosome, unbiased proteomics studies revealed a dramatic impact of loss of CLN3 function on the lysosomal proteome. The two proteins with the greatest change in homozygous Cln3∆ex7/8/∆ex7/8 microglia, at both time points analyzed, were PTTG1IP and TTYH3, which were both previously identified as likely novel lysosomal membrane transporters by biochemical studies and by their co-localization with lysosomal markers when overexpressed as tagged proteins40. TTYH3 is predicted to be a calcium-activated chloride channel and PTTG1IP is an integral membrane protein that was also shown to be induced by TFEB, the master regulator of lysosomal biogenesis64,65. Unfortunately, there are no validated antibodies available that recognize these proteins, hampering our ability to readily follow up on these findings; however, this is of high importance for future work to better understand the potential role of these two proteins in CLN3 protein function and CLN3 disease pathophysiology. Accompanying these changes in the lysosomal proteome were changes in levels of the mTORC1 signaling complex, a major signaling hub associated with the lysosome and metabolic regulation39. Prior reports of altered mTORC1 signaling in CLN3-deficiency models support these findings10,12,66.
In addition to the lysosomal proteomic signature detected in CLN3-deficient microglia, other pathways were also highlighted that prompted our follow-up functional studies. These included changes in cytoskeletal proteins and cell motility/migration pathways, sphingolipid/glycolipid metabolism pathways, as well as in myelin component and myelin-associated proteins and lipid-related proteins. Microglia are highly dependent on their ability to migrate towards sites of infection and injury and on their ability to phagocytose damaged or foreign debris/pathogens to neutralize these danger signals. Herein, we found that CLN3-deficient microglia were unable to perform either of these processes in an efficient manner, strongly suggesting CLN3-deficent microglia cannot serve their normal neuroprotective role. In addition to reducing microglial motility, CLN3-deficiency also led to abnormalities in the phagocytic uptake and turnover of myelin. More specifically, myelin uptake was slightly reduced while its degradation was delayed in the CLN3-deficient microglia. Interestingly, defects in phagocytosis have also been implicated in other CLN3 deficiency model systems67,68. In a recent study by Tang et al., CLN3-deficient retinal pigment epithelial cells (RPE) were less efficient in their ability to bind and take up photoreceptor outer segments (POS)68. Together, these studies support a broader role for CLN3 in the phagocytic turnover of cellular debris, supporting the need for further investigation of phagocytic processes in CLN3 disease pathophysiology.
CLN3-deficient microglia were unable to efficiently store and clear lipids following myelin uptake, including cholesterol, strongly implicating lipid handling as a major defective pathway upon loss of CLN3 function. Consistent with these findings, a recent study of CLN3 patient brain showed a dramatic lysosomal accumulation of cholesterol compared to age-matched controls69, and CLN3 was previously implicated as a genetic regulator of LD formation70. Another recent study implicated CLN3 in phospholipid catabolism. Using an elegant lysosomal enrichment technique, coupled with untargeted metabolite profiling of CLN3-deficient lysosomes, Laqtom et al. identified a unique and robust lysosomal accumulation of glycerophosphodiesters (GPDs) in the absence of CLN3, together with an increase in lysophospholipids, the metabolite upstream of GPDs in the phospholipid catabolism pathway 21. As a result of these findings and further CLN3 complementation and transport assays in cell culture, Laqtom et al. hypothesized CLN3 is required for the efflux of GPDs from the lysosome, a key process at the final stages of phospholipid catabolism21.
Given the importance of lipids and lipoprotein metabolism to microglial function48, these cells may be especially vulnerable to the loss of CLN3 function and its impact on phospholipid catabolism and cholesterol handling. Lipids serve as essential building blocks for cells and are an important energy source for microglia48, but they also have the potential to act as pro-inflammatory stimuli. Excessive cellular accumulation of lipids can lead to toxicity and trigger apoptosis, emphasizing the delicate balance required for maintaining cellular health71,72. Although we have not specifically tested the impact of defective lipid handling and phospholipid catabolism on the survival of CLN3-deficient microglia in this study, a prior study showed a significant decrease specifically in the numbers of surviving autofluorescent-positive microglia following acute isolation from Cln3∆ex7/8 mice at 9 and 18 months of age, which was accompanied by elevated apoptosis markers, suggesting accumulation of toxic lipid species and/or an altered metabolic capacity may predispose CLN3-deficient microglia to cell death35.
In summary, the high level of CLN3 expression in microglia, together with the early onset of lysosomal storage, disrupted lipid metabolism, and inflammatory signatures in Cln3∆ex7/8/∆ex7/8 microglia suggests that CLN3 exerts a cell-autonomous function in microglia and that microglial pathology is not solely triggered by a neurodegenerative environment. Moreover, deficient microglial function likely contributes to and/or exacerbates the dysfunction of other cell types in CLN3 disease given the importance of cell-cell interactions in the brain microenvironment. Interestingly, a comprehensive investigation into microglia from a mouse model of another neurodegenerative lysosomal storage disorder, Nieman-Pick Type C disease (NPC), caused by NPC1 deficiency (Npc1−/−)73, identified defects in myelin turnover and an inflammatory signature that were similar to what has been described here for homozygous Cln3∆ex7/8 mouse microglia. However, in the case of Npc1−/− mouse microglia, phagocytic uptake was enhanced and lysosomal degradation was preserved, yet myelin turnover and lipid handling, particularly of cholesterol, were impaired33. Indeed, in an Ingenuity Pathways Analysis (IPA) comparison of the proteomic signatures in Cln3∆ex7/8/∆ex7/8 and Npc1−/− microglia33, there are clear similarities in the impacted canonical signature pathways and upstream activators, including in CLEAR Signaling and Sphingolipid Metabolism pathways (Supplementary Figs. 8 and 9, Supplementary Data 2). Comparison with IPA analysis of microglial proteomic signatures from the APPPS1 mouse model of Alzheimer’s disease (AD) pathology74,75 also showed some overlap with the CLN3 and NPC signatures, albeit to a lesser degree than the two lysosomal storage diseases (Supplementary Figs. 8 and 9, Supplementary Data 2). Microglial lipid metabolism has also been highlighted as an important pathway in AD and other neurodegenerative diseases including multiple sclerosis and amyotrophic lateral sclerosis (ALS)48,76. Therefore, further studies to expand our understanding of CLN3 and microglial function will provide critical new insights into the role of these cells in CLN3 disease and other neurodegenerative diseases, which will be fundamental for the development of novel therapies in the future.
Materials and methods
Antibodies
The following antibodies were used: anti- Iba-1 [Western blot (WB): 1:1000, Wako 016-20001], anti- Iba-1 [immunofluorescence (IF): 1:500, Wako 019-19741], anti-Tuj-1 (WB: 1:1000, BioLegend MMS-435P), anti- GFAP (WB: 1:1000, Cell Signalling 3670S), anti-mCln3p (WB: 1:1000,10), anti- Apoe (WB/IF: 1:250, Invitrogen 16H22L18), anti- PLP [WB: 1:1000, immunohistochemistry (IHC): 1:300, Abcam ab28486], anti- MBP (IHC: 1:1000, Sigma Aldrich SMI-99), anti- actin (WB: 1:1000, Santa Cruz Biotech sc-81178), anti- subunit c (IF: 1:300, Abcam ab181243 (ATP Synthase C [EPR13907]) and11), anti- Lamp1 (WB: 1:1000, IF: 1:100, Santa Cruz Biotech sc-19992), anti- Lamp2a (WB: 1:1000, IF: 1:500, Abcam ab18528), anti- GM130 (IF: 1:100, BD Transduction 610822), anti- GRP175 (IF: 1:100, Stressgen SPS-825), anti- LRP1 (WB: 1:50000, Abcam ab92544), anti- Galectin 3 (WB: 1:500, Santa Cruz Biotech sc-32790), anti- PDI (IF: 1:100, StressGen SPA-891), anti- Ganglioside GM1 (IF: 1:50, Abcam ab23943). Anti-Ganglioside GM2 (IF: 1:100) and anti-Ganglioside GM3 (IF: 1:50) were generous gifts from Dr. Kostantin Dobrenis, Albert Einstein College of Medicine. Secondary antibody goat anti-rat AlexaFluor 568 (1:800, Thermo Fisher Scientific Inc), secondary antibody donkey anti-rabbit AlexaFluor 488 (1:500, Thermo Fisher Scientific Inc) or 555 (1:800, Thermo Fisher Scientific Inc), secondary antibody donkey anti-mouse Alexa Fluor 488 (1:500, Thermo Fisher Scientific Inc) or 555 (1:800, Thermo Fisher Scientific Inc).
Reagents
The following compounds were used in this study. ML-SA5 (10 uM) (Cat no: AOB11116 (Aobious INC)), Verapamil (10 uM) (Cat no: CAS 152-11-4 (Santa Cruz)), Lovastatin (10 uM) (Cat no: S2061 (Selleck Chemicals)), LipiDyeII (1 uM) (DiagnoCine FNK-FDV-0027), Filipin III (Sigma- Aldrich SAE0087), LysoTracker™ Deep Red (Invitrogen L12492), TrueBlack® Lipofuscin Autofluorescence Quencher (Biotium), ProLong™ Gold Antifade Mountant with DNA Stain DAPI (Invitrogen P36931), Tissue-Tek® O.C.T. Compound, ProLong™ Gold Antifade Mountant (P36930), PKH67 Green Fluorescent Cell Linker Kit for General Cell Membrane Labeling (Sigma-Aldrich Cat no: PKH67GL).
Mice
Cln3∆ex7/8 mice have been previously described and are publicly available at The Jackson Laboratory (Strain #: 017895)34. Cln3∆ex7/8 mice were maintained on a C57BL/6 J congenic background (>20 generations) in the AAALAC-accredited rodent facility at MGH. Heterozygous matings were set up to generate wild-type, heterozygous, and homozygous littermate experimental animals (males and females), which were aged to the indicated time points for microglia isolation or tissue collection for histology. Mice were euthanized by inhalation of CO2-bottled gas, in accordance with current guidelines from the American Veterinary Medical Association. Mice collected for Histology studies were transcardially perfused, as described below. All procedures used for this study were reviewed and received ethical approval by the Subcommittee on Research Animal Care at MGH to ensure compliance with government regulations and animal welfare guidelines (Protocol # 2008N000013). We have complied with all relevant ethical regulations for animal use.
Histology
Mice (N = 3, 20-month old mice per genotype) were transcardially perfused with 4% PFA/0.1 M PBS, followed by brain dissection and six hours post-fixation with 4% PFA/0.1 M PBS at 4 °C. Brain hemispheres were placed into a scintillation vial and filled with 10%sucrose/ 1X PBS. After 24 h (24 h), the sunken tissues at the bottom of the vial were transferred to 20%Sucrose/ 1X PBS. The next day, tissues were transferred to 30% sucrose/ 1X PBS. After 24 h, the sunken tissues at the bottom of the vial were frozen in O.C.T. A Leica CM3050 S Cryostat was used to make 40 μm coronal brain sections collected in cryoprotectant solution (25% Ethylene Glycol/ 25% Glycerol/ 50% 1X PBS). Free-floating brain sections were processed for antigen retrieval in 1 mM sodium citrate buffer and microwaving for 10 min (min) at 10% power level. Brain sections were then blocked for 1 h in 1X PBS/ 0.1% Triton X-100/ 1% Glycine/ 2% Bovine Serum Albumin (BSA)/ 10% Normal Horse Serum (NHS). Next, sections were incubated for two days at 4 °C with primary antibody diluted in 1X PBS/ 2% BSA/ 10% NHS. After washing three times with 1X PBS/ 0.05% Tween 20, corresponding secondary antibodies were diluted in 1X PBS/ 2% BSA/ 10% NHS and incubated for 2 h at room temperature (RT). Following secondary antibody incubation, sections were washed three times with 1X PBS/ 0.05% Tween 20 and incubated with Hoechst (1:10,000) for 10 min. Finally, sections were washed three more times in 1X PBS and subsequently mounted onto microscope slides in a bath of 1X PBS and allowed to dry overnight (O/N). Slides with the mounted sections were then coverslipped with mounting media and stored at 4 °C in the dark until imaging. Imaging was performed on a Zeiss Celldiscoverer 7 microscope using a 20× / 0.95 Autocorr Objective.
Primary microglia
Cerebellum, olfactory bulb, and brain stem were removed from dissected mouse brains. Meninges was dissected away from the remaining cerebrum in ice-cold HBSS under a dissection microscope. The cerebrum was then cut into 16 pieces and dissociated using the Neural Tissue Dissociation Kit (P) (Miltenyi Biotec), according to the manufacturer’s protocol. Microglial cells were isolated away from the rest of the cells in the suspension by incubation with CD11b microbeads (Miltenyi Biotec), which were then pulled down using a magnetic-activated cell sorting (MACS) separation LS column (Miltenyi Biotec). Samples prepared for mass spectrometry or biochemical analysis (e.g. immunoblotting) were further washed twice with PBS and frozen for subsequent analysis without culturing. The purity of the microglial fraction was confirmed by immunoblot with cell-type specific antibody markers (e.g. Iba1, microglia; Tuj1, neurons; GFAP, astrocytes). For cell culture experiments, isolated microglia were resuspended in microglial culture medium [DMEM/F12 (Gibco), 5% fetal bovine serum (FBS, Sigma Aldrich), 1% PenStrep (Gibco), and 10 ng/ml GM-CSF (R&D System)] and plated onto 12 mm glass coverslips. 50 µl, containing 8 × 104 cells, was plated onto each coverslip placed inside of a well of a 24-well tissue culture plate. Cells were then left to attach for 30’ at 37 °C, 5% CO2. After the 30 min incubation, 450 µl of culture media were added to each well containing the coverslips. The following day, media was exchanged for fresh media, and the cells were incubated for a further five days at 37 °C, 5% CO2. After day five, cultured microglia were further processed for immunostaining, or labeling with LipiDyeII [1 µM overnight (O/N) incubation at 37 °C, 5% CO2] or Lysotracker (500 nM for 30 min). For fixation, cells were washed three times with 1X PBS and fixed with 4% PFA for 15 min at RT. Alternatively, cells were fixed with ice-cold methanol: acetone (1:1) at −20 °C for GRP75 and LAMP 1 immunostaining.
Immunostaining
Fixed coverslips were further processed for immunostaining by first blocking and permeabilizing with 0.05% Triton X-100 in 10% NGS/PBS. To quench autofluorescence, coverslips were then treated with 1X TrueBlack solution for 30 s, followed by three washes with 1X PBS. Coverslips were then incubated with primary antibody diluted in 10% NGS/PBS O/N at 4 °C. After the O/N incubation, coverslips were washed three times in 1X PBS, followed by a 1 h RT incubation with secondary antibody diluted in 10% NGS/PBS (and where indicated, 100 µg/ml Filipin dye for cholesterol staining was also added to the secondary diluted antibody solution). For LAMP 2 A staining, 1% BSA/ 2% NHS/ 0.05% Saponin/ 1X PBS was used for blocking and antibody dilutions. Cells were washed and mounted with or without DAPI using Prolong Gold anti-fade mounting media, according to manufacturer guidelines. Samples were imaged on a Zeiss Epifluorescent Microscope equipped for digital capture of fluorescent images.
Sample preparation for mass spectrometry
The microglial proteome of Cln3Δex7/8/Δex7/8 mice was compared with those of littermate control heterozygous (Cln3Δex7/8/+) or wildtype (Cln3+/+) mice at 3 (Cln3Δex7/8/Δex7/8 n = 5 vs controls n = 7) and 12 months (Cln3Δex7/8/Δex7/8 n = 4 vs controls n = 5) of age. Microglia were acutely isolated using MACS with CD11b microbeads (Miltenyi Biotec) as above and as previously described33. Sample preparation and mass spectrometric measurements using data independent acquisition were performed as previously described75. Data analysis was performed with Spectronaut 12.0.20491.14.21367 using a microglia spectral library generated in Sebastian Monasor et al.75. Briefly, peptide and protein FDR were set to 1%. Protein label free quantification (LFQ) required at least two peptides. The statistical data analysis was performed in Perseus version 1.6.15.077. Protein LFQ intensities were log2 transformed and two-sided Student’s T-tests were performed between Cln3Δex7/8/Δex7/8 and controls at 3 and 12 months to evaluate the significance of protein abundance changes. Only proteins with at least three valid values per group were considered for statistical testing. Additionally, a permutation-based FDR was used for multiple hypothesis correction78 (p = 0.05, s0 = 0.1).
The gene ontology enrichment for cellular component (GOTERM_CC_FAT) and biological process (GOTERM_BP_FAT) were carried out with the webtool DAVID79. Proteins with an absolute log2 fold change larger than 0.5 and a p-value less than 0.05 were used as a target list and were compared with the list of all relatively quantified proteins as background.
Immunoblot analysis
Acutely isolated microglia were lysed in lysis buffer (50 mM Tris, 150 mM NaCl, 0.2% Triton X-100) containing protease inhibitors (cOmplete Mini, Sigma Aldrich). Cells were completely lysed by passing the lysates through a 25 G needle ten times followed by incubation on ice for 10 min. Lysates were then cleared by centrifugation at maximum speed in a Microfuge® 18 (Beckman) inside a 4 oC cold room. Supernatants were collected and lysate protein content was quantified using a Pierce™ BCA Protein Assay Kit according to the manufacturer’s protocol. 10–20 µg of total protein per sample was separated on a bis-tris acrylamide gel, followed by Western transfer and immunoblotting on nitrocellulose membrane (Bio-Rad). Immunoblots were developed using horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) and the Western Lightning® Plus-ECL reagent (Perkin Elmer). Actin was used as a loading control. Developed immunoblots were imaged using an iBright FL1500 Imaging system (ThermoFisher) or on X-ray films. Immunoblot densitometry (n ≥ 3 replicate samples) was performed using the gel analyzer tool in ImageJ (NIH).
Crude myelin preparation and phagocytic assays
Dissected three-month-old wildtype mouse brains were sonicated in 10 mM HEPES/ 5 mM EDTA/ 0.3 M Sucrose/ Protease inhibitors buffer. A sucrose gradient was prepared in ultracentrifuge tubes with 0.85 M sucrose (pH 7.4) on the bottom and 0.32 M sucrose (pH 7.4) on top. The brain homogenate was added to the sucrose gradient and centrifuged at 75,000 g for 30 min with a SW41 Ti rotor. The myelin fraction from the interface was collected in a new ultracentrifuge tube, and osmotic shock was applied with ultra-pure water by centrifuging the tubes at 75,000 g for 15 min three times. The yield of myelin was quantified by BCA assay, and some of the myelin was labelled with the green-fluorescent cell linker kit (PKH67 labeling kit, Sigma), according to manufacturer’s specifications.
For myelin phagocytosis, unlabeled or green fluorescent labeled myelin was resuspended with a 27 G needle in microglial medium. Primary microglia were then incubated with 10 µg/ml of the resuspended myelin; for monitoring myelin uptake, cells were incubated with green fluorescent myelin for 2 h in a 37 °C incubator33, followed by three 1X PBS washes, and fixation with 4%PFA at RT for 15 min at the indicated timepoints. Myelin (green) phagocytosis inside phagocytic Lamp2a-positive vesicles (red) was monitored by fluorescent microscopy. For unlabeled myelin turnover studies, microglia were incubated with 10 µg/ml of the resuspended myelin for 6 h at 37 °C, 5% CO2. Myelin containing media was then removed, cells were washed three times with 1X PBS, and fresh microglial medium was added. Further incubation for the indicated times was followed by fixation and further processing for labeling and imaging studies. For microglia rescue experiments, after unlabeled myelin washout, microglia were incubated for an additional 48 h, and for the last 24 h, small molecule compounds (10 µM concentration) were added (with or without LipiDyeII (1 µM), as indicated). At the end of 48 h, cells were fixed with 4% PFA at RT for 15 min and further processed for immunostaining. ML-SA5 was confirmed to have activity on the autophagy marker, LC3 by immunoblot analysis of LC3-II levels (Supplementary Fig. 6).
Transmission electron microscopy (TEM) of isolated microglia
Microglial cells were isolated as described above, the only exception being that the flow through from the MACS separation LS column was again loaded on the MACS separation MS column (Miltenyi Biotec) to ensure highest purity of the microglial fraction for TEM imaging. Brains from three 12-month-old mice/genotype were collected for three independent microglial preps/genotype for the TEM studies (N = 3/genotype). The microglia pellets were collected and resuspended (1 h at RT with gentle rotation) in 4% glutaraldehyde in cacodylate buffer. Fixed isolated cell suspensions were stored at 4 °C until they were ready to be processed and sectioned for imaging.
Fixed isolated cell suspensions were centrifuged (3500 rpm for 20 min @ 4 °C) to re-form a pellet aggregate, which was then resuspended for 1 h at RT on a gentle rotator. Samples were rinsed multiple times in 0.1 M cacodylate buffer, centrifuging to re-pellet as needed. Following supernatant removal, pellets were stabilized in 2% agarose in PBS, and the agarose-embedded pellets were then dehydrated using a graded series of ethanols to 100%, followed by a brief (10 min) dehydration in 100% propylene oxide. Dehydrated samples were then pre-infiltrated O/N in a 1:1 mix of propylene oxide:Eponate resin (Ted Pella, Redding, CA) on a gentle rocker at RT. The following day, samples were placed into fresh 100% Eponate, which was allowed to infiltrate for several hours before embedding with additional 100% Eponate in flat molds. Resin was then allowed to polymerize 24–48 h at 60 °C. Thin (70 nm) sections were cut on a Leica EM UC7 ultramicrotome using a diamond knife (Diatome U.S., Fort Washington, PA) and collected onto formvar-coated grids. Sections were then contrast-stained with 2% uranyl acetate and Reynold’s lead citrate, followed by examination on a JEOL JEM 1011 transmission electron microscope at 80 kV. Images were collected using an AMT digital camera and imaging system with proprietary image capture software (Advanced Microscopy Techniques, Danvers, MA).
For the purposes of morphometric quantification a minimum of 38 nucleated cells/biological replicate sample were captured at 20,000X magnification. The number of storage bodies and lipid structures in each cell was then counted from intact cells in the images (i.e. full cell must have been visible in the image) until a total of 38 nucleated cells/biological replicate sample was reached (totaling 114 cells/genotype), and quantification data were further plotted and analyzed using GraphPad Prism.
TEM of optic nerves
Mice were euthanized with CO2 (according to the guidelines by the State Office of Health and Social Affairs Berlin), blood was removed by transcardial perfusion with PBS containing heparin and tissue was fixed by perfusion with 4% PFA and 2% glutaraldehyde in cacodylate buffer. Optic nerves were collected, postfixed overnight, osmicated, and processed for light and electron microscopy as previously described31,32; morphometric quantification of neuropathological alterations was performed using a LEO906 E electron microscope (ZEISS) and corresponding software iTEM v.5.1 (Soft Imaging System). On average, 5 regions of interest (approximately 1000 fibers per individual optic nerve) were analyzed per optic nerve per mouse. The percentages of fibers showing myelin aberrations were identified individually by their characteristic morphological features in electron micrographs and related to the number of all investigated fibers per optic nerve per mouse. Images were processed (rotation, cropping, addition of symbols and pseudocolor) to generate figures using Photoshop CS6 and Illustrator CS6 (Adobe).
Motility assay
20,000 microglial cells were seeded per 96 Well Black/Clear Bottom Plate (TC Surface, cat number 165305). Microglial cells were imaged on a Incucyte® Live-Cell Analysis System (Sartorius), at 20X, every 15 min, for 5 h. Image sets were exported from the software as 8-bit .tif files, imported into FIJI image analysis software, and an image stack file was created. As described previously, the center of the nucleus on each image for every cell during the time course was tracked using the Manual Tracking plugin of ImageJ, and the velocity (µm/min) of each cell was quantified using the Chemotaxis Tool plugin of ImageJ and a Rose diagram graph data presentation80.
Image analysis
For intensity comparisons, color images were converted to gray, and using CellProfiler, both the primary (nucleus) and the secondary (cells) objects were identified. Each cell’s specific channel intensity was thresholded and measured using the MeasureObjectIntensity module to analyze the total intensity of a specific channel in each cell.
For the lysosomal diameter quantification, the method used by Cao et al.81 was used with the following specific details. 32 cells were analyzed for wild type microglia and 40 cells were analyzed for homozygous Cln3∆ex7/8 microglia. Using ImageJ, all of the lysosomes from cells containing a minimum of three lysosomes with diameters bigger than 2 µm were measured. The minimum size of a quantified vesicle diameter was set to 0.25 µm and the average vesicle diameter per cell was plotted.
To quantify the number of lysotracker positive vesicles per cell, nuclei were identified as primary objects and expanded to identify cells using CellProfiler. The IdentifyPrimaryObjects module was used to identify and threshold vesicles from the identified cells’ images. The RelateObjects module was used to identify the vesicles and their numbers in each cell.
For the Golgi compactness calculation, the primary (nucleus; A), secondary (cell; B), and tertiary (Golgi; B-A) objects were identified using CellProfiler. The MeasureObjectSizeShape module was used to see each cell’s compactness value for the Golgi82,83.
For the mitochondrial fragmentation and length analysis, the primary (nucleus; A), secondary (cell; B), and tertiary (mitochondria; B-A) objects were identified using CellProfiler. Mitochondria features were suppressed and thresholded, and the MorphologicalSkeleton module was used to get the mitochondrial skeleton. Primary objects were identified from the skeleton image to identify each mitochondria in each cell. Mitochondrial primary objects were shrunk, and endpoints were added by the Morph module. Primary objects were identified and shrunk from the endpoint data. Lastly, the MeasureObjectskeleton module was used to have the values for mitochondrial fragmentation and the length of each mitochondria from every cell84.
For the LD analysis, primary objects were identified and expanded to identify cells using CellProfiler. The IdentifyPrimaryObjects module was used again to identify LDs from the identified cells’ images. The RelateObjects module was used to identify the LDs of each cell and calculate their numbers in every single cell.
For MBP and PLP fluorescent intensity measurement, tile scan images captured using the Zeiss Celldiscoverer 7 microscope were initially processed through the ImageJ “Make Binary” tool for binary image conversion. Subsequently, the region of interest was delineated using the ImageJ “Freehand Selections” tool on the binary image. The region of interest was the outline of the tissue section, with folds in the tissue excluded. The ImageJ “Measure” tool was employed to directly assess the mean intensity values for MBP and PLP fluorescence by utilizing the “mean grey value” readout. Mean MBP and PLP fluorescent intensities were measured in three separate brain sections per animal, which were then averaged for subsequent statistical analysis, as done previously85.
Statistical analysis
An unpaired Student’s t-test with a two-tailed distribution was used to compare the two groups (Figs. 1, 2, 3, 4a–c, 6) (Supplementary Figs. 3, 4, 7). One-way ANOVA followed by Tukey’s multiple comparisons test was used to compare multiple groups (Fig. 5) (Supplementary Fig. 6). Two-way ANOVA followed by Sidac’s multiple comparison test was used to compare the mean differences between groups split into two independent variables (Fig. 4e) (Supplementary Fig. 5). GraphPad Prism 10.0.2 was used for all. A value of P ≤ 0.05 was considered significant (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Ethics approval and consent to participate
Studies involving use of mice underwent thorough review and approval by the Subcommittee on Research Animal Care at MGH, ensuring adherence to government regulations and guidelines for animal welfare (Protocol # 2008N000013).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the source data supporting the findings of this study are available within the paper and the associated supplementary information files (Supplementary Figs., Supplementary Data 1–3). All other data are available from the corresponding author on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the name “Proteomics of CLN3-deficient murine microglia identifies a disease associated phenotype with lysosomal alterations” and with the dataset identifier PXD048550.
References
Mole SE, Williams RE, Goebel HH. The Neuronal Ceroid Lipofuscinoses (Batten Disease) Second Edition ed. Oxford Univ. Press, Oxford; 2011.
Butz, E. S., Chandrachud, U., Mole, S. E. & Cotman, S. L. Moving towards a new era of genomics in the neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165571 (2020).
Mole, S. E., Williams, R. E. & Goebel, H. H. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics 6, 107–126 (2005).
Radke, J., Stenzel, W. & Goebel, H. H. Human NCL Neuropathology. Biochim. Biophys. Acta 1852 (10 Pt B), 2262–2266 (2015).
Williams, R. E. & Mole, S. E. New nomenclature and classification scheme for the neuronal ceroid lipofuscinoses. Neurology 79, 183–191 (2012).
Fossale, E. et al. Membrane trafficking and mitochondrial abnormalities precede subunit c deposition in a cerebellar cell model of juvenile neuronal ceroid lipofuscinosis. BMC Neurosci. 5, 57 (2004).
Luiro, K. et al. Interconnections of CLN3, Hook1 and Rab proteins link Batten disease to defects in the endocytic pathway. Hum. Mol. Genet. 13, 3017–3027 (2004).
Luiro, K. et al. Batten disease (JNCL) is linked to disturbances in mitochondrial, cytoskeletal, and synaptic compartments. J. Neurosci. Res 84, 1124–1138 (2006).
Schultz, M. L., Tecedor, L., Stein, C. S., Stamnes, M. A. & Davidson, B. L. CLN3 deficient cells display defects in the ARF1-Cdc42 pathway and actin-dependent events. PLoS ONE 9, e96647 (2014).
Cao, Y. et al. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J. Biol. Chem. 281, 20483–20493 (2006).
Cao, Y. et al. Distinct Early Molecular Responses to Mutations Causing vLINCL and JNCL Presage ATP Synthase Subunit C Accumulation in Cerebellar Cells. PLoS ONE 6, e17118 (2011).
Vidal-Donet, J. M., Carcel-Trullols, J., Casanova, B., Aguado, C. & Knecht, E. Alterations in ROS activity and lysosomal pH account for distinct patterns of macroautophagy in LINCL and JNCL fibroblasts. PLoS ONE 8, e55526 (2013).
Chandrachud, U. et al. Unbiased Cell-based Screening in a Neuronal Cell Model of Batten Disease Highlights an Interaction between Ca2+ Homeostasis, Autophagy, and CLN3 Protein Function. J. Biol. Chem. 290, 14361–14380 (2015).
Pearce, D. A., Nosel, S. A. & Sherman, F. Studies of pH regulation by Btn1p, the yeast homolog of human Cln3p. Mol. Genet. Metab. 66, 320–323 (1999).
Gachet, Y., Codlin, S., Hyams, J. S. & Mole, S. E. btn1, the Schizosaccharomyces pombe homologue of the human Batten disease gene CLN3, regulates vacuole homeostasis. J. Cell Sci. 118, 5525–5536 (2005).
Holopainen, J. M., Saarikoski, J., Kinnunen, P. K. & Jarvela, I. Elevated lysosomal pH in neuronal ceroid lipofuscinoses (NCLs). Eur. J. Biochem. 268, 5851–5856 (2001).
Persaud-Sawin, D. A., McNamara, J. O. I., Vandongen, R. S. & Boustany, R. A galactosylceramide binding domain is involved in trafficking of CLN3 from golgi to rafts via recycling endosomes. Pediatr. Res. 56, 449–463 (2004).
Tecedor, L. et al. CLN3 loss disturbs membrane microdomain properties and protein transport in brain endothelial cells. J. Neurosci. 33, 18065–18079 (2013).
Yasa, S. et al. CLN3 regulates endosomal function by modulating Rab7A-effector interactions. J. Cell Sci. 133, jcs234047 (2020).
Yasa, S., Sauvageau, E., Modica, G. & Lefrancois, S. CLN5 and CLN3 function as a complex to regulate endolysosome function. Biochem J. 478, 2339–2357 (2021).
Laqtom, N. N. et al. CLN3 is required for the clearance of glycerophosphodiesters from lysosomes. Nature 609, 1005–1011 (2022).
Rietdorf K, et al. Cardiac pathology in neuronal ceroid lipofuscinoses (NCL): More than a mere co-morbidity. Biochim Biophys Acta Mol Basis Dis. 1866, 165643 (2020).
International Batten Disease Consortium. Isolation of a novel gene underlying Batten disease, CLN3. The International Batten Disease Consortium. Cell 82, 949–957 (1995).
Eliason, S. L. et al. A knock-in reporter model of Batten disease. J. Neurosci. 27, 9826–9834 (2007).
Margraf, L. R. et al. Tissue expression and subcellular localization of CLN3, the Batten disease protein. Mol. Genet Metab. 66, 283–289 (1999).
Pontikis, C. C., Cotman, S. L., MacDonald, M. E. & Cooper, J. D. Thalamocortical neuron loss and localized astrocytosis in the Cln3Deltaex7/8 knock-in mouse model of Batten disease. Neurobiol. Dis. 20, 823–836 (2005).
Burkovetskaya, M. et al. Evidence for aberrant astrocyte hemichannel activity in Juvenile Neuronal Ceroid Lipofuscinosis (JNCL). PLoS One 9, e95023 (2014).
Parviainen, L. et al. Glial cells are functionally impaired in juvenile neuronal ceroid lipofuscinosis and detrimental to neurons. Acta Neuropathol. Commun. 5, 74 (2017).
Xiong, J. & Kielian, T. Microglia in juvenile neuronal ceroid lipofuscinosis are primed toward a pro-inflammatory phenotype. J. Neurochem 127, 245–258 (2013).
Burkovetskaya, M., Karpuk, N. & Kielian, T. Age-dependent alterations in neuronal activity in the hippocampus and visual cortex in a mouse model of Juvenile Neuronal Ceroid Lipofuscinosis (CLN3). Neurobiol. Dis. 100, 19–29 (2017).
Groh, J. et al. Sialoadhesin promotes neuroinflammation-related disease progression in two mouse models of CLN disease. Glia 64, 792–809 (2016).
Groh, J., Berve, K. & Martini, R. Fingolimod and Teriflunomide Attenuate Neurodegeneration in Mouse Models of Neuronal Ceroid Lipofuscinosis. Mol. Ther. 25, 1889–1899 (2017).
Colombo, A. et al. Loss of NPC1 enhances phagocytic uptake and impairs lipid trafficking in microglia. Nat. Commun. 12, 1158 (2021).
Cotman, S. L. et al. Cln3Δex7/8 knock-in mice with the common JNCL mutation exhibit progressive neurologic disease that begins before birth. Hum. Mol. Genet. 11, 2709–2721 (2002).
Burns, J. C. et al. Differential accumulation of storage bodies with aging defines discrete subsets of microglia in the healthy brain. Elife 9, e57495 (2020).
Codlin, S. & Mole, S. E. S. pombe btn1, the orthologue of the Batten disease gene CLN3, is required for vacuole protein sorting of Cpy1p and Golgi exit of Vps10p. J. Cell Sci. 122, 1163–1173 (2009).
Lojewski, X. et al. Human iPSC models of neuronal ceroid lipofuscinosis capture distinct effects of TPP1 and CLN3 mutations on the endocytic pathway. Hum. Mol. Genet. 23, 2005–2022 (2014).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Sengupta, S., Peterson, T. R. & Sabatini, D. M. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 (2010).
Chapel, A. et al. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell Proteom. 12, 1572–1588 (2013).
Bu, G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 10, 333–344 (2009).
Uusi-Rauva, K. et al. Neuronal ceroid lipofuscinosis protein CLN3 interacts with motor proteins and modifies location of late endosomal compartments. Cell Mol. Life Sci. 69, 2075–2089 (2012).
Getty, A. L., Benedict, J. W. & Pearce, D. A. A novel interaction of CLN3 with nonmuscle myosin-IIB and defects in cell motility of Cln3(-/-) cells. Exp. Cell Res 317, 51–69 (2011).
Somogyi, A. et al. Altered Expression of Ganglioside Metabolizing Enzymes Results in GM3 Ganglioside Accumulation in Cerebellar Cells of a Mouse Model of Juvenile Neuronal Ceroid Lipofuscinosis. Int J. Mol. Sci. 19, 625 (2018).
Schmidtke, C. et al. Lysosomal proteome analysis reveals that CLN3-defective cells have multiple enzyme deficiencies associated with changes in intracellular trafficking. J. Biol. Chem. 294, 9592–9604 (2019).
El-Sitt, S. et al. Developmental Comparison of Ceramide in Wild-Type and Cln3Δex7/8 Mouse Brains and Sera. Front Neurol. 10, 128 (2019).
Gaultier, A. et al. Lowdensity lipoprotein receptor-related protein 1 is an essential receptor for myelin phagocytosis. J. Cell Sci. 122, 1155–1162 (2009).
Loving, B. A. & Bruce, K. D. Lipid and Lipoprotein Metabolism in Microglia. Front Physiol. 11, 393 (2020).
Cantuti-Castelvetri, L. et al. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science 359, 684–688 (2018).
Petcherski, A. et al. An Autophagy Modifier Screen Identifies Small Molecules Capable of Reducing Autophagosome Accumulation in a Model of CLN3-Mediated Neurodegeneration. Cells 8, 1531 (2019).
Wünkhaus, D. et al. TRPML1 activation ameliorates lysosomal phenotypes in CLN3 deficient retinal pigment epithelial cells. Sci. Rep. 14, 17469 (2024).
Hughes, A. N. & Appel, B. Microglia phagocytose myelin sheaths to modify developmental myelination. Nat. Neurosci. 23, 1055–1066 (2020).
Djannatian, M. et al. Myelination generates aberrant ultrastructure that is resolved by microglia. J. Cell Biol. 222, e202204010 (2023).
Safaiyan, S. et al. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat. Neurosci. 19, 995–998 (2016).
Jadav, R. H. et al. Magnetic Resonance Imaging in Neuronal Ceroid Lipofuscinosis and its Subtypes. Neuroradiol. J. 25, 755–761 (2012).
Palmieri, M. et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 8, 14338 (2017).
Weimer, J. M. et al. Visual deficits in a mouse model of Batten disease are the result of optic nerve degeneration and loss of dorsal lateral geniculate thalamic neurons. Neurobiol. Dis. 22, 284–293 (2006).
Hickman, S., Izzy, S., Sen, P., Morsett, L. & El Khoury, J. Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369 (2018).
Xu, T. et al. The roles of microglia and astrocytes in myelin phagocytosis in the central nervous system. J. Cereb. Blood Flow. Metab. 43, 325–340 (2023).
Bosch, M. E. & Kielian, T. Neuroinflammatory paradigms in lysosomal storage diseases. Front Neurosci. 9, 417 (2015).
Song, J. W. et al. Lysosomal activity associated with developmental axon pruning. J. Neurosci. 28, 8993–9001 (2008).
Llavero Hurtado, M. et al. Proteomic mapping of differentially vulnerable pre-synaptic populations identifies regulators of neuronal stability in vivo. Sci. Rep. 7, 12412 (2017).
Kielar, C. et al. Molecular correlates of axonal and synaptic pathology in mouse models of Batten disease. Hum. Mol. Genet 18, 4066–4080 (2009).
Sardiello, M. et al. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009).
Suzuki, M., Morita, T. & Iwamoto, T. Diversity of Cl(-) channels. Cell Mol. Life Sci. 63, 12–24 (2006).
Zhong, Y. et al. Loss of CLN3, the gene mutated in juvenile neuronal ceroid lipofuscinosis, leads to metabolic impairment and autophagy induction in retinal pigment epithelium. Biochim Biophys. Acta Mol. Basis Dis. 1866, 165883 (2020).
Wavre-Shapton, S. T. et al. Photoreceptor phagosome processing defects and disturbed autophagy in retinal pigment epithelium of Cln3Δex1-6 mice modelling juvenile neuronal ceroid lipofuscinosis (Batten disease). Hum. Mol. Genet 24, 7060–7074 (2015).
Tang, C. et al. A human model of Batten disease shows role of CLN3 in phagocytosis at the photoreceptor-RPE interface. Commun. Biol. 4, 161 (2021).
Chen, J. et al. Juvenile CLN3 disease is a lysosomal cholesterol storage disorder: similarities with Niemann-Pick type C disease. EBioMedicine 92, 104628 (2023).
Marschallinger, J. et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208 (2020).
Zeng, X. et al. Pyroptosis in NLRP3 inflammasome-related atherosclerosis. Cell Stress 6, 79–88 (2022).
Lee, J., Dimitry, J. M. & Song, J. H. Microglial REV-ERBα regulates inflammation and lipid droplet formation to drive tauopathy in male mice. Nat. Commun. 14, 5197 (2023).
Loftus, S. K. et al. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277, 232–235 (1997).
Radde, R. et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7, 940–946 (2006).
Sebastian Monasor, L. et al. Fibrillar Aβ triggers microglial proteome alterations and dysfunction in Alzheimer mouse models. Elife 9, e54083 (2020).
Estes, R. E., Lin, B., Khera, A. & Davis, M. Y. Lipid Metabolism Influence on Neurodegenerative Disease Progression: Is the Vehicle as Important as the Cargo? Front Mol. Neurosci. 14, 788695 (2021).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Murray, M. Y. et al. Macrophage migration and invasion is regulated by MMP10 expression. PLoS One 8, e63555 (2013).
Cao, Q. et al. Calcium release through P2X4 activates calmodulin to promote endolysosomal membrane fusion. J. Cell Biol. 209, 879–894 (2015).
Mejia, I., Chen, Y. C. & Díaz, B. Analysis of Golgi Morphology Using Immunofluorescence and CellProfiler Software. Methods Mol. Biol. 2557, 765–784 (2023).
Patrick, E. et al. A cortical immune network map identifies distinct microglial transcriptional programs associated with β-amyloid and Tau pathologies. Transl. Psychiatry 11, 50 (2021).
Panusatid, C., Thangsiriskul, N. & Peerapittayamongkol, C. Methods for mitochondrial health assessment by High Content Imaging System. MethodsX 9, 101685 (2022).
Sun, L. O. et al. Spatiotemporal Control of CNS Myelination by Oligodendrocyte Programmed Cell Death through the TFEB-PUMA Axis. Cell 175, 1811–1826.e21 (2018).
Acknowledgements
This work was supported by a Neurodegeneration Research Award (S.T. and S.L.C.) from NCL Stiftung and an NCL Research Award (E.B. and S.L.C.) from NCL Stiftung, funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy– ID 390857198) (S.F.L.), the Centers of Excellence in Neurodegeneration (CoEN6005) (S.F.L.), an Alzheimer’s Association Grant (S.T.) through the AD Strategic Fund (ADSF-21-831226-C), the Federal Ministry of Education and Research (BMBF, TreatHSP.net, Project 8, 01GM2209E (J.G.), and by philanthropic support (S.L.C.). We thank Diane Capen for assistance in TEM studies, which were performed in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology at MGH, which is partially supported by an Inflammatory Bowel Disease Grant (DK043351) and a Boston Area Diabetes and Endocrinology Research Center (BADERC) Award (DK057521). We also thank Emily Beckett and Brigitte Demelo for their assistance in mouse husbandry and Anna Berghofer for technical assistance.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.Y., E.B., J.G., S.T., S.L.C.; Methodology: S.Y., E.B., A.C., S.D.S., J.G., S.T., S.L.C.; Validation: S.Y., J.G., S.T., S.L.C.; Formal analysis: S.Y., E.B., A.C., U.C., L.M., S.Tsc., M.P., S.A.M., J.G., S.F.L., S.T., S.L.C.; Investigation: S.Y., E.B., A.C., U.C., L.M., S.A.M., S.F.L.; Writing - original draft: S.Y., S.L.C.; Writing - review & editing: S.Y., E.B., A.C., U.C., L.M., S.Tsc., M.P., S.D.S., S.A.M., J.G., S.F.L., S.T., S.L.C.; Supervision: J.G., S.F.L., S.T., S.L.C.; Project administration: S.L.C.; Funding acquisition: E.B., J.G., S.T., S.L.C
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Kalipada Pahan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Fereshteh Nugent and Joao Sousa Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yasa, S., Butz, E.S., Colombo, A. et al. Loss of CLN3 in microglia leads to impaired lipid metabolism and myelin turnover. Commun Biol 7, 1373 (2024). https://doi.org/10.1038/s42003-024-07057-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07057-w
This article is cited by
-
Gene therapy ameliorates neuromuscular pathology in CLN3 disease
Acta Neuropathologica Communications (2025)
-
Genetic pleiotropy underlying obesity and autoimmune disorders: a large-scale cross-trait gwas analysis in European ancestry populations
Journal of Translational Medicine (2025)
-
Neuronal ceroid lipofuscinosis: underlying mechanisms and emerging therapeutic targets
Nature Reviews Neurology (2025)