Abstract
Photosynthetic organisms harvest light for energy. Some eukaryotic algae have specialized in harvesting far-red light by tuning chlorophyll a absorption through a mechanism still to be elucidated. Here, we combined optically detected magnetic resonance and pulsed electron paramagnetic resonance measurements on red-adapted light-harvesting complexes, rVCP, isolated from the freshwater eustigmatophyte alga Trachydiscus minutus to identify the location of the pigments responsible for this remarkable adaptation. The pigments have been found to belong to an excitonic cluster of chlorophylls a at the core of the complex, close to the central carotenoids in L1/L2 sites. A pair of structural features of the Chl a403/a603 binding site, namely the histidine-to-asparagine substitution in the magnesium-ligation residue and the small size of the amino acid at the i-4 position, resulting in a [A/G]xxxN motif, are proposed to be the origin of this trait. Phylogenetic analysis of various eukaryotic red antennae identified several potential LHCs that could share this tuning mechanism. This knowledge of the red light acclimation mechanism in algae is a step towards rational design of algal strains in order to enhance light capture and efficiency in large-scale biotechnology applications.

Similar content being viewed by others
Introduction
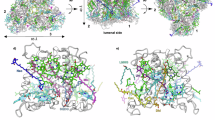
Photosynthetic eukaryotes rely mainly on the proteins of the light-harvesting complex family (LHCs) to perform the important role of harvesting and transferring energy to the photosynthetic reaction centres. LHCs are characterised by a conserved structural blueprint consisting of three transmembrane α-helices binding chlorophylls (Chls) and carotenoids (Cars)1. An essential part of their architecture consists of a left-handed coiled-coil helix pair constituted by the two homologous α-helices A and B, which generate two conserved carotenoid binding sites, labelled L1 and L2 in the light harvesting complex II (LHCII) of higher plants (Fig. 1a)2. An eight-pigment cluster, constituted by these two Cars each strongly interacting with an excitonically-coupled cluster of three Chls (see Fig. 1), has been shown to be pivotal in both the light-harvesting3,4 and photoprotective5,6,7,8,9 properties of these LHCs. Pigment binding sites of helices A and B are strongly conserved among different LHCs, with modifications limited to substitutions of individual chlorophyll/carotenoids. Pigment binding sites closer to helix C are less conserved2,10,11,12.
Lumenal view of a Phaeodactylum tricornutum FCP (PDB ID: 6A2W10), b Spinacia oleracea LHCII (PDB ID: 1RWT2), and c Pisum sativum Lhca4 (PDB ID: 7DKZ80), focusing on the conserved pigments close to the L1/L2 sites. Green sticks, Chls a; cyan sticks, Chl b; blue sticks, Chls c; orange sticks, luteins, violaxanthin, and fucoxanthins; pink sticks, dipalmitoyl-phosphatidylglycerol (DPG); white cartoons, polypeptide chain.
One aspect of photosynthetic organisms that holds both evolutionary and technological interest is the ability to adapt to different light conditions. LHC proteins provide flexible scaffolds for binding of diverse pigment mixtures. An example is the fucoxanthin-chlorophyll a/c protein complex of diatoms (FCP, Fig. 1b), which achieves excellent light-harvesting efficiency in the blue-green part of the spectrum through the utilization of Chl c and the carotenoid fucoxanthin. This is optimised for survival in the water column, where the red part of the spectrum is strongly depleted through absorption of water13. This approach has been also emulated in vitro by attaching artificial pigments to light-harvesting complexes to enhance their absorption in the green gap14,15.
Another adaptation to light limitation is the expansion of the chlorophyll absorption to the far-red part of the light spectrum. This has been most studied in the cyanobacteria synthesising specific low-energy chlorophyll species, Chl d16 or Chl f 17. While these pigments are specifically cyanobacterial, Chl a-to-Chl d substitution has been performed in LHCII producing a red-shifted LHC complex18. The promise of this research direction lies in harvesting the portion of light most available in dense canopies19,20.
Under natural conditions, eukaryotes achieve the red-shift of LHC absorbance without the need to produce specialized pigments by relying on pigment-pigment and pigment-protein interactions21,22,23,24,25,26,27,28,29, such as in the case of the antennae of photosystem I (like Lhca4, see Fig. 1c). Some of the redmost LHCs have been identified in algae belonging to the class Eustigmatophyceae21,24,25,26. Eustigmatophyte LHCs bind solely Chl a and carotenoids violaxanthin and vaucheriaxanthin, and are therefore called Violaxanthin-Chlorophyll a Proteins (VCP)30,31,32,33. Red-shifted VCP (rVCP) has been so far isolated from two eustigmatophyte species24,25, including the strain CCALA 838 known as Trachydiscus minutus (Tm)34 that is further investigated in this report, and it has been speculated that the red-shifted absorption is the result of an excitonic coupling of pigments located on the interface between subunits of an oligomeric LHC complex35.
In the present study, we employed photogenerated triplet states as internal probes in a magneto-optical spectroscopic investigation that, along with new knowledge of the rVCP peptide sequences of T. minutus, allowed us to distinguish and localise the different Chl pools, aiming to understand the mechanism of the colour tuning in rVCP.
Results
Pigment and protein composition of rVCP in T. minutus
rVCP binds 19 violaxanthin and 10 vaucheriaxanthin per 100 Chl a35, differing from VCP in the almost complete lack of esterified vaucheriaxanthin (10% of the vaucheriaxanthin pool) and in the pigment ratio (3.4:1 Chl a:Car) that is more similar to plant LHCII2 than to VCP30,31,35.
The composition of purified Tm rVCP protein was analysed using tandem mass spectrometry (MS/MS): the sample was predominantly composed of three polypeptides (see Supplementary Table S1) referred to as DN2982, DN29098, and DN6201 (following the IDs of the sequence contigs in the transcriptome assembly). The rVCP polypeptide sequences were aligned with sequences of antenna proteins from the diatom Phaeodactylum tricornutum (Pt), i.e. Lhcf4, the canonical FCP protein, and Lhcf15, the building block of the diatom red-shifted antenna36,37,38,39 (Fig. 2).
Polypeptide sequences of the LHCs from Trachydiscus minutus (Tm) and Phaeodactylum tricornutum (Pt)65,66 were used for the alignment. Yellow background signifies a transmembrane helix (derived from the crystallographic structures 6A2W10). Chl binding residues in the sequences are colour-coded: green, Chl side chain ligand; light green, Chl backbone ligand via a water molecule; red, Chl red-shifting GxxxN binding motif. Note that in the case of the a404 site, a suitable ligand is found at the i-4 position in the Tm sequences (similarly to the binding of a604 in LHCII2), which is proposed to replace the ligation in Pt.
The sequence alignment revealed a clear conservation of the amino acid residues involved in the binding of several pigments (see Fig. 2 and Supplementary Fig. S1 for Chls and Cars, respectively). Five Chls a (namely, a402, a403, a406, a407, and a408) appear strictly conserved in rVCP (we will refer to the Chl-binding sites named c403 and c408 by Wang et al. 10 as a403 and a408, since rVCP binds only Chl a35). Note that in two of the three Tm rVCP sequences, the ligand of the Chl a403 site is an asparagine (Asn), substituting the histidine (His) commonly present at this site2,10, and Asn in this position was connected to the red-shifting of Lhca3-4 in plants27,40,41.
The a405 site seems to be retained in two of the three Tm sequences (DN2982 and DN29098). Regarding the a404 site, a direct binding as observed in Pt FCP can be excluded, but direct binding from the residue at the position i-4, similarly to Gephyrocapsa (=Emiliania) huxleyi E-FCP42, can be proposed. The retainment of a401 is difficult to assess due to its binding mode: a phosphoryl group of a lipid in the case of LHCII, and a QPP motif in a loop in the case of FCPs10,43. A binding mode like the latter can be proposed to take place also in rVCPs, due to the conservation of an FP pair of amino acid residues at a similar position.
Carotenoid binding sites L1 and L2 are definitely conserved in rVCP (see Supplementary Fig. S1), congruently with their known structural role in LHCs44. Therefore, given the 3.4:1 Chl a:Car ratio determined via HPLC35, the binding of at least 7 Chls a and 2 Cars is expected for rVCP, pointing towards the likely conservation of some of the Chl binding sites discussed above (likely, a404 and a405). This minimal number is most probably to be expanded to 3 carotenoids and about 10 Chls a, in agreement with closely related LHCs35.
The higher sequence similarity of the three Tm sequences with Pt Lhcf4 (21–26%), when compared to the one with LHCII (16–17%), suggests the use of the Pt FCP structure as a more adequate structural model for rVCP in the following analyses.
Identification of the site energy of the unquenched 3Chls a
Illumination of rVCP at cryogenic temperatures leads to the formation of 3Chl a, which can be detected by Fluorescence Detected Magnetic Resonance (FDMR). The transitions between the triplet sublevels of the 3Chl a (namely, |D|−|E| and |D|+|E|, see Fig. 3a) are detected by monitoring the emission of the sample as a function of the frequency of a microwave radiation (650–1050 MHz)45. The presence of unquenched 3Chl a is a common finding in LHCs despite the presence of photoprotective carotenoids5,46,47,48 and has been previously reported also for VCP5,46.
a Jablonski diagram of the main electronic states of Chls and Cars in LHCs (in green and orange, respectively). The states are arranged vertically by energy (not in scale), and horizontally by spin multiplicity. Absorption (A), fluorescence (F), and ISC are indicated by straight grey arrows; singlet-singlet (S-S) and triplet-triplet (T-T) energy transfers by curved dashed grey arrows; the transitions between the spin sublevels by double-pointed black arrows. For readability, only the transitions more relevant for the discussion of the results are drawn. The triplet sublevels are highlighted by dashed boxes for the two molecules. The relative populations of the triplet sublevels are indicated by the thickness of the level bars. At the bottom of the panel, the molecular structure of chlorophyll a (Chl a) and lutein (Car) with the directions of the zero-field splitting principal axes (zfs, the axes of the dipolar interaction between the two unpaired electrons) are reported (in blue arrows). b Absorption (black line) and fluorescence emission (red line, excitation wavelength 481 nm) spectra of rVCP24. Temperature 77 K. c FDMR spectra (black lines) of the 3Chl |D|−|E| and |D|+|E| transitions at different wavelengths in the 680–760 nm range, as indicated. Amplitude modulation frequency 33 Hz, time constant 100 ms, temperature 1.8 K. The spectra are vertically shifted for better comparison. Reconstruction (green lines) of the experimental spectra as a sum of Gaussian components (blue and red lines). The fitting parameters are reported in Supplementary Table S2. d T-S spectra of 3Chl a. Resonance frequencies: 733 MHz (black line), 945 MHz (red line), and 1000 MHz (blue line). Amplitude modulation 33 Hz, time constant 1 s, temperature 1.8 K.
The optical properties of rVCP Chls a are dominated by a pool of red-shifted Chls (see Fig. 3b and Supplementary Fig. S2), which contribute to a third of the integrated area of the Qy 0-0 absorption band at room temperature (Supplementary Fig. S3), and are the prevalent emitters when it comes to fluorescence, particularly at cryogenic temperatures24 (Fig. 3b). However, the largest intensities are found at bluer (680–700 nm) wavelengths (Fig. 3c) when the wavelength-dependence of the 3Chl a FDMR spectra is analysed. Strong suppression of the ΔF/F ratio for wavelengths higher than 700 nm is an indication that the unquenched 3Chls a are not significantly connected to the redmost Chls a and that the latter are instead effectively photoprotected. Two 3Chl a populations contribute to the FDMR spectra at all wavelengths, except at 680 nm. The two contributions are well resolved in the |D|+|E| transitions and can be disentangled due to their different wavelength dependence (see Fig. 3c).
Further data on the pool of unquenched rVCP 3Chl have been collected by an ODMR variant, absorption detected magnetic resonance, by monitoring microwave-induced changes in the sample absorption. The spectra (Triplet minus Singlet, T-S) result from a wavelength sweep while fixing the microwaves at a frequency in resonance with a zero-field splitting (zfs) magnetic transition of 3Chl (either |D|−|E| or |D|+|E|, see Fig. 3a). In good agreement with the 3Chl FDMR results, the 3Chl T-S signal is also arising from the bluer pools of Chls, as shown by the narrow bleaching peaking at 680 nm (Fig. 3d), corresponding to the Qy 0-0 absorption band of the 3Chls, and the flat profile for wavelengths longer than 690 nm, a clear indication of a lack of interaction of the unquenched 3Chls with redmost chlorophylls. Note that the selectivity of ODMR towards the triplet state allows the detection of unquenched 3Chl even at very low concentration, as evident from the lack of marked fluorescence from their parent excited singlet states24.
The absence of signals attributable to triplet states localised on the redmost Chls a, which dominate the fluorescence spectra and are expected to be the terminal collectors of the excitation in the antenna system, implies an efficient triplet quenching mechanism on these Chls provided by nearby carotenoids. To characterise these photoprotective pathways, analogous 3Car ODMR experiments were performed.
3Car ODMR reveals that the redmost Chls are effectively photoprotected
Although carotenoids are non-fluorescent molecules, their FDMR transitions can be indirectly detected via the emission of Chls coupled by energy transfer pathways45. The change of the steady state population of the 3Car levels, induced by a resonant microwave field, is detected as an intensity change in the fluorescence of nearby coupled Chl45. Therefore, the wavelength dependence of 3Car FDMR signals can be utilised to selectively investigate the optical properties of just the coupled Chls.
Figure 4a shows the FDMR spectra detected irradiating around the main resonance transition of 3Cars45 (2|E|, 210–250 MHz), at various emission wavelengths24. The lack of signal in the 690–700 nm region points to a weak or absent coupling of the triplet-carrying carotenoid to the bluer chorophylls, which were found to mostly contribute to the unquenched 3Chl pools (seen in Fig. 3b). On the contrary, a strong FDMR signal is found from 710 nm on, a clear indication that 3Cars are coupled to a pool of red Chls a. The zfs parameters of 3Car (|D| = 0.0387 cm−1, |E| = 0.0038 cm−1) were determined from the 2|E|, |D|−|E| and |D|+|E| transitions at 740 nm. These values are close to the main 3Car component observed in VCP (|D| = 0.0393 cm−1, |E| = 0.0039 cm−1)5.
a FDMR spectra of the 3Car 2|E| transition detected at different wavelengths in the 690–760 nm range, as indicated. Amplitude modulation 333 Hz, time constant 100 ms, temperature 1.8 K. The spectra are vertically shifted for better comparison. The vertical dotted line highlights the 2|E| peak position. b FDMR spectrum of 3Car 2|E|, |D|−|E|, and |D|+|E| transitions at 740 nm. Amplitude modulation 333 Hz, time constant 100 ms, temperature 1.8 K. c T-S spectrum of 3Car, obtained with a resonance frequency of 230 MHz (3Car 2|E| transition, see panels a and b). Amplitude modulation 333 Hz, time constant 1 s, temperature 1.8 K.
The 3Car T-S spectrum (Fig. 4c), obtained with the microwave frequency set at the maximum of the 3Car 2|E| transition (230 MHz), is dominated by an intense band corresponding to the triplet-triplet absorption of 3Car (512 nm). Its remarkably narrow bandwidth (full width at half maximum, FWHM = 640 cm−1) suggests the presence of a single 3Car component. The negative signals at 460 nm and 485 nm are due to the bleaching of the carotenoid S2 singlet-singlet absorption bands. The T-S spectrum shows a bleaching at 711 nm, i.e. in the region where no carotenoid signals are expected. This feature, commonly observed in antenna proteins, originates from the electronic coupling of the Car carrying the triplet state with the proximal Chls, as recently explained by Migliore et al.49. The far-red wavelength of the interaction peak in rVCP is similar to that in plant Lhca450, confirming the strong interaction of the 3Car with the redmost pool of Chls a in agreement with the 3Car FDMR spectra. This Car-Chl a interaction peak has a FWHM of 160 cm−1, similar to those for the interaction of the luteins in site L1/L2 with the excitonic clusters of a610-a612-a611 and a602-a603 in plant LHCII9. Whereas for Chls a with weaker coupling with nearby Chls a, such as in dinoflagellate PCP47 and LHC48, and diatom FCPs43,51, the corresponding FHWMs were found to be smaller, in a 90–120 cm−1 range.
These results show that far-red absorbing chlorophylls are in close proximity to the carotenoids that populate the triplet state. To determine the mutual orientation of the Chl-Car triplet-triplet energy transfer (TTET) pair identified from the 3Car T-S spectrum, we measured the 3Car using pulse electron paramagnetic resonance (EPR).
Identification of the TTET pathways by means of 3Car pulse EPR
EPR techniques have proven to be an invaluable asset in obtaining structural information regarding the TTET acceptor-donor pair6,52, since the initial spin polarisation pattern of 3Car immediately following TTET is inherited from that of the 3Chl donor during TTET and depends on the relative pigment arrangement inside the protein scaffold. The determination of the initial spin polarisation pattern of the 3Cars, before the onset of the anisotropic relaxation of its triplet spin sublevels53,54, requires a light-induced field-swept electron spin echo (FS-ESE) sequence, which selectively suppresses the contribution of 3Chl a5,6, for the strong anisotropic relaxations of porphyrin scaffolds55. In light-induced FS-ESE, laser photoexcitation is followed by two nanosecond microwave pulses obtaining an ESE whose integrated intensity is recorded as a function of the intensity of a static magnetic field removing the degeneracy of the triplet sublevels (see Fig. 5a).
a Scheme of the energies of the triplet sublevels of a 3Car (D > 0 and E < 0) as a function of an external magnetic field, B0, aligned with the 3Car zfs axis Y. Whenever the energy of the microwave radiation matches the energy gap between T0 and either T+1 or T−1, a transition can be observed (XI or XII for B0 parallel to Y, respectively). The transitions can be either emissive (E) or absorptive (A) depending on the relative populations of the high-field triplet sublevels involved, indicated by the thickness of the level bars. b FS-ESE spectrum of rVCP (black) and Chl a dissolved in Triton X-100 micelles (green) at 50 K. The difference (orange curve) between the FS-ESE spectrum of rVCP and the FS-ESE spectrum of Chl a, which corresponds to the ‘pure’ 3Car spectrum, has been vertically translated for clarity. The simulation of the 3Car spectrum for Fx303/Fx305 (blue line) is calculated considering a population of the triplet state by means of TTET starting from the triplet state of the closest conserved Chl a (Chls a403 and a408, respectively). The polarizations of the simulated 3Car components were determined on the basis of atomic coordinates for the acceptor-donor pairs derived from the crystallographic structure10 and an initial donor 3Chl polarisation (Px:Py:Pz = 0.375:0.425:0.200)5, resulting in a 3Car polarization of (Px:Py:Pz = 0.41:0.20:0.39) for both Fx303 and Fx305 (a molecular scheme of the acceptor-donor pair with the zfs tensors of the two molecules is reported at the bottom of the panel). The simulated 3Car spectra were calculated using the following parameters: D = −41.0 mT; E = −4.1 mT; linewidths (lwx, lwy, lwz) = (2.0, 2.0, 2.5) mT. Canonical transitions discussed in the text have been highlighted in the low-field half of the spectra. A = absorption, E = emission.
Residual contributions from 3Chl a were still present in the FS-ESE spectrum of rVCP (Fig. 5b – black), which were subtracted using the FS-ESE of Chl a dissolved in Triton X-100 (Fig. 5b – green). The resulting 3Car FS-ESE spectrum of rVCP is characterised by an EEAEAA polarisation pattern (Fig. 5b – orange), as already observed in eustigmatophyte VCP5 and plant LHCII6. When considering the polarizations for the possible couples of Chl and Car, calculated on the basis of atomic coordinates for the acceptor-donor pairs derived from the crystallographic structure10, the symmetrically related pairs Chl a408-L1 and Chl a403-L2 best fitted the experimental spectrum. These polarizations were compatible also with time-resolved EPR spectra reported in the SI (Supplementary Fig. S4).
The assignment of Chl a408-L1 and Chl a403-L2 as partners in TTET aligns with these being the closest Chl a-Car pairs in terms of π-π and centre-centre distances, both in the LHCII6 and in the Pt FCP43 structures. Together with the finding derived from the analysis of the 3Car T-S bleaching at 711 nm (Fig. 4c), and the presence of Asn as the ligand of Chl a403 (Fig. 2), known to lead to red-shifted excitonic states in Lhca3-4 in plants27,40,41, we can assign the red exciton to the cluster Chl a402-a403-a406. The present observation that the development of the low-energy states in rVCP does not entail major changes in pigment organisation from regular LHC agrees with previous conclusions derived from an analysis of the singlet excitation energy transfer dynamics35.
Discussion
The ODMR and EPR data combined with the sequence analysis point out the location of the red-shifted Chl a cluster and suggest a mechanism for the red shift in rVCP. To verify whether these features are shared by other systems, we conducted a phylogenetic analysis and explored the sequences and the structures of LHCs.
The phylogenetic analysis of the antenna sequences unsurprisingly places Tm rVCP close to the main antenna proteins of the eustigmatophyte Nannochloropsis, Lhcv1/2, within the main group of FCP-like LHC proteins (Supplementary Fig. S5). This placement supports earlier proposals that low-energy Chl a forms evolved independently in various algal groups and that this evolution requires only minor changes in the protein framework. Focusing on the Chl a-binding sites, the multiple alignment of helix B sequences (Fig. 6) shows that in two of the three proteins forming the rVCP complex, the ligand at the a403 (a603 in plant LHCII) site is an Asn, instead of the typical His. This amino acid exchange is responsible for the development of the red state in plant Lhca427,40. The role of Asn appears to be twofold. Primarily, the smaller volume of the sidechain compared to His residue brings the pigment into closer contact with the pigment bound at the a406 (b609 in plant LHCII) site. Secondly, as shown recently41, the Asn sidechain forms a hydrogen bond to the a406 pigment (see Fig. 7), likely stabilising the conformation of the a403-a406 chlorophyll dimer. The resulting molecular orbital overlap of the paired Chls a leads to the development of low-energy states of a mixed excitonic-charge-transfer (CT) nature41. In rVCP, the presence of CT states was not demonstrated directly but was inferred from the broadening of the low-temperature emission spectra24 (see Fig. 3b). One of the three proteins identified in rVCP contains the His-ligand to Chl a at the a403 position and thus could in principle be lacking the red-Chl a forms. However, we detected no Chl a absorbing around 680 nm in contact with carotenoids (Fig. 4c). This suggests that the sample consists of heterooligomers, all of which contain the a403-Asn protein(s), and hence the excitation energy is always efficiently transferred to the lowest-lying Chl a state.
Highlighted is the Chl-binding site i = a403/a603 (H or N) and the corresponding position at i-4 (see text for explanation). Detailed version of the phylogenetic tree used to classify the proteins into groups given on the left is presented in Supplementary Fig. S5. Sequences of proteins that have been purified in a red-shifted Chl a-containing form are shown in red.
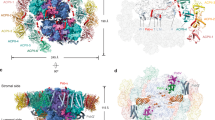
Detailed view of the chlorophyll-binding site a403(a603) in different LHC proteins (a: FCP: pdb 6A2W, b LHCII: 1RWT, c, d Lhca3, 4: 7DKZ, e Lhcr1: 5ZGH). f Plot of excitonic coupling between Chl a at sites a403 and a406 according to the Chl a ligand (i = N or H). Black diamonds, H-ligation; red diamonds, N-ligation; grey, green, and white diamonds were employed to highlight LHCII, CP29, and FCP, respectively. g Plot of excitonic coupling vs. the residue at i-4 position. Residue sidechain volumes are given on the top horizontal axis. All chlorophylls in the respective sites were parametrized as Chl a for the purpose of coupling computation.
The plant photosystem I antenna is so far the only system for which this particular mechanism has been unequivocally demonstrated experimentally and its extrapolation to other systems, while parsimonious, is not granted. Since the a403-a406 dimer consists of molecules bound to different transmembrane helices of the LHC structure, the coupling ultimately depends on the tertiary structure of the whole protein and could be influenced by factors beyond an exchange of a single amino acid residue. A case in point appears to be the main antenna protein of the alveolate Chromera (C.) velia (19753_CLH), which also contains Asn at the a403 position while not exhibiting the spectroscopic signature of a red-shifted chlorophyll56. As seen in Fig. 6, a recurring theme of the Chl a binding of the red-shifted systems is the presence of a small-sidechain residue (Gly/Ala) in the position i−4 from the chlorophyll ligand, resulting in a [A/G]xxxN binding motif. Two of the three Tm sequences show a conserved glycine at this position, similarly to plant Lhca3 and red algal Lhcr1. As visible from the comparison in Fig. 7a–e, Gly or Ala in position i-4 give more space to the Chl in the a403 site, so it can be pulled by the shorter Asn ligation without steric clashes between the carbonyl group in position 131 of the isocyclic ring and the side chain of the amino acid in i-4 position. Remarkably, in the C. velia CLH a bulky Lys is located at i-4, which is expected to prevent the proposed mechanism (see Fig. 6). To illustrate the point in a more quantitative manner, a plot of the excitonic coupling between a403 and a406 for a range of LHC proteins is given in Fig. 7f. For this analysis, all pigments were parametrized as Chl a. Despite the data scattering, an overall trend towards a stronger coupling for the Asn ligand is present. Relating the excitonic coupling to the volume of the i-4 residue sidechain57 suggests a trend towards a decreasing coupling with an increasing volume (Fig. 7g), even for the His-coordinated systems. This can originate from the capacity of His to form the hydrogen bond with the carbonyl group in position 131 of Chl a406 (see LHCII in Fig. 7b). Such an H-bond would be influenced as well by the size of the residue at position i-4. In order to verify this model, a mutation analysis of these positions, in analogy with previous studies40,58, would be highly informative.
FCP stands out from the trend outlined in Fig. 7g. However, this is the only LHC in which the chlorophyll at the a403 (a603) position is natively a Chl c, hence the simple dipole/site energy parametrisation to Chl a while retaining the Chl c geometry might not be fully adequate. However, if the estimated large coupling is correct, the Asn for His replacement in such a system assembled with only Chl a would lead to the formation of a red-shifted antenna complex. Remarkably, the diatom Lhcf15 also contains the Gly at the i-4, forming a GxxxN binding motif. This protein has been identified as the origin of the red-shifted antenna in P. tricornutum38,39. Lhcf15 thus fits the proposed mechanism, should c403 be occupied by Chl a. Pending further study, we note that this agrees with the (previously unexplained) elevated content of Chl a relative to Chl c in Lhcf15 antenna compared to FCP59.
The third representative of a far-red antenna is the redCLH of the alveolate C. velia formed by the proteins 1646_Red-CLH-1 and 532_Red-CLH-2 (Fig. 6). These proteins do not follow the present pattern since they possess a VCP/FCP-like binding motif [I/V]xxxH, but according to the phylogenetic analysis these proteins belong to a different LHC class than rVCP (Supplementary Fig. S5), and more data are required to investigate the origin of the low-energy states in these LHCs.
Concluding remarks
In the present work, we located the site of the red-shifted pigments of the eustigmatophyte antenna to the conserved core of the LHC protein. The primary factor for the red-shifted light adaptation appears to be a single amino acid residue exchange (Asn for His) of the Chl a ligand, supported by the presence of a small-sidechain residue at i-4 position. The proposed [A/G]xxxN motif in the helix B of LHC proteins thus emerges as a marker of a red-shifted antenna complex and can potentially serve as a genomic marker of the physiology adapted to the survival in shaded environments, in particular when present in an antenna complex not associated with photosystem I.
A yet unresolved general issue is the extent of the contribution of the tertiary and quaternary structure to direct pigment-pigment interactions. Resolving this in the red-shifted LHC complexes might bring important insights into other unsolved problems of the structure-function relationship in LHCs, such as the still elusive mechanisms of switching between the light-harvesting and the photoprotective conformations.
In perspective, understanding the factors governing the spectral tuning of LHCs could lead to the rational design of optimized light-harvesting systems for industrial cultivation of algae19,20, without need of introducing the metabolic pathways necessary to produce red-shifted chlorophylls d or f.
Experimental methods
Sample preparation
Cultures of Trachydiscus minutus CCALA 838 and its biologically identical subculture CCALA 931 were used for obtaining transcriptome data, and biochemical and biophysical analyses, respectively. Cells of Trachydiscus minutus CCALA 931 were used as a source material for obtaining rVCP samples. T. minutus was batch-cultured in 5 L Erlenmeyer flasks in a freshwater WC medium60 at 20 °C. The cell cultures were stirred and bubbled with filtered air. Illumination was provided by a common halogen light bulb as a red-enhanced light source (intensity of 20 µmol photons m−2 s−1), following a rectangular wave cycle of 15 h light and 9 h dark38. rVCP was purified from thylakoid membranes solubilized with n-dodecyl-β-D-maltoside (2% w/v)24 by a combination of sucrose gradient centrifugation (0.1-1.1 M, 100,000 × g, 17 h) and size exclusion chromatography (Superdex 200 10/300 GL (GE Healthcare))35. The base buffer for these procedures as well as for rVCP sample dilution during spectroscopy measurements was 50 mM HEPES, pH 7.5, 2 mM KCl, 0.02% (w/v) n-dodecyl-β-D-maltoside.
Transcriptome sequencing and assembly
T. minutus CCALA 838 was cultivated in liquid Bold’s basal medium (BBM)61. Total RNA was isolated using TRI Reagent® (TR 118) (Molecular Research Center, Inc., Cincinnati, USA), following standard procedures. Transcriptome sequencing was performed by the Institute of Applied Biotechnologies a.s. (Olomouc, Czech Republic) with the TruSeq Stranded Total RNA library preparation protocol, the Illumina NextSeq 550 platform, and the pair-end sequencing strategy. The obtained sequence data (36,062,624 reads) were quality trimmed and adapter clipped with Trimmomatic v0.3962. De novo transcriptome assembly was performed using Trinity v2.1.163 and protein sequences were predicted with TransDecoder v5.5.064 (https://github.com/TransDecoder/TransDecoder).
Protein identification by MS/MS
Tandem mass spectroscopy (MS/MS) was used to identify the protein composition of purified rVCP33 at the proteomics service facility of the University of South Bohemia. In short, the samples were first analysed by denaturing polyacrylamide gel electrophoresis stained with Coomassie Blue, visible bands were excised from the gel, digested with trypsin, and analysed on a nano-scale UPLC coupled online to an ESI-Q TOF Premier Mass spectrometer (Waters, USA). Raw MS/MS data were processed and resulting peptides were subjected to a database search using PLGS2.3 software (Waters) against a custom database of T. minutus LHC protein sequences recovered from the T. minutus CCALA 838 transcriptome-derived protein sequence set using a blastp search (with N. oceanica VCP Lhcv1 sequence33 as a query).
Sequence analysis
Comparison of LHCs amino acid sequences from Pt (Pt|Lhcf4, uniProt: B7FRW2; Pt|Lhcf15, uniProt: B7G8Q1)65,66 and Tm (DN2982, DN29098, and DN6201, inferred from the contigs TRINITY_DN2982_c0_g1_i1, TRINITY_DN29098_c0_g1_i1, and TRINITY_DN6201_c1_g1_i1, respectively, from the transcriptome assembly de novo generated for T. minutus, see above) was carried out by means of a multiple sequence alignment built using MAFFT (version 7)67. For phylogenetic analysis a set of LHC sequences was gathered by homology searches and literature survey. Specifically, candidate LHC sequences encoded by T. minutus were identified by searching the sequence set predicted from the de novo generated transcriptome assembly with hmmsearch (HMMER 3.0 package)68 as a query using a profile HMMM derived from the seed alignment of the Pfam protein family PF00504 (“Chlorophyll A-B binding protein”). Hits above the inclusion threshold were evaluated by blastp searches against the NCBI non-redundant protein sequence database and only evident members of the LHC family retained (excluding the divergent LHC-like LIL1 type). Some of the sequences proved truncated due to incomplete assembly of the respective transcripts. Virtually all of them could be completed by manually joining two or more separate contigs exhibiting perfect or near-perfect overlaps. The assembly was aided by a draft genome assembly69 and a separate alternative transcriptome assembly from T. minutus made available at Figshare (https://doi.org/10.6084/m9.figshare.25706553). The previously published sets of LHC sequences (excluding LIL1) encoded by the Nannochloropsis oceanica and Microchloropsis (=Nannochloropsis) gaditana genomes33,70 were retrieved from the respective databases and further refined by identification of additional, previously missed LHC family members and replacing some of the incorrect gene models with accurate protein sequences derived from transcriptome assemblies. The eustigmatophyte LHC sequences (with technical details listed in Supplementary Data 1) were combined with previously identified LHC sequences from the diatom P. tricornutum71 and a subset of LHC sequences from the alveolate C. velia, red algae (Cyanidioschyzon merolae, Galdieria sulphuraria, and Porphyridium purpureum), the green algal Chlamydomonas reinhardtii, and the plant Arabidopsis thaliana. The sequences were aligned with hmmalign (HMMER package) using the PF00504-derived profile HMM as the template and the --trimm option to remove the unaligned flanking (non-conserved) regions. The phylogenetic reconstruction was performed using the ETE3 3.1.2 pipeline72 as implemented on the GenomeNet (https://www.genome.jp/tools/ete/). Columns with more than twenty percent of gaps were removed from the alignment using trimAl v1.4.rev673 and a maximum likelihood tree was inferred using IQ-TREE 1.5.5 ran with ModelFinder and tree reconstruction74, with PMB + F + R5 selected as the best-fit substitution model according to BIC. Branch support was tested by SH-like aLRT with 1000 replicates. For presentation purposes, the tree was visualised and adjusted using iTOL75.
Optically detected magnetic resonance
ODMR spectra were acquired in a home-built set-up described in detail previously45,76. In short, the light from a halogen lamp (250 W, Philips) were focused on the sample cell, which was immersed in a bath helium cryostat (all measurements were carried out at a temperature of 1.8 K), after being filtered through either a 5 cm CuSO4 solution (FDMR spectra) or a 10 cm water filter (Triplet minus Singlet, T-S, absorption-detected spectra). In FDMR experiments, the fluorescence was detected through bandpass filters (characterised by a full width at half maximum of about 10 nm) using a photodiode placed at 90° with respect to the excitation light direction, while in absorption-detected experiments, the light transmittance was detected with straight geometry through a monochromator (Jobin Yvon, mod. HR250). By sweeping the microwave frequency (MW source HP8559b, sweep oscillator equipped with a HP83522s plug-in and amplified by a TWT Sco-Nucletudes mod 10-46-30 amplifier) while detecting the fluorescence changes at specific wavelengths, the resonance transitions between spin sublevels of the triplet states can be determined. The microwaves were on/off amplitude modulated for selective amplification and the signal from the detector was demodulated and amplified using a Lock-In amplifier (EG&G, mod 5210). The microwave resonator, where the sample cell is inserted, consists of a slow pitch helix. FDMR spectra are presented as ΔF/F versus microwave frequency, where ΔF is the fluorescence change induced by the resonant microwave field and F is the steady-state fluorescence.
Microwave-induced T-S spectra can be collected by fixing the microwave frequency at a resonant value and sweeping the absorption detection wavelength. Compared to optical time-resolved absorbance spectroscopy on the triplet state, the ODMR technique allows selection (by the resonant microwave field) of specific triplet populations present in the sample, and in this way well resolved T-S spectra associated with specific chromophores can be obtained.
Pulse and time-resolved EPR
The rVCP samples were concentrated to a concentration of about 350 μg/mL of Chl a. Glycerol, previously degassed by several cycles of freezing and pumping, was added (60% v/v) just before freezing to obtain a transparent matrix. The sample of Chl a in Triton X-100 micelles was obtained by adding a few microliters of a concentrated solution of the pigment (SIGMA) dissolved in methanol to 1 ml of 1 mM Triton X-100, as previously reported48.
Pulse EPR experiments were performed on a Bruker ELEXSYS E580 spectrometer, equipped with a dielectric cavity (Bruker ER 4117-DI5, TE011 mode), an Oxford CF935 liquid helium flow cryostat, and an Oxford ITC4 temperature controller. The microwave frequency was measured using a frequency counter (HP5342A). The temperature was controlled in a helium-flow and all experiments were conducted at 50 K, disabling magnetic field modulation and using pulsed sample photo-excitation from a Nd:YAG pulsed laser (Quantel Brilliant) equipped with second and third harmonic modules and an optical parametric oscillator (OPOTECH) (λ = 440 nm, pulse length = 5 ns, E/pulse ≅ 1.5 mJ, 10 Hz repetition time). The pulse EPR experiments were recorded using a two-pulse Electron Spin Echo (ESE) sequence (flash-delay after flash-π/2-τ-π-τ-echo), where the echo intensity was registered as a function of the magnetic field. The microwave π/2-pulse was of 16 ns and the delay τ was set at 300 ns.
The TR-EPR spectra were performed on the same instrument, where the EPR direct-detected signal was recorded with a LeCroy 9300 digital oscilloscope, triggered by the laser pulse. The temperature was controlled in a helium-flow and all experiments were conducted at 50 K and 120 K. For every field position, 300 transient signals were averaged. To eliminate the laser background signal, transients accumulated at off-resonance field positions were subtracted from those on resonance.
Simulations of the powder spin-polarised triplet spectra were performed using a program written in Matlab®, with the aid of the EasySpin routine (ver. 5.2.25)77, based on the full diagonalisation of the triplet state spin Hamiltonian, including the Zeeman and electron-electron magnetic dipole interactions, considering a powder distribution of molecular orientations with respect to the magnetic field direction. Input parameters are the triplet state sublevel populations, the zfs parameters, the linewidth, and the isotropic g value.
Calculations of the sublevel triplet state populations of the acceptor (Car), starting from those of the donor (Chl), were performed using a home-written program in Matlab® software previously described in great detail6,43,51, utilising the X-ray coordinates of Pt FCP10. In short, the orientation of the zfs axes of the Chls a molecules are considered passing through the methines bridging the pyrrole rings, following Vrieze et al.78, whereas, in case of the carotenoids, the zfs axes are identified using principal components analysis, using the MatLab function named “pca”. In fact, due to the orientation of the Car’s zfs axes, the solution of the covariance matrix of the position of the carbon atoms constituting the conjugated system gives the axes Z, X, and Y (in this order of decreasing eigenvalue). The relationship between the populations of acceptor (A) and donor (D), in zero magnetic field, is the following: \({{{{\rm{P}}}}}_{i}^{{{{\rm{A}}}}}=\,{\sum}_{j}\cos {\theta }_{{ij}}^{2}{{{{\rm{P}}}}}_{j}^{{{{\rm{D}}}}}\), where \({\theta }_{{ij}}\) is the angle between the principal ZFS axis j of the donor (D) and the axis i of the acceptor (A). The relative population rates of the acceptor, which actually determine the shape of the spin-polarised triplet spectra, can be calculated directly from the above reported equation, using the relative \({{{{\rm{P}}}}}_{j}^{{{{\rm{D}}}}}\) values (Px:Py:Pz =0.375:0.425:0.200)5.
Dipole coupling calculation
The inter-pigment coupling was computed using a point-dipole approximation. The direction of the Chl a transition dipole was assumed to correspond to the NB-ND direction (in the pdb notation), following Madjet et al. 79 in defining the direction of the Qy transition dipole moments by the position of the nitrogen atoms of the pyrrole rings A and C of the Chl a molecular structure. The effective size of the dipole was taken to be 4 D with dielectric constant of the environment set to 1. For details see the code deposited at https://github.com/dbina/CDC and retrievable at https://doi.org/10.5281/zenodo.13939211.
Visualization
OriginPro 2015 (OriginLab Corporation) was used for plotting the graphs. All structure figures were prepared using PyMol 0.99 (The PyMOL Molecular Graphics System, http://www.pymol.org).
Statistics and reproducibility
Each subsection of the methods contains detailed explanations of various statistical models used to test the veracity of the data presented in this paper.
Data availability
Raw sequencing reads from Trachydiscus minutus CCALA 838 are available as NCBI BioProject PRJNA1078143. The primary T. minutus transcriptome assembly, the full set of protein sequences inferred from it, an alternative transcriptome assembly and a draft genome assembly from the same species, and sequence data used to reconstruct the LHC phylogeny are available from Figshare (https://doi.org/10.6084/m9.figshare.25706553). The mass spectrometry proteomics data have been deposited to the MassIVE repository with the dataset identifier MSV000096023. All data that support the findings of this study are available in the article, Supplementary information and Supplementary Data files. Source data for graphs can be found in Supplementary Data 2.
Code availability
The code employed to calculate the inter-pigment coupling is available at https://github.com/dbina/CDC and retrievable at https://doi.org/10.5281/zenodo.13939211.
References
Green, B. R. & Kühlbrandt, W. Sequence conservation of light-harvesting and stress-response proteins in relation to the three-dimensional molecular structure of LHCII. Photosynth. Res. 44, 139–148 (1995).
Liu, Z. et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 (2004).
Renger, T., Madjet, M. E., Knorr, A. & Müh, F. How the molecular structure determines the flow of excitation energy in plant light-harvesting complex II. J. Plant Physiol. 168, 1497–1509 (2011).
Novoderezhkin, V., Marin, A. & van Grondelle, R. Intra- and inter-monomeric transfers in the light harvesting LHCII complex: the Redfield–Förster picture. Phys. Chem. Chem. Phys. 13, 17093 (2011).
Carbonera, D. et al. Photoprotective sites in the violaxanthin–chlorophyll a binding Protein (VCP) from Nannochloropsis gaditana. Biochim. Biophys. Acta - Bioenerg. 1837, 1235–1246 (2014).
Di Valentin, M., Biasibetti, F., Ceola, S. & Carbonera, D. Identification of the sites of chlorophyll triplet quenching in relation to the structure of LHC-II from higher plants. Evidence from EPR spectroscopy. J. Phys. Chem. B 113, 13071–13078 (2009).
Lampoura, S. S., Barzda, V., Owen, G. M., Hoff, A. J. & Van Amerongen, H. Aggregation of LHCII leads to a redistribution of the triplets over the central xanthophylls in LHCII. Biochemistry 41, 9139–9144 (2002).
Mozzo, M., Dall’Osto, L., Hienerwadel, R., Bassi, R. & Croce, R. Photoprotection in the antenna complexes of photosystem II: Role of individual xanthophylls in chlorophyll triplet quenching. J. Biol. Chem. 283, 6184–6192 (2008).
Agostini, A. et al. Altering the exciton landscape by removal of specific chlorophylls in monomeric LHCII provides information on the sites of triplet formation and quenching by means of ODMR and EPR spectroscopies. Biochim. Biophys. Acta Bioenerg. 1862, 148481 (2021).
Wang, W. et al. Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 363, eaav0365 (2019).
Su, X. et al. Antenna arrangement and energy transfer pathways of a green algal photosystem-I–LHCI supercomplex. Nat. Plants 5, 273–281 (2019).
Pi, X. et al. Unique organization of photosystem I–light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc. Natl Acad. Sci. 115, 4423–4428 (2018).
Papagiannakis, E., H.M. van Stokkum, I., Fey, H., Büchel, C. & van Grondelle, R. Spectroscopic Characterization of the Excitation Energy Transfer in the Fucoxanthin–Chlorophyll Protein of Diatoms. Photosynth. Res. 86, 241–250 (2005).
Gundlach, K., Werwie, M., Wiegand, S. & Paulsen, H. Filling the “green gap” of the major light-harvesting chlorophyll a/b complex by covalent attachment of Rhodamine Red. Biochim. Biophys. Acta Bioenerg. 1787, 1499–1504 (2009).
Hancock, A. M. et al. Enhancing the spectral range of plant and bacterial light-harvesting pigment-protein complexes with various synthetic chromophores incorporated into lipid vesicles. J. Photochem. Photobiol. B Biol. 237, 112585 (2022).
Miyashita, H. et al. Chlorophyll d as a major pigment. Nature 383, 402–402 (1996).
Chen, M. et al. A Red-Shifted Chlorophyll. Science 329, 1318–1319 (2010).
Elias, E., Liguori, N., Saga, Y., Schäfers, J. & Croce, R. Harvesting Far-Red Light with Plant Antenna Complexes Incorporating Chlorophyll d. Biomacromolecules 22, 3313–3322 (2021).
Ort, D. R. et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl Acad. Sci. 112, 8529–8536 (2015).
Walter, J. & Kromdijk, J. Here comes the sun: How optimization of photosynthetic light reactions can boost crop yields. J. Integr. Plant Biol. 64, 564–591 (2022).
Wolf, B. M. & Blankenship, R. E. Far-red light acclimation in diverse oxygenic photosynthetic organisms. Photosynth. Res. 142, 349–359 (2019).
Kotabová, E. et al. Novel type of red-shifted chlorophyll a antenna complex from Chromera velia. I. Physiological relevance and functional connection to photosystems. Biochim. Biophys. Acta Bioenerg. 1837, 734–743 (2014).
Bína, D. et al. Novel type of red-shifted chlorophyll a antenna complex from Chromera velia: II. Biochemistry and spectroscopy. Biochim. Biophys. Acta Bioenerg. 1837, 802–810 (2014).
Litvín, R. et al. Red-shifted light-harvesting system of freshwater eukaryotic alga Trachydiscus minutus (Eustigmatophyta, Stramenopila). Photosynth. Res. 142, 137–151 (2019).
Wolf, B. M. et al. Characterization of a newly isolated freshwater Eustigmatophyte alga capable of utilizing far-red light as its sole light source. Photosynth. Res. 135, 177–189 (2018).
Niedzwiedzki, D. M., Wolf, B. M. & Blankenship, R. E. Excitation energy transfer in the far-red absorbing violaxanthin/vaucheriaxanthin chlorophyll a complex from the eustigmatophyte alga FP5. Photosynth. Res. 140, 337–354 (2019).
Morosinotto, T., Breton, J., Bassi, R. & Croce, R. The Nature of a Chlorophyll Ligand in Lhca Proteins Determines the Far Red Fluorescence Emission Typical of Photosystem I. J. Biol. Chem. 278, 49223–49229 (2003).
Romero, E. et al. The Origin of the Low-Energy Form of Photosystem I Light-Harvesting Complex Lhca4: Mixing of the Lowest Exciton with a Charge-Transfer State. Biophys. J. 96, L35–L37 (2009).
Passarini, F., Wientjes, E., van Amerongen, H. & Croce, R. Photosystem I light-harvesting complex Lhca4 adopts multiple conformations: Red forms and excited-state quenching are mutually exclusive. Biochim. Biophys. Acta Bioenerg. 1797, 501–508 (2010).
Basso, S. et al. Characterization of the photosynthetic apparatus of the Eustigmatophycean Nannochloropsis gaditana: Evidence of convergent evolution in the supramolecular organization of photosystem I. Biochim. Biophys. Acta Bioenerg. 1837, 306–314 (2014).
Keşan, G. et al. Efficient light-harvesting using non-carbonyl carotenoids: Energy transfer dynamics in the VCP complex from Nannochloropsis oceanica. Biochim. Biophys. Acta Bioenerg. 1857, 370–379 (2016).
Sukenik, A., Livne, A., Apt, K. E. & Grossman, A. R. Characterization of a gene encoding the light-harvesting violaxanthin-chlorophyll protein of Nannochloropsis sp. (Eustigmatophyceae). J. Phycol. 36, 563–570 (2000).
Litvín, R., Bína, D., Herbstová, M. & Gardian, Z. Architecture of the light-harvesting apparatus of the eustigmatophyte alga Nannochloropsis oceanica. Photosynth. Res. 130, 137–150 (2016).
Přibyl, P., Eliáš, M., Cepák, V., Lukavský, J. & Kaštánek, P. Zoosporogenesis, morphology, ultrastructure, pigment composition, and phylogenetic position of Trachydiscus minutus (Eustigmatophyceae, Heterokontophyta). J. Phycol. 48, 231–242 (2012).
Bína, D. et al. Energy transfer dynamics in a red-shifted violaxanthin-chlorophyll a light-harvesting complex. Biochim. Biophys. Acta Bioenerg. 1860, 111–120 (2019).
Gundermann, K., Schmidt, M., Weisheit, W., Mittag, M. & Büchel, C. Identification of several sub-populations in the pool of light harvesting proteins in the pennate diatom Phaeodactylum tricornutum. Biochim. Biophys. Acta Bioenerg. 1827, 303–310 (2013).
Lepetit, B. et al. Spectroscopic and Molecular Characterization of the Oligomeric Antenna of the Diatom Phaeodactylum tricornutum. Biochemistry 46, 9813–9822 (2007).
Herbstová, M. et al. Molecular basis of chromatic adaptation in pennate diatom Phaeodactylum tricornutum. Biochim. Biophys. Acta Bioenerg. 1847, 534–543 (2015).
Wang, L. et al. LHCF15 facilitates the absorption of longer wavelength light and promotes growth of Phaeodactylum tricornutum under red light. Algal Res. 75, 103249 (2023).
Wientjes, E., Roest, G. & Croce, R. From red to blue to far-red in Lhca4: How does the protein modulate the spectral properties of the pigments? Biochim. Biophys. Acta Bioenerg. 1817, 711–717 (2012).
Sláma, V. et al. Origin of Low-Lying Red States in the Lhca4 Light-Harvesting Complex of Photosystem I. J. Phys. Chem. Lett. 14, 8345–8352 (2023).
Agostini, A., Bína, D., Carbonera, D. & Litvín, R. Conservation of triplet-triplet energy transfer photoprotective pathways in fucoxanthin chlorophyll-binding proteins across algal lineages. Biochim. Biophys. Acta Bioenerg. 1864, 148935 (2023).
Agostini, A., Büchel, C., Di Valentin, M. & Carbonera, D. A distinctive pathway for triplet-triplet energy transfer photoprotection in fucoxanthin chlorophyll-binding proteins from Cyclotella meneghiniana. Biochim. Biophys. Acta Bioenerg. 1862, 148310 (2021).
Paulsen, H., Rümler, U. & Rüdiger, W. Reconstitution of pigment-containing complexes from light-harvesting chlorophyll a/b-binding protein overexpressed in Escherichia coli. Planta 181, 204–211 (1990).
Carbonera, D., Giacometti, G. & Agostini, G. FDMR of Carotenoid and Chlorophyll triplets in light-harvesting complex LHCII of spinach. Appl. Magn. Reson. 3, 859–872 (1992).
Khoroshyy, P. et al. Quenching of chlorophyll triplet states by carotenoids in algal light-harvesting complexes related to fucoxanthin-chlorophyll protein. Photosynth. Res. 135, 213–225 (2018).
Di Valentin, M. et al. Triplet-triplet energy transfer in Peridinin-Chlorophyll a-protein reconstituted with Chl a and Chl d as revealed by optically detected magnetic resonance and pulse EPR: Comparison with the native PCP complex from Amphidinium carterae. Biochim. Biophys. Acta Bioenerg. 1787, 168–175 (2009).
Di Valentin, M. et al. Triplet–triplet energy transfer in the major intrinsic light-harvesting complex of Amphidinium carterae as revealed by ODMR and EPR spectroscopies. Biochim. Biophys. Acta Bioenerg. 1797, 1759–1767 (2010).
Migliore, A., Corni, S., Agostini, A. & Carbonera, D. Unraveling the electronic origin of a special feature in the triplet-minus-singlet spectra of carotenoids in natural photosystems. Phys. Chem. Chem. Phys. 25, 28998–29016 (2023).
Carbonera, D., Agostini, G., Morosinotto, T. & Bassi, R. Quenching of chlorophyll triplet states by carotenoids in reconstituted Lhca4 subunit of peripheral light-harvesting complex of photosystem I. Biochemistry 44, 8337–8346 (2005).
Di Valentin, M. et al. Triplet–triplet energy transfer in fucoxanthin-chlorophyll protein from diatom Cyclotella meneghiniana: Insights into the structure of the complex. Biochim. Biophys. Acta Bioenerg. 1827, 1226–1234 (2013).
Di Valentin, M., Ceola, S., Salvadori, E., Agostini, G. & Carbonera, D. Identification by time-resolved EPR of the peridinins directly involved in chlorophyll triplet quenching in the peridinin–chlorophyll a–protein from Amphidinium carterae. Biochim. Biophys. Acta Bioenerg. 1777, 186–195 (2008).
Carbonera, D. et al. Energy transfer and spin polarization of the carotenoid triplet state in synthetic carotenoporphyrin dyads and in natural antenna complexes. Appl. Magn. Reson. 13, 487–504 (1997).
Bittl, R., Schlodder, E., Geisenheimer, I., Lubitz, W. & Cogdell, R. J. Transient EPR and Absorption Studies of Carotenoid Triplet Formation in Purple Bacterial Antenna Complexes. J. Phys. Chem. B 105, 5525–5535 (2001).
Kay, C., Elger, G. & Mobius, K. The photoexcited triplet state of free-base porphycene: a time-resolved EPR and electron spin echo investigation. Phys. Chem. Chem. Phys. 1, 3999–4002 (1999).
Tichy, J. et al. Light harvesting complexes of Chromera velia, photosynthetic relative of apicomplexan parasites. Biochim. Biophys. Acta Bioenerg. 1827, 723–729 (2013).
Harpaz, Y., Gerstein, M. & Chothia, C. Volume changes on protein folding. Structure 2, 641–649 (1994).
Remelli, R., Varotto, C., Sandonà, D., Croce, R. & Bassi, R. Chlorophyll binding to monomeric light-harvesting complex. A mutation analysis of chromophore-binding residues. J. Biol. Chem. 274, 33510–33521 (1999).
Herbstová, M., Bína, D., Kaňa, R., Vácha, F. & Litvín, R. Red-light phenotype in a marine diatom involves a specialized oligomeric red-shifted antenna and altered cell morphology. Sci. Rep. 7, 11976 (2017).
Guillard, R. R. L. & Lorenzen, C. J. Yellow-green algae with chlorophyllide c. J. Phycol. 8, 10–14 (1972).
Bischoff, H. W. & Bold, H. C. Phycological studies IV. Some soil algae from Enchanted Rock and related algal species. Univ. Tex. Publ. 6318, 1–95 (1963).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Haas, B. J. https://github.com/TransDecoder/TransDecoder.
Premvardhan, L., Robert, B., Beer, A. & Büchel, C. Pigment organization in fucoxanthin chlorophyll a/c2 proteins (FCP) based on resonance Raman spectroscopy and sequence analysis. Biochim. Biophys. Acta Bioenerg. 1797, 1647–1656 (2010).
Bowler, C. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 (2008).
Katoh, K. & Standley, D. M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780 (2013).
Eddy, S. R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 7, e1002195 (2011).
Yurchenko, T. et al. A gene transfer event suggests a long-term partnership between eustigmatophyte algae and a novel lineage of endosymbiotic bacteria. ISME J. 12, 2163–2175 (2018).
Alboresi, A. et al. Conservation of core complex subunits shaped the structure and function of photosystem I in the secondary endosymbiont alga Nannochloropsis gaditana. N. Phytol. 213, 714–726 (2017).
Kumazawa, M. et al. Molecular phylogeny of fucoxanthin‐chlorophyll a/c proteins from Chaetoceros gracilis and Lhcq/Lhcf diversity. Physiol. Plant. 174, e13598 (2022).
Huerta-Cepas, J., Serra, F. & Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 33, 1635–1638 (2016).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res 52, W78–W82 (2024).
Agostini, A., Palm, D. M., Paulsen, H. & Carbonera, D. Optically Detected Magnetic Resonance of Chlorophyll Triplet States in Water-Soluble Chlorophyll Proteins from Lepidium virginicum: Evidence for Excitonic Interaction among the Four Pigments. J. Phys. Chem. B 122, 6156–6163 (2018).
Stoll, S. & Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 178, 42–55 (2006).
Vrieze, J. & Hoff, A. J. The orientation of the triplet axes with respect to the optical transition moments in (bacterio) chlorophylls. Chem. Phys. Lett. 237, 493–501 (1995).
Madjet, M. E., Abdurahman, A. & Renger, T. Intermolecular coulomb couplings from ab initio electrostatic potentials: Application to optical transitions of strongly coupled pigments in photosynthetic antennae and reaction centers. J. Phys. Chem. B 110, 17268–17281 (2006).
Wang, J. et al. Structure of plant photosystem I−light harvesting complex I supercomplex at 2.4 Å resolution. J. Integr. Plant Biol. 63, 1367–1381 (2021).
Acknowledgements
The skilled technical assistance of Ivana Hunalová, František Matoušek, and Sabrina Mattiolo is gratefully acknowledged. We extend our thanks to Peter Konik for his work on the proteomic identification of LHC proteins. The Czech Science Foundation (20-01159S to D. Bína and 21-19664S to M.E.), the MEMOVA project (EU Operational Programme Research, Development and Education No. CZ.02.2.69/0.0/0.0/18_053/0016982), the University of Padova (P-DiSC#02BIRD2020-UNIPD to M.B.), and the MUR (support to D.C and A.A. under the National Recovery and Resilience Plan (NRRP/PNRR), Mission 4, Component C2, Investment 1.1, Call for tender No. 104 of the 02/02/2022 by the Italian Ministry of University and Research (MUR), funded by the European Union –NextGenerationEU– Project Title “Extending the red limit of oxygenic photosynthesis: basic principles and implications for future applications (20224HJWMH)” – CUP C53D23004620006 Grant Assignment Decree No. 1017 adopted on 07/07/2023 by MUR) financially supported this work. Institutional support RVO: 60077344 is also acknowledged.
Author information
Authors and Affiliations
Contributions
A.A., D. Bína, and R.L. conceived and designed the research and coordinated the project. D. Bína extracted and purified rVCP and analysed its composition by tandem mass spectrometry. A.A., M.B., and D.C. performed the EPR and ODMR experiments and analysed the data. D. Barcytė and M.E. obtained the T. minutus transcriptome. A.A., D. Bína, D. Barcytė, and M.E. analysed the LHCs polypeptide sequences. A.A. wrote the original draft of the paper. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Emilie Wientjes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Xiaoling Xu and David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Agostini, A., Bína, D., Barcytė, D. et al. Eustigmatophyte model of red-shifted chlorophyll a absorption in light-harvesting complexes. Commun Biol 7, 1406 (2024). https://doi.org/10.1038/s42003-024-07101-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07101-9