Abstract
Cyclic di-adenylate monophosphate (cyclic di-AMP) is an important second messenger in microorganisms. Cyclic di-AMP regulates bacterial cell volume and turgor via control of potassium and compatible solute transport but is also involved in many other processes, including the activation of the metazoan innate immune response to bacterial infections. We compare the activity of full-length membrane-embedded CdaA, the enzyme that synthesizes cyclic di-AMP, with the water-soluble catalytic domain CdaA-DAC. Purified CdaA from L. lactis was studied in the detergent-solubilized state, and in lipid nanodiscs and vesicles. We show that CdaA is tetrameric and the membrane-bound complex has more than 2-orders of magnitude higher activity than soluble CdaA-DAC. CdaA activity increases with pH but does not strongly depend on the salt or lipid content, factors that are crucial for the control of osmoregulatory transporters. Cryo-EM and in-silico structure prediction of CdaA show that the two DAC dimers engage in a head-to-head interaction, leading to cyclic-di-AMP formation. The inhibitor phosphoglucomutase prevents this active conformation. We observe dynamic flexibility between the catalytic and membrane domains, even in the presence of ATP or non-hydrolyzable substrate ApCpp. This is the first comprehensive functional and structural characterization of a full-length cyclic di-AMP-specific cyclase.
Similar content being viewed by others
Introduction
Bacteria flourish in diverse, fluctuating environments and cope with rapid variations in e.g. external osmolality. They have acquired mechanisms to counter or adapt to these fluctuations by adjusting the activity of enzymes and membrane transporters and thereby tune the amounts of cytoplasmic osmolytes. Among these mechanisms, signalling pathways with cyclic nucleotide-based second messengers, like cyclic di-AMP, take a prominent role in the response and adaptation to environmental challenges. Cyclic di-AMP has been established as a master regulator of bacterial cell volume due to its ability to control the activity of ion and compatible solute transporters1. In general, binding of cyclic di-AMP to potassium and compatible solute importers down-regulates their activity, whereas binding of cyclic di-AMP to potassium exporters up-regulates their activity. Thus, cyclic di-AMP prevents over-accumulation of abundant osmolytes. During osmotic upshift (hyperosmotic conditions) cyclic di-AMP levels decrease, and as a result, potassium and compatible solute import increases to allow the cytoplasm to rehydrate and increase in volume. If compatible solutes are not available in the extracellular environment some can also be synthesized de novo, however at a greater energetic cost and at longer timescales than by active transport.
Cyclic di-AMP is synthesized by specific diadenylate cyclases (DACs) from two ATP molecules. CdaA (also referred to as DacA in some bacteria) is the most ubiquitous cyclase, as it is found in most bacterial phyla and present in almost all Firmicutes2. It is a membrane protein consisting of (i) an extracellular N-terminal region that may interact with wall components; (ii) three transmembrane helices; (iii) a short amphipathic linker region; (iv) an intracellular catalytic DAC domain; and (iv) a flexible cationic C-terminal region.
While the truncated, soluble catalytic DAC domain of CdaA (CdaA-DAC) has been characterised, both structurally and biochemically, the full-length membrane protein has not. Studies of CdaA-DAC from L. monocytogenes3,4,5, B. subtilis6,7, S. aureus8 and L. lactis9 allowed (i) a structural description of the active site and the identification of the residues required for metal ion coordination and substrate binding4,5,10,11; showed (ii) that the enzyme activity is dependent on the presence of Mn2+, Co2+ or Mg2+ and inhibited by the phosphoglucomutase GlmM; and suggested (iii) that the stable, inactive dimer requires transient formation of higher order oligomeric structures for activity. This leaves open the key questions: how is the activity of CdaA controlled and why is CdaA a membrane-embedded enzyme?
Here, we present the first structural and biochemical analysis of a full-length CdaA-type cyclase from Lactococcus lactis. We contrast our findings to the truncated form, in which the transmembrane helices have been removed (CdaA-DAC), to facilitate a comparison with published work and to extract the importance of membrane localisation on the oligomeric state of the protein, its enzymatic activity and regulation. We reconstituted CdaA in lipid nanodiscs and lipid vesicles (proteoliposomes), and analysed the activity at various lipid compositions, and divalent ions, ionic strength and pH conditions. Attempts to determine a high-resolution structure of full-length CdaA were hindered by the inherent inter-domain flexibility. However, a combination of our cryo-EM data with the computational prediction tool Alphafold12,13 provide structural insights into CdaA topology.
Results
Expression and purification of CdaA and CdaA-DAC
We characterised the full-length membrane-bound CdaA protein (llmg0448 – aa. 0 to 293) and a truncated soluble version of the protein in which the transmembrane helices were removed (hereafter referred to as CdaA-DAC – aa. 98 to 293) of L. lactis (subsp. cremoris NZ9000) (Fig. 1A). Attempts to purify stable full-length CdaA from E. coli was unsuccessful, as the majority of the protein could not be recovered from the insoluble fraction. Instead, we constructed a nisin-inducible cdaA expression vector for the endogenous host, L. lactis, where a 10x histidine-tag for metal affinity chromatography and a 3 C cleavage site for subsequent elution were introduced to the C-terminus. No cytotoxic effects of cdaA overexpression were observed during pH-controlled growth (Supplementary Fig. 1A).
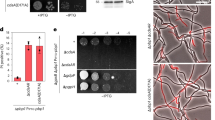
A Schematic of full-length CdaA. The protomers of full-length CdaA and CdaA-DAC are, respectively, 300 amino acids (33.3 kDa) and 196 amino acids (22.4 kDa) in length without the histidine tag. B Representative SEC analysis of purified CdaA, with a peak fraction at 10.5 to 11 mL elution volume, plus pop-out showing a representative SDS-PAA gel (full SDS-PAA gel in Supplementary Fig. 2A). C Representative SEC analysis of purified CdaA-DAC, with a peak fraction at 13.5 to 14.5 mL elution volume, plus pop-out showing a representative SDS-PAA gel (full SDS-PAA gel in Supplementary Fig. 2B). M represents the protein marker (kDa) and E represents the elution fraction used for SEC analysis. D SEC-MALLS analysis of DDM-solubilised CdaA in 50 mM HEPES-NaOH pH 7.5, 200 mM KCl, 5% (w/v) glycerol plus 0.04% (w/w) DDM. For SEC-MALLS, the red line represents weighted-average mass of the CdaA protein (calculated to be 135 kDa) and its size distribution over the peak; similarly, the blue line represents the weighted-average mass of the DDM-bound to CdaA, calculated to be 100 kDa; and the grey line represents the weighted-average mass of the DDM detergent micelle-protein complex calculated to be 235 kDa; black line represents UV absorbance at 280 nm.
Next, we performed a detergent screen to optimise the CdaA solubilisation and stability. We tested the detergents DDM, DM, LMNG, DMNG and CYMAL-5/6, which differ in carbon chain length, branching, micelle size and critical micellar concentration, with DDM showing the best results (Fig. 1B, Supplementary Fig. 1C). We purify approximately 1 mg of monodisperse CdaA from 120 mg of membrane vesicles, corresponding to 10 L of L. lactis cells (A600nm ~ 7). The molecular weight of CdaA estimated from the SDS-PAA gel deviated from the expected monomeric size of 33 kDa, which is not unexpected for membrane proteins as they often do not unfold completely in the presence of SDS (Fig. 1B, Supplementary Fig. 2A). We then performed size-exclusion chromatography (SEC) and find that the elution volume of CdaA is suggestive of a higher order complex (dimer or tetramer).
By contrast to full-length CdaA, we succeeded in expressing and purifying CdaA-DAC from E. coli using an IPTG-inducible expression vector (Fig. 1C, Supplementary Fig. 2B). We purify approximately 200 mg of CdaA-DAC from 6 L of E. coli cells. A 10x histidine-tag and 3 C cleavage site were introduced to the C-terminus of the protein. The molecular weight of CdaA-DAC estimated from the SDS-PAA gel matches the predicted monomeric size of 22 kDa, corroborating with the later observed SEC elution volume, suggestive of a dimeric state (Fig. 1C). Aggregated protein in the void volume suggests that CdaA-DAC is less stable than full-length CdaA. For subsequent activity assays and cryo-EM analysis, the affinity tags from both CdaA and CdaA-DAC were removed. We did this to eliminate the possibility of artefacts e.g. higher-order oligomeric structures driven by the presence of the affinity tag14.
Membrane-embedded CdaA forms a tetramer
To investigate the oligomeric state of the membrane-embedded full-length CdaA, we performed SEC-MALLS (Size-exclusion chromatography multi-angle laser light scattering) analysis. SEC-MALLS determines the absolute molar mass and size of a protein complex in solution, by accounting for the mass of surrounding detergent, which varies for each membrane protein and type of amphiphile used. To ensure a monodisperse sample, we used the peak elution fractions of purified CdaA and removed the affinity tag for SEC-MALLS analysis. We obtain a protein mass of ~135 kDa for CdaA (Fig. 1D), which almost perfectly corresponds to a tetrameric structure (predicted tetrameric mass is 132 kDa).
Membrane-embedded CdaA is two orders of magnitudes more active than CdaA-DAC
Membrane proteins are often unstable when removed from their native lipid environment, and their structure and activity can be influenced by lipids. To better mimic a native-like environment we reconstituted purified CdaA into lipid nanodiscs using the scaffold protein MSP1E3D1. If not otherwise stated, we used a synthetic lipid mixture composed of DOPC (12 mol%), DOPE (50 mol%), DOPG (38 mol%), which supports high activity of several membrane proteins tested in our laboratory15, and we find a molar ratio of CdaA:scaffold protein:lipid of 1:2:40 to be optimal. An efficient separation of the CdaA-containing nanodiscs from empty nanodiscs was obtained via a second metal-affinity chromatography step, followed by elution using 3 C cleavage (Supplementary Fig. 3).
Functional activity assays of the water-soluble CdaA-DAC were conducted in absence of detergent or nanodiscs. The concentrations of CdaA and CdaA-DAC used in the following activity assays were calculated assuming that the active structure is tetrameric, as determined by our SEC-MALLS analysis. Remarkably, membrane-embedded full-length CdaA shows more than two orders of magnitude higher activity than soluble CdaA-DAC, which bares only the catalytic domains (Fig. 2A, B). We then determined the apparent Km of CdaA (Fig. 2A) and CdaA-DAC (Fig. 2B) for ATP, revealing a similar binding affinity for both constructs, with a Km of CdaA for ATP of 173 (± 19 µM SD, R2 of 0.96), and of CdaA-DAC of 174 (± 16 µM SD, R2 of 0.90). The Hill coefficients (n) for CdaA (n = 1.9 ± 0.3) and CdaA-DAC (n = 2.8 ± 0.6) indicate cooperativity between protein subunits, most likely between two dimeric DAC domains. Unless stated otherwise, all subsequent assays were performed at an ATP concentration of 0.4 mM.
Activity assays were performed using 1 μM and 16 μM (monomer concentration) of CdaA and CdaA-DAC, respectively; kcat values were calculated on the assumption that the active enzyme is tetrameric). Buffer was composed of 50 mM HEPES-NaOH pH 8.0, 1 mM MnCl2 plus 200 mM KCl in a final reaction volume of 0.2 or 0.3 mL. Bars, lines and axes coloured red and blue correspond to full-length CdaA and CdaA-DAC, respectively. All error bars represent the standard error of the mean (SEM) for ≥3 biologically independent replicates. A ATP titration fitted with the Hill equation for CdaA. n = ≥3 biologically independent samples. B ATP titration fitted with the Hill equation for CdaA-DAC. n = ≥3 biologically independent samples. C Testing metal ion-dependent activity of CdaA and CdaA-DAC. Mn2+, Co2+, Mg2+, Ni2+ and Cu2+ were added to a final concentration of 1 mM. A negative control was performed using the same conditions but without the addition of any metal (not shown). For CdaA n = 3 and for CdaA-DAC n = 2 biologically independent samples. D pH-dependent activity of CdaA and CdaA-DAC. Buffer was composed of 25 mM MES-NaOH, 25 mM HEPES-NaOH, 1 mM MnCl2 plus 200 mM KCl set to the appropriate pH using 4 N NaOH. pHs above 8.0 were not tested due to formation of metal-based precipitates. For CdaA n = 3 and for CdaA-DAC n = 2 biologically independent samples. E Testing activity of CdaA dependent on the concentrations of ATP and divalent metal ion in buffer composed of composed of 50 mM HEPES-NaOH pH 8.0, 1 mM MnCl2, 200 mM KCl plus the indicated concentrations of MgCl2 and ATP. Experiments were also repeated at pH 6.5 with the same result. n = 3 biologically independent samples. F GlmM-dependent inhibition of CdaA and CdaA-DAC. 32 µM GlmM was added to 16 µM CdaA-DAC. 20 µM GlmM was added to 1 µM CdaA. For CdaA n = 3 and for CdaA-DAC n = 3 biologically independent samples.
Divalent metal ion preference, pH-dependence and inhibition by GlmM
Next, we determined the dependence of CdaA and CdaA-DAC activity on divalent metal ions, ionic strength, pH and presence of the inhibitory interaction partner GlmM. Despite, the difference in turnover rate, CdaA and CdaA-DAC show a similar dependency on Mn2+ and Co2+ (Fig. 2C). A low activity was also observed in the presence of 1 mM MgCl2, however, within the short time-course of our experiments a rate could not be accurately determined. No activity was observed with other metals tested: Ni2+, Zn2+ and Cu2+.
Figure 2D shows that CdaA and CdaA-DAC have the highest turnover at pH 8, with an activity drop of ~10-fold and ~7-fold at pH 6.5, respectively. We assume that the pH-dependence might be a result of a change in the protonation state of conserved histidine residues located in the active site. Supplementary Fig. 4 shows a sequence alignment of CdaA homologs from lactobacillaceae and the highly conserved His-183 in the L. lactis CdaA. The corresponding His-170 in L. monocytogenes CdaA-DAC has been shown to coordinate the divalent metal cation in the active site and be essential for enzyme activity5. The pKa of the γ-phosphate of ATP is ~6.516,17 and the availability of Mn-ATP2- relative to Mn-ATP1- increases by ~2-fold in the pH range from 6.5 to 8.0 (Supplementary Fig. 5A), which is less than the observed ~10-fold increase in kcat of CdaA. We thus conclude that the pH-dependent activity is a property of the enzyme rather than a change in substrate availability. All subsequent reactions were performed at pH 8.0.
Next, we investigated the effect of metal-chelating molecules on the enzymatic activity of CdaA, as they could potentially remove Mn2+ from the active site. PPi4- and ATP chelate metal ions to form (Metal3PO4)2 and ATP-Metal complexes, respectively. ATP has a dissociation constant for magnesium and manganese of ~71 µM18,19,20 and ~77 µM21, respectively (Supplementary Fig. 5B), while PPi4- has a dissociation constant for magnesium and manganese of ~26 µM and ~4 µM, respectively (Supplementary Fig. 5B). We determined the activity of CdaA as a function of ATP and MgCl2 (Fig. 2E). For this purpose, all reactions were performed in 50 mM HEPES-NaOH pH 8.0 with 1 mM MnCl2 plus 200 mM KCl, supplemented with varying concentrations of ATP and MgCl2. When ATP and MgCl2 were added at equimolar concentrations (both 10 mM) the catalytic rate decreased ~2-fold. When the MgCl2 concentration (10 mM) exceeded the ATP concentration (0.4 mM), the catalytic rate decreased ~2.5-fold. The reduction in activity in the presence of 10 mM MgCl2 likely represents some exchange of Mn2+ for Mg2+ in the active site of CdaA. If the ATP concentration (10 mM) exceeded the metal concentration (no MgCl2 added), the decrease in activity was much stronger. We propose that an excess of unbound ATP in the absence of Mg2+ chelates the available Mn2+ and thus remove the metal ion from the active site of CdaA, thereby reducing the activity of the enzyme. During synthesis of cyclic di-AMP, two ATP are consumed to form one cyclic di-AMP plus two PPi4-. However, given the catalytic rate of CdaA, the quantity of PPi4- formed in our time course assays is always negligible when compared to the concentration of divalent metal ion present. Only when the concentration of unbound chelating compounds (e.g., ATP) exceeds the concentration of divalent metal ions, synthesis of cyclic di-AMP is significantly inhibited. The intracellular concentrations of ATP22 and magnesium23 in bacteria are generally in the millimolar range, and most ATP has Mg2+ bound, hence, the scenario described heretofore is physiological less relevant.
Finally, we could reproduce the inhibitory effect of GlmM, which had been reported from studies on soluble DAC domains from other species3,7,9. Both, CdaA and CdaA-DAC are inhibited by GlmM, ~13-fold and ~7-fold, respectively (Fig. 2F).
Cryo-EM analysis of full-length CdaA in lipid nanodiscs
To gain a better understanding of the overall architecture and the catalytic mechanism of CdaA, we sought to resolve the structure of the tetrameric membrane-embedded CdaA in nanodiscs using cryo-electron microscopy. The 2D classes confirm the formation of the tetramer, both in the presence and absence of substrate and cofactors. Here, two membrane-embedded domains with flexible soluble domains, or alternatively two dimers of DAC domains can be visibly identified (Fig. 3A bottom, Supplementary Fig. 6, Supplementary Fig. 7). However, despite several image processing strategies, we were unable to resolve the structure of the full-length CdaA complex. We attribute this to the inherent flexibility in the position of the catalytic domains with respect to the transmembrane domains, which originates from the connecting region consisting of the α-helix ( ~ 16 amino acids) plus the linker ( ~ 8 amino acids) (Supplementary Fig. 4) (Fig. 3A).
A Alphafold 2 multimer prediction of full-length CdaA in surface representation. The side view of the predicted tetrameric full-length CdaA complex. The lipid environment is indicated with thick grey lines. The flexibility of each inactive DAC-dimer is indicated with arrows and transparent hemispheres. The bottom view highlights the opposing inactive DAC-dimers in proximity while attached to the transmembrane domain via a linker. The proximal active sites of the opposing DAC-dimers (teal) are facing towards each other. Representative 2D class averages of each stage are displayed below each panel. B Alphafold 3 prediction of the full-length CdaA in the presence of ligands (Mn-AMP). The ‘proximal’ active sites are engaged head-to-head orienting the two AMPs in a conformation that closely resembles cyclic di-AMP (dashed line). C Alphafold 2 multimer prediction of the full-length CdaA complex inhibited by GlmM. Representation is the same as in (A). The GlmM protomers are displayed in light brown and gray respectively. Representative 2D class averages of each stage are displayed below each panel. D Cryo-EM density map of the inactive DAC-dimer of CdaA bound to GlmM with the respective model (PDB:9G69) represented in ribbons. The two protomers of the catalytic DAC domain are coloured in teal and green respectively for clarity. Similarly, promoters of the GlmM complex are shown in light brown and light grey. The flexible domain 4 of GlmM is indicated in a grey ellipse. E A closer view of the inactive DAC-dimer of CdaA is displayed in ribbons. The active sites are situated in opposing sides of the inactive DAC-dimer (indicated by arrows). F A closer view of the active site of the catalytic domain (teal). The catalytic residues, D184GA and R216HR are coloured in yellow and orange respectively. A density (mesh) that corresponds to bound Mn-ATP is present in the active site (ligand fitted from homologue (PDB:4RV7) from L. monocytogenes).
In order to further interpret the empirical data obtained from the cryo-EM data, we made use of the newest AI-based structure predictions tools, including their ability to predict oligomers and protein-protein interactions12,13,24. To this end, we used AF2 for an in silico prediction of the tetrameric CdaA complex. The overall architecture of the predicted complex falls in line with the cryo-EM data and allow a more detail description of the complex. The transmembrane domain (TMD) forms a membrane-embedded tetrameric interface between the TMH-3 of each protomer (Supplementary Fig. 8). The TMH-1,2 enclose the tetrameric interface from each side with a characteristic V-shape. The four soluble catalytic DAC domains, in turn, form two ‘side-to-side’ dimers, which resemble the previously determined dimeric structure obtained for the soluble DAC-domains from B. subtilis7, L. monocytogenes4 and M. ovipneumoniae25 (Fig. 3A). These dimers are inactive and insufficient for the synthesis of cyclic-di-AMP, as their respective active centres face opposite sites (Fig. 3A, E). With respect to the position of the two DAC-dimers to each other and the transmembrane region, we name the catalytic sites facing the symmetry axis ‘proximal’ sites, and the two facing to the periphery the ‘distal’ sites (Fig. 3A). Intriguingly, AF3 structure prediction of CdaA with the addition of Mn-AMP resulted in a model displaying a head-to-head engagement of the ‘proximal’ active sites. The co-ordination of the two AMPs in the engaged ‘proximal’ active sites closely aligns with the formation of cyclic-di-AMP (Fig. 3B).
Cryo-EM analysis of inhibited full-length CdaA in lipid nanodiscs
In order to limit the movement of the catalytic domains we extended our cryo-EM analysis by incubating CdaA with different inhibitors at physiologically relevant conditions prior to sample preparation. We attempted to capture the complex of CdaA in the “substrate-bound” state by using the non-hydrolysable ATP analog ApCpp (α,β-Methyleneadenosine-5’-triphosphate). ApCpp has been shown to competitively inhibit cyclase activity8,10,26. However, the catalytic domains of CdaA show the same flexibility with respect to the transmembrane domains as observed before (Supplementary Fig. 7C, D). The small molecular weight of the DAC domains combined with their inherent flexibility prevented a reconstruction of the substrate-bound complex. A strategy to combat these pitfalls in cryo-EM is the use of binding partners, e.g. sybodies and nanobodies27, which increase the mass of the sample acting as fiducials and might limit the flexibility of the target protein/complex.
GlmM is a known inhibitor of CdaA, which has been reported to bind the DAC domains in a 1:1 stoichiometric ratio with each GlmM dimer (98 kDa) binding a dimer of DAC-domains7. In an attempt to limit the flexibility of the catalytic domains and increase their resolvability, samples of CdaA with Mn-ATP in the presence of GlmM were prepared. The addition of GlmM was already noticeable from the 2D classification/3D classification (Fig. 3C, Supplementary Fig. 9B). Further image processing resulted in a 4.8 Å global reconstruction of the soluble region of the complex showing the DAC dimer bound to a dimer of GlmM (Fig. 3D). The DAC domains appear to form the same catalytically inactive dimer as seen in the apo state, with their active centres facing opposite sites (Fig. 3E). These findings corroborate previous structural studies of GlmM and the soluble DAC domains from B. subtilis7, where the catalytic DAC domains were found to bind on the interface between the two GlmM dimers. Importantly, and for the first time seen in a DAC, a density corresponding to the bound ligand (ATP) is present in the active site of CdaA, signifying that ATP can bind upon inhibition of the complex by GlmM (Fig. 3F).
Ionic strength and lipid-dependence of CdaA
We established that the activity of membrane-bound CdaA is more than two orders of magnitude higher than that of soluble CdaA-DAC. This led us to a key question: does the membrane serve only as a platform to form a tetramer and thereby increase the local CdaA concentration and the concomitant higher probability of DAC active sites interacting to synthesize cyclic-di-AMP, or does CdaA form interactions with the membrane that serve a regulatory function, e.g., via changes in ionic strength as a consequence of osmotic stress or binding of specific membrane lipids? We have previously shown that the osmoregulatory membrane transport protein, OpuA, is activated in an ionic strength-dependent manner via protein-lipid interactions and that this interaction requires anionic lipids like PG (or PS) and cationic residues on the sensing domain of OpuA28,29,30. A multiple sequence alignment of the best-studied homologs of CdaA (Supplementary Fig. 4) shows that CdaA contains several conserved clusters of cationic residues.
We first tested the effect of salt or ionic strength on the activity of CdaA-containing nanodiscs composed of DOPC (12 mol%), DOPE (50 mol%), DOPG (38 mol%), and on the activity of soluble CdaA-DAC by varying the concentration of KCl. The activities of CdaA and CdaA-DAC show a light ionic strength-dependency. When the KCl concentration is lowered from 200 mM to 50 mM KCl, the activity drops 2-fold and 3-fold for CdaA and CdaA-DAC, respectively (Fig. 4A). In the assay buffer this corresponds to a decrease in ionic strength from ~0.25 to ~0.1 M. However, above 200 mM KCl, up to 700 mM KCl (ionic strength ~0.75 M), the activity of CdaA and CdaA-DAC is not significantly affected.
Proteoliposomes with 200 mM KCl inside and 50 mM KCl outside were used to simulate a hypotonic stress, and proteoliposomes with 50 mM KCl inside and 200 mM KCl outside simulate hypertonic stress. We emphasize that these vesicles do not maintain much of an osmotic pressure difference, and the hypertonicity will result in shape changes and invagination of the vesicles (30). We also compared these osmotic conditions with vesicles with equimolar concentrations of KCl inside and outside, either at 50 mM or 200 mM. All assays were performed using 1 μM CdaA. All error bars represent the SEM for ≥ 3 biologically independent replicates. A Ionic strength-dependent activity of CdaA and CdaA-DAC. Assays were performed using CdaA-containing nanodiscs composed of 38 mol% DOPG, 12 mol% DOPC plus 50 mol% DOPE in 50 mM HEPES-NaOH pH 8.0 containing 1 mM MnCl2 plus the indicated concentration of KCl. For CdaA n = 3 and for CdaA-DAC n = 2 biologically independent samples. B DOPG dependence of CdaA in nanodiscs. Nanodisc composition is the indicated DOPG mol% plus the required mol% DOPC, and the buffer composition was 50 mM HEPES-NaOH pH 8.0 containing 1 mM MnCl2 plus 200 mM KCl. n = 3 biologically independent samples. C DOPE dependence of CdaA in nanodiscs. Nanodisc composition is the indicated DOPE mol% plus the required mol% DOPC and the buffer composition was the same as for (B). n = 3 biologically independent samples. D Testing of osmotic effect on activity of CdaA-containing vesicles. The liposome assays only measures CdaA oriented inside-out, as shown in the pop-out where the grey circle represents the lipid vesicle. n = 3 biologically independent samples. E Testing of effect of membrane potential on activity of CdaA-containing vesicles. Membrane potential was generated using valinomycin-mediated K+ diffusion potential. For an inside-negative/outside-positive potential the vesicles were loaded with 200 mM KCl in buffer and suspended in buffer containing 200 mM NaCl, whereas for an inside-positive/outside-negative potential the liposomes were loaded with 200 mM NaCl and then suspended in buffer with 200 mM KCl. n = 3 biologically independent samples.
To study the effect of the lipid composition and that of the membrane surface charge on CdaA, we tested its activity reconstituted in nanodiscs composed of DOPC (zwitterionic, bilayer-forming lipid) only; DOPC plus DOPG (anionic bilayer-forming lipid) or DOPC plus DOPE (zwitterionic, non-bilayer-forming lipid) (Fig. 4B, C). We used a KCl concentration of 200 mM (ionic strength in assay ~0.25 M). The activity of CdaA decreases with an increasing fraction of DOPG (Fig. 4B), whereas the activity of CdaA increases with an increase in DOPE (Fig. 4C). However, the highest activity was observed in DOPC alone (100 mol%), which is a eukaryotic lipid and not found in bacteria. Therefore, anionic lipids have no important role in the synthesis of cyclic di-AMP, but the stimulation by the non-bilayer type of lipid phosphatidyl ethanolamine (DOPE) may be physiologically relevant.
We then determine the effect of osmotic pressure on CdaA. To this end we reconstituted CdaA into vesicles (proteoliposomes) composed of E. coli polar lipids/EggPC with varying concentrations of KCl inside and outside, and measured the response to osmotic stress of inside-out oriented CdaA. (Fig. 4D). While the measured differences between any of these conditions were small, all of the samples in which the catalytic site of CdaA was exposed to a higher concentration of KCl on the outside showed a higher activity. This corroborates our observation that increasing the concentration of KCl stimulates CdaA activity.
We also tested the activity of CdaA in lipid vesicles by loading them with different concentrations of KCl/NaCl in the presence of valinomycin to generate a membrane potential (inside negative or inside positive) (Fig. 4E). We did not find a significant effect of the membrane potential, but did see that exposure of the catalytic domain of CdaA to KCl stimulated the activity more than exposure to NaCl.
Discussion
We present the first functional and structural characterisation of a full-length membrane-embedded CdaA, the main enzyme for synthesis of the second messenger cyclic di-AMP. We used a truncated, soluble version of CdaA (named CdaA-DAC) to benchmark the role of the membrane domain on the cyclase activity of the protein. We note that each DAC domain binds one ATP, and that two DAC dimer domains have to interact in a head-to-head orientation for the synthesis of cyclic-di-AMP.
Among the most striking findings, we show that membrane-bound CdaA forms a tetrameric structure, which has more than 2-orders of magnitude higher activity than the soluble catalytic CdaA-DAC. We attribute the increase in activity to the net effects of localisation, orientational restriction and volume exclusion on the oligomeric state of CdaA31, factors that favour higher order oligomerization of membrane-associated proteins. Hence, the TMDs of CdaA function to constrain the catalytic domains from a 3D to 2D environment of the membrane, thereby increasing their interaction probability.
A combination of empirical cryo-EM data with in silico prediction tools allowed us to further describe the architecture of CdaA (Fig. 3, Supplementary Fig. 8, Supplementary Fig. 9). Despite the confinement effect via membrane association, the DAC domains are connected to the TMDs via a flexible linker and show a high level of flexibility relative to the position of the TMDs. We observe the formation of stable DAC dimers3,4,7,8, which bind side-to-side with the catalytic centres facing opposite sides. In this configuration the formed dimers are incapable of synthesizing cyclic di-AMP and require the association to another dimeric DAC. Since, the formed dimers are stable and the linker does not allow a 180-degree rotation of the DAC dimer, only an interaction of the proximal catalytic sites facing the symmetry axis will lead to the synthesis of cyclic-di-AMP within the tetramer (Fig. 3B, Fig. 5A, B). In solution, the equilibrium of soluble CdaA-DAC is towards the dimeric state and only a very small fraction of active tetramers may be formed3,4,7,8. Analogously, almost none of our cryo-EM datasets contained particles in which two catalytic DAC dimers are interacting, suggesting that the interaction is transient (Fig. 3A, right). Thus, we conclude that the observed flexibility of the catalytic domains likely allows sampling of the cellular environment for substrate capture or physicochemical signalling, while the membrane embedding creates a high local concentration of the catalytic domain dimers, increasing the rates of self-association required for efficient synthesis.
The transmembrane segments create an oligomerization interface that associates the two inactive DAC-dimers to the membrane. A The flexible inactive DAC-dimers can diffuse in relative proximity to the membrane and bind ATP. B The proximal active sites of the opposing inactive DAC dimers transiently engage in a head-head confirmation which enables the formation of c-di-AMP. C Inhibition by GlmM allosterically prevents the enzymatic activity by hindering the proximal active sites of the opposing DAC dimers to engage.
We could recapitulate the inhibitory effect of GlmM on the full-length membrane-embedded CdaA (Fig. 2F) and verify its interaction by cryo-EM, where each catalytic domain dimer is interacting with a dimer of GlmM3,7,9 (Fig. 3C, D). Interestingly, the catalytic DAC sites can bind ATP while bound to GlmM, indicating that inhibition by GlmM occurs by sterically preventing the opposing DAC dimers and the respective proximal catalytical sites to engage (Fig. 5C). Additionally, GlmM binding would also prevent the association of the distal catalytical sites to an adjacent CdaA. Thus, while the protein’s flexibility poses a problem for the structural analysis of the tetrameric complex, this feature is likely a necessity for its function (efficient substrate sampling) and its regulation via binding to GlmM (Fig. 5).
In L. monocytogenes, two truncated and soluble CdaA mutants have been investigated4, one mutant in which the N-terminal transmembrane regions were deleted (named Δ80CdaA), and one in which the transmembrane regions, linker sequence and adjacent amphipathic regions were deleted (named Δ100CdaA). The activity of Δ100CdaA was approximately 6-fold higher in activity than Δ80CdaA, indicating the possibility of a regulatory role for the highly conserved amphipathic region. Extrapolating from this result to the full-length protein is difficult since the truncated, soluble protein may be in a deregulated state. However, the amphipathic nature of this sequence may allow it to act as a regulatory element, where CdaA interacts with the membrane.
Our comprehensive functional assays, allowed us to further dissect its kinetics and functional dependencies. Both CdaA-containing nanodiscs and CdaA-DAC display the highest activity in the presence of manganese, with a ~ 3-fold lower activity in the presence of cobalt. We also observed residual activity in the presence of magnesium, but the activity was too low to accurately measure the kcat. The divalent metal ion dependencies of CdaA from L. lactis are in agreement with the findings of other CdaA-DAC homologs, albeit with differences in their preference for specific divalent metal cations3,4,7. Further, we could show that while the association to the membrane has an enormous effect on the turnover rate, CdaA and CdaA-DAC have a similar apparent Km for ATP of ~170 µM, which is well below the concentration of ATP in growing cells. Hence, the synthesis of cyclic di-AMP will not be limited by the availability of ATP, at least not in actively metabolizing cells. The Hill coefficient is well above one, corroborating that ATP binding occurs at multiple sites with positive cooperativity, which is in line with a model consisting of two catalytic domain dimers, each carrying their own ATP molecule. While synthesis of cyclic di-AMP requires interaction between the proximal active sites it remains unclear what the function of the distal active sites might be (Fig. 5). One possibility is that these distal active sites form interactions with distal active sites of neighbouring CdaA tetramers, leading to formation of transient CdaA concatemers on the membrane, but we have no evidence for this suggestion. We note that the environment of an in vitro enzyme activity assay does not fully recapitulate the complex (crowded) milieu of the bacterial cytoplasm, and higher-order oligomeric structures may have gone unnoticed. Finally, we did not find cyclic-di-GMP formation from GTP.
The activity of CdaA increases with pH from 6.5 to 8.0. The pH-dependence is most likely due to the protonation state of the histidine residues that coordinate the divalent metal cation located within the active site of the catalytic domain. From our work, it is clear that a change in intracellular pH will have a dramatic effect on the activity of CdaA, which likely affects host-pathogen interactions, such as with S. aureus32 and Streptococcus mitis33. The pH dependence also suggests a connection to acid stress response, as seen for mutant strains of B. subtilis34, L. lactis35,36 and L. monocytogenes37 with high levels of cyclic di-AMP. These mutants are more resistant to acid stress, but the effect was not observed in S. pneumoniae38,39. The role of cyclic di-AMP in acid stress has not been defined on the molecular level40.
Intracellular levels of manganese vary considerably between bacterial species (at least 3-orders of magnitude)41. However, some lactic acid bacteria are known to contain millimolar concentrations of manganese where it is used to combat reactive oxygen species42,43. Magnesium homeostasis has been connected to cyclic di-AMP signalling44,45,46, suggesting that the intracellular ratios of divalent metal ions and metal chelating compounds like ATP must be carefully balanced. Cyclic di-AMP is bound by the CBS domain of the Mg2+ importer MgtE of B. subtilis47. However, this is not the case for all Firmicutes since MgtE of S. aureus does not bind cyclic di-AMP48. The transport of essential divalent metal ions may link magnesium homeostasis and cyclic di-AMP signalling; however, more work is required to understand the relationship.
Studies using L. lactis, L. monocytogenes, S. aureus and L. plantarum cells indicate that CdaA-dependent synthesis of cyclic di-AMP is osmoregulated49. Here, synthesis of cyclic di-AMP is inhibited in cells placed in a hypertonic solution, using either ionic solutes (NaCl or KCl) or non-ionic solutes (sorbitol and sucrose). We observe that decreasing the salt concentration from 200 to 10 mM, which in the assay buffer corresponds to a decrease in ionic strength from ~0.25 to ~0.06 M, result in a 2-fold and 3-fold decrease in the rate of cyclic di-AMP synthesis by CdaA and CdaA-DAC, respectively. However, increasing the KCl concentration from 200 to 700 mM, which in the assay buffer corresponds to an increase in ionic strength from ~0.25 to 0.75 M had little effect on the rate of cyclic di-AMP synthesis. Since the KCl concentration in the bacterial cytoplasm is estimated to be 250 mM (E. coli), 300 mM (B. subtilis) and 500 mM (C. glutamicum and L. lactis)50,51,52,53, the salt-dependent activation of CdaA at these elevated KCl concentrations may physiologically be less relevant, even when part of the ions is bound to macromolecules. We observe that cyclic di-AMP synthesis is stimulated more effectively by K+ than Na+ ions both in CdaA-containing nanodiscs and lipid vesicles.
Osmoregulatory transport proteins, such as OpuA, BetP and ProP, require interaction with specific lipids in order to function correctly. In the case of OpuA, the activity of OpuA in nanodiscs increases linearly with increasing concentrations of DOPE and a decoupling between ionic strength gating and substrate transport is observed when the levels of anionic lipids like DOPG are below ~20 mol%54. In these proteins, key sensing residues form electrostatic interactions with negatively charged head groups of the inner leaflet of the membrane, which forms the basis for ionic strength gating of osmoregulatory transporters. We tested the lipid and ionic strength dependency of CdaA. We find that DOPE also stimulates the activity of CdaA, but we find no evidence that CdaA is regulated by ionic strength-dependent interactions of a protein domain with anionic lipids.
While using nanodiscs has advantages over vesicles for structural studies and activity measurements (all catalytic sites are accessible), nanodiscs do not allow the formation of a genuine membrane compartment. Hence, osmotic effects and the role of electrochemical ion gradients like the membrane potential cannot be determined. In order to overcome this limitation, we embedded CdaA within lipid vesicles. We did not see any significant effect on cyclic di-AMP synthesis when changing the osmotic pressure or generating a membrane potential across the lipid membrane. We conclude that the nanodisc system captures all the (membrane) features that are important for the activity and regulation of CdaA.
In summary, we show that membrane-embedded CdaA has more than 2-orders of magnitude higher cyclase activity than the soluble catalytic domain alone. The strongest physicochemical effect on CdaA is elicited by protons, where activity decreases almost 10-fold when the (intracellular) pH is decreased from 8.0 to 6.5. We show that the membrane-embedded state is required for bringing the catalytic domains of CdaA together, but the membrane is not playing a direct role in ionic strength-dependent gating as observed in osmoregulatory transporters. Our cryo-EM studies show that two DAC dimers have to interact in a head-to-head orientation for cyclase activity of CdaA.
Materials and Methods
Strains and cloning
All strains, vectors and primers used in this study are presented in Supplementary Table 1, Supplementary Table 2 and Supplementary Table 3, respectively.
Construction of E. coli expression vectors
The full-length cdaA ORF (CdaA) and the truncated cdaA sequence (CdaA-DAC) were amplified via colony PCR. The primers include overhangs containing SapI restriction sites making them compatible for handling by FX cloning55. The DNA sequences were inserted into the pINITIAL_cat vector following the standard protocol. The initial vectors were named ‘pINIT_CdaA_CmR’ and ‘pINIT_CdaA-DAC_CmR’ for the full-length and truncated sequences, respectively. For expression in E. coli CdaA-DAC, ‘pINIT_CdaA-DAC_CmR’ was digested and ligated with the pBxC3H expression vector of the FX cloning kit. The final construct was named ‘pBxC3H_CdaA-DAC_AmpR’. The vector is designed to add to the ORF a C-terminal 3 C cleavage site and 10x histidine tag for affinity purification.
Chemically competent E. coli MC1061 was transformed with ‘pBxC3H_CdaA-DAC_AmpR’. Transformants were selected by plating on LB plates containing 1% agar plus 100 µg/mL ampicillin. Successful transformants were confirmed by plasmid isolation, restriction digestion and Sanger DNA sequencing (Eurofins).
Construction of L. lactis expression vectors
The FX cloning kit does not contain an expression vector for L. lactis. Therefore, we constructed a new vector to enhance the workflow, which is also useful for future experiments. First, empty ‘pREXC3H’ vector (gift from; Raimund Dutzler & Eric Geertsma (Addgene plasmid #47078; RRID:Addgene_47078) was ligated with pERL56 to remove unwanted SapI sites and to make it compatible with FX cloning. Next, the oroP and erythromycin genes and their respective promoter/terminator sequences from pCS196657 were inserted into the ‘pREXC3H-pERL’ intermediate vector. Addition of these sequences allows multiple selection criteria (orotate import and erythromycin resistance). The resultant FX cloning compatible expression vector was named ‘pNZxC3H’. The ‘pINIT_CdaA_CmR’ vector was then digested and ligated with ‘pNZxC3H’ by following the standard FX cloning procedure55. The final construct, named ‘pNZ_CdaA_CmR’, was used for nisin-inducible overexpression of cdaA in L. lactis. This vector included the addition of a 3 C cleavage site and a C-terminal 10x histidine tag to the C-terminus of the ORF.
Electrocompetent L. lactis strain NZ9000 was transformed with the ‘pNZ_CdaA_CmR’ vector via standard electroporation procedures. Transformants were selected by plating on M17 plates containing 1% bacteriological agar, 1% (w/v) glucose and 5 µg/mL chloramphenicol. Successful transformants were confirmed by plasmid isolation, restriction digestion and Sanger DNA sequencing (Eurofins).
The L. lactis expression vector for glmM was constructed using the same method as described for cdaA. In short, the ORF of glmM (llmg0451) was amplified via colony PCR and inserted into the ‘pINITIAL_cat’ vector to form ‘pINIT_GlmM_CmR’. This vector and ‘pNZxC3H’ were digested and ligated to form the expression vector ‘pNZ_GlmM_CmR’. This construct was transformed into electrocompetent L. lactis and successful transformants were confirmed by plasmid isolation, restriction digestion and Sanger DNA sequencing (Eurofins).
Protein expression
CdaA-DAC expression in E. coli
E. coli MC1061 containing ‘pBxC3H_CdaA-DAC_AmpR’ was grown aerobically in LB medium containing 100 µg/mL ampicillin at 37 °C in 5 L baffled flasks with shaking ~200 rpm. At an A600 of 0.5 the temperature was lowered to 16 °C and expression was induced by addition of 0.001% (w/v) L-arabinose. The cells were grown for a further 18 h at 16 °C at which point they were harvested by centrifugation at 6000 g for 15 min at 4 °C (Supplementary Fig. 2B). The pellet from 6 L of culture was resuspended in ice-cold 100 mM KPi pH 7.5 before the centrifugation step was repeated and the cells were resuspended in 50 mM KPi pH 7.5 plus 10% (w/v) glycerol and stored at −80 °C in two aliquots of ~50 mL A600 ~ 240.
50 mL cells were thawed at room temperature and then diluted to a final volume of 100 mL and final buffer composition of 50 mM KPi pH 7.5, 200 mM NaCl and 5% (w/v) glycerol. Next, the cells were broken via two passes at 1500–2000 Bar via a high-pressure device (Maximator) in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM MgSO4 plus 100 µg/mL deoxyribonuclease (DNase). Immediately after breaking of the cells, 5 mM sodium-EDTA pH 8.0 was added and the mixture was centrifuged at 18,000 g for 30 min at 4 °C. The supernatant (100 mL crude lysate) was isolated and flash frozen in liquid nitrogen and stored at −80 °C in aliquots of 2 mL. The total protein concentration was determined using the BCA (bicinchoninic acid assay) assay.
CdaA expression and preparation of crude membrane vesicles of L. lactis
L. lactis NZ9000 containing pNZ_CdaA_CmR was grown semi-anaerobically at 30 °C in rich medium containing 2% (w/v) gistex, 65 mM sodium phosphate, 1% (w/v) glucose plus 5 µg/mL chloramphenicol. Growth was carried out in a 10 L pH-controlled bioreactor. pH was maintained at ~6.8 using 4 M KOH. At an A600 of 2, expression of cdaA was induced by addition of 1:2000 (v/v) dilution of a nisin A extract (derived from nisin-producing strain NZ9700). After 2 hours of induction the cells were harvested by centrifugation at 6000 g for 15 minutes at 4 °C. The pellet was resuspended in ice-cold 100 mM KPi pH 7.5 and the centrifugation step was repeated, after which the cells were resuspended in 50 mM KPi pH 7.5 plus 10% (w/v) glycerol and stored at −80 °C in six aliquots of ~50 mL A600 ~ 200.
150 mL of cells were quickly thawed at room temperature and then diluted with 50 mM KPi pH 7.5, 200 mM NaCl and 5% (w/v) glycerol with a final volume of 300 mL. The cells were broken via two passes at 2000–3000 bars in a high-pressure device (Maximator) in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM MgSO4 plus 100 µg/mL deoxyribonuclease (DNase). Immediately after breaking, 5 mM sodium-EDTA pH 8.0 was added and the cells were centrifuged at 15,000 g for 20 min at 4 °C. The supernatant was isolated and subjected to ultracentrifugation at 135,000 g for 2 h 15 min at 4 °C. The supernatant was discarded and the cell pellet was resuspended in 50 mM KPi pH 7.5, 200 mM NaCl plus 20% (w/v) glycerol, after which the cells were centrifuged again. The pellet was resuspended to a final volume of 10 mL in 50 mM KPi pH 7.5, 200 mM NaCl plus 10% (w/v) glycerol before flash freezing in liquid nitrogen and storage at −80 °C in aliquots of 2 mL. The total protein concentration was determined using the BCA (bicinchoninic acid) assay as CdaA has low UV absorbance.
GlmM expression in L. lactis
L. lactis NZ9000 containing pNZ_GlmM_CmR was grown semi-anaerobically at 30 °C in rich medium containing 2% (w/v) gistex, 65 mM sodium phosphate, 1% (w/v) glucose plus 5 µg/mL chloramphenicol. Growth was carried out in a 2 L pH-controlled bioreactor. pH was maintained at ~6.8 using 4 M KOH. At an A600 of 2, expression of glmM was induced by addition of 1:2000 (v/v) dilution of a nisin A extract (derived from nisin-producing strain (NZ9700)). After 2 h of induction the cells were harvested by centrifugation at 6000 g for 15 min at 4 °C. The pellet was resuspended in 100 mL of ice-cold 50 mM HEPES-KOH pH 7.5, 200 mM KCl plus 15% (w/v) glycerol, after which the cells were centrifuged again and resuspended in 50 mL of the same buffer to an Abs600 ~ 240. The cells were broken via two passes at 1500–2000 bar via a high-pressure device (Maximator) in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM MgSO4 plus 100 µg/mL deoxyribonuclease (DNase). Immediately after breaking of the cells, 5 mM sodium-EDTA pH 8.0 was added and the mixture was centrifuged at 18,000 g for 1 h at 4 °C, using the 45Ti rotor. The supernatant (50 mL crude lysate) was removed and flash frozen in liquid nitrogen and stored at −80 °C in aliquots of two aliquots of 25 mL. The total protein concentration was determined using the BCA (bicinchoninic acid) assay.
Protein purification
Purification of CdaA-DAC
0.5 mL CV of Ni2+-Sepharose resin was equilibrated with 10 CVs of ddH2O followed by 2 CVs of wash buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol plus 50 mM histidine). 2 mL of the E. coli crude lysate was incubated with the equilibrated resin at 4 °C for 1 h with nutation. The resin-protein solution was then allowed to drain by gravity flow before washing with 20 CVs of wash buffer. The outlet of the column was closed before addition of 0.4 mL elution buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl and 5% (w/v) glycerol) plus 0.1 mL 3 C protease (final concentration 6 mg/mL) and incubation for 2 h at 4 °C with nutation. The protein was then allowed to elute by gravity flow before addition of 1 mM EDTA and centrifugation at 21,000 x g for 5 min at 4 °C. The eluted protein had a volume of ~0.8 mL and the concentration was ~1.1 mg/mL using the BCA assay. The purified protein was then aliquoted in volumes 0.1–0.2 mL and flash frozen in liquid nitrogen before storage at −80 °C.
Purification of CdaA
8 mL crude membrane vesicles of CdaA were diluted in solubilisation buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol plus 1% (w/w) DDM) to a total protein concentration of 5 mg/mL. CdaA was solubilised at 4 °C for 1 h with nutation. Solubilised membranes were centrifuged at 335,000 g for 20 min at 4 °C, during which time 0.5 – 1.0 mL CV of Ni2+-Sepharose resin was equilibrated with 10 CVs of doubly distilled H2O followed by 2 CVs of wash buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol, 0.04% (w/w) DDM plus 50 mM histidine). The supernatant was incubated with the equilibrated resin at 4 °C for 1 h with nutation. The resin-protein solution was then allowed to drain by gravity flow before washing with 20 CVs of wash buffer.
Elution using 3 C protease
The resin-protein solution was washed with 4 CVs of elution buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol, 0.04% (w/w) DDM) without 3 C protease to remove excess histidine. The column outlet was closed before addition of 0.4 mL elution buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol, 0.04% (w/w) DDM) plus 0.1 mL 3 C protease (final concentration 6 mg/mL) and incubation for 2 h at 4 °C with nutation. The protein was then allowed to elute by gravity flow. Elution buffer without 3 C protease was added in fractions of 0.25 mL to recover more protein. Fractions with the highest protein concentrations were pooled before addition of 1 mM EDTA and centrifugation at 21,000 x g for 5 min at 4 °C. Protein concentration was determined, using the BCA assay. The eluted protein had a volume of ~0.8 mL and the concentration was determined to be ~0.9 mg/mL using the Nanodrop, with an extinction coefficient at 280 nm of 18,450 and monomeric molecular weight of 34.81 kDa (with his-tag). The purified protein was then directly used for SEC or cryo-EM.
Initially, we separated the CdaA-containing nanodiscs from empty nanodiscs, using size exclusion chromatography, e.g. when the deca-histidine tag of CdaA was removed via 3 C cleavage during the purification procedure (Supplementary Fig. 3). The. In later experiments, we eluted CdaA from the affinity column with 200 mM histidine instead of via 3 C cleavage.
Elution using histidine
The protein was eluted using an initial volume of 0.5 mL of elution buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol, 0.04% (w/w) DDM plus 200 mM histidine), followed by fractions of 0.25 mL elution buffer. The column was allowed to drain by gravity flow and the fractions were collected. Fractions with the highest protein concentrations were pooled before addition of 1 mM EDTA and centrifugation at 21,000 x g for 5 min at 4 °C. The eluted protein had a volume of ~0.9 mL and the concentration was ~1 mg/mL, using BCA assay. The purified protein was then directly used for SEC or reconstitution into nanodiscs.
Purification of GlmM
1 mL CV of Ni2+-Sepharose resin was equilibrated with 10 CVs of doubly distilled H2O followed by 2 CVs of wash buffer (50 mM HEPES-NaOH pH 7.5, 200 mM KCl, 15% (w/v) glycerol plus 50 mM imidazole). 25 mL of the E. coli crude lysate was incubated with 10 mM imidazole and the equilibrated resin at 4 °C for 1 h with nutation. The resin-protein solution was then allowed to drain by gravity flow before washing with 20 CVs of wash buffer. The protein was eluted using an initial volume of 0.5 mL of elution buffer (50 mM HEPES-NaOH pH 7.5, 200 mM KCl, 15% (w/v) glycerol plus 500 mM imidazole) followed by fractions of 0.25 mL elution buffer. The column was allowed to drain by gravity flow and the fractions were collected. Fractions with the highest protein concentrations were pooled before addition of 1 mM EDTA and centrifugation at 21,000 x g for 5 min at 4 °C. The eluted protein had a volume of ~1 mL and the concentration was ~2.3 mg/mL, using the Nanodrop, with an extinction coefficient of 22,350 at 280 nm and monomeric molecular weight of 50 kDa. The imidazole was then removed by buffer exchange with 50 mM HEPES-NaOH pH 7.5, 200 mM KCl plus 15% (w/v) glycerol. The purified protein was aliquoted in volumes of 0.1 mL before flash freezing in liquid nitrogen and storage at −80 °C.
Size-exclusion chromatography and SEC-MALLS analysis
CdaA-DAC was purified as described above, and loaded onto the SEC column (Superdex 200 10/300 GL - GE Healthcare) equilibrated with 50 mM HEPES-NaOH pH 7.5, 200 mM NaCl plus 5% (w/v) glycerol. The 500 µL elution fractions were collected and the concentration of the appropriate peak fraction was determined via BCA assay.
CdaA was purified as in section ‘Purification of CdaA’ and then loaded onto the SEC column (Superdex 200 10/300 GL - GE Healthcare) equilibrated with 50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol plus 0.04% (w/w) DDM. The sample was separated by SEC and detected via MALLS analysis (DawnHeleos II-Optilab T-rEX). Aldolase (derived from rabbit muscle) was used for instrument calibration and analysis.
SEC-MALLS differentiates between the molecular mass of the protein and that of the detergent micelle58 by measuring the UV absorbance at a wavelength of 280 nm, light scattering and refractive index. Changes in these readouts are measured over time as the size separated sample elutes from the SEC column. Since DDM does not absorb at 280 nm, the changes in absorbance at 280 nm and the refractive index are dependent on the protein concentration, whereas the light scatter reports the mass of the total protein-detergent micelle complex. This allows the weight-averaged molecular mass of the protein to be calculated independently from the mass contribution of the detergent micelle.
Reconstitution of CdaA into nanodiscs
Preparation of synthetic lipid mixtures
25 mg/mL stock solutions of DOPC, DOPE and DOPG were prepared in chloroform. Individual stock solutions were then mixed in the appropriate volumes to give the desired synthetic lipid mixture. Lipids were then dried and chloroform was removed via evaporation in a rotary evaporator device. During evaporation the lipids were incubated at 40 °C using a water bath and were placed under vacuum until all liquid was removed. After evaporation, diethyl ether was added to dissolve the lipids. Once completely dissolved, lipids were dried using the same procedure as for the evaporation of chloroform. After drying the lipids were dispersed in 50 mM KPi pH 7.0. The sample was then subjected to tip-sonication (Vibra-Cell VCX 130 Sonicator, tip diameter 6 mm) in an ice-water bath using the following settings: 15 s on; 45 s off; amplitude 70%; 4 min. The resulting solution was used to prepare 250 µL aliquots of lipid vesicles at a concentration of 25 mg/mL, which were subjected to three freeze-thaw cycles using liquid nitrogen and slow thawing at room temperature. The samples were then flash frozen and stored in liquid nitrogen until required for reconstitution.
Nanodisc reconstitution of CdaA
First, 250 µL of 25 mg/mL synthetic lipid mixture was diluted to 6.25 mg/mL in a 15 mL tube using 50 mM KPi pH 7.5. The synthetic lipid mixture was composed of DOPG (38 mol%), DOPC (12 mol%), DOPE (50 mol%) in 50 mM KPi pH 7.0. The same procedure was used for other lipid compositions. The solution was subjected to tip-sonication, while in an ice-water bath, using the following settings: 15 s on; 45 s off; amplitude 70%; 2 min. 900 µL lipids were transferred to a glass vial and 100 µL of 10% (w/w) DDM was added. The appropriate volumes of purified CdaA, purified MSP1E3D1 and the synthetic lipid mix (containing DDM) were added to obtain a protein:MSP:lipid ratio of 1:2:40 in a final reaction volume of 700 µL in a 1.5 mL tube. Purification of MSP1E3D1 was performed as described previously30. The sample was incubated for 1 h at 4 °C with nutation before 500 mg of Biobeads SM-2 were added and incubation was continued overnight at 4 °C with nutation. The following day a needle and syringe were used to isolate the nanodisc containing solution by aspirating the solution away from the Biobeads. The solution was then centrifuged at 21,000 g for 10 min at 4 °C to remove aggregates. The supernatant was transferred to a new 1.5 mL centrifuge tube.
At this point, dependent on if the protein was eluted in the previous steps via histidine or 3 C cleavage, separation of CdaA-containing nanodiscs from empty nanodiscs was carried out using two different methods. In experiments using 3 C cleaved protein (without affinity tag) the sample was loaded directly on the SEC column, as described above for CdaA. In experiments using histidine-eluted protein (with affinity tag) a second round of affinity purification was carried out as described above. Purified CdaA-containing nanodiscs were then loaded on the SEC column or directly used in activity assays.
Affinity purification of CdaA-containing nanodiscs
After separation of reconstituted CdaA-containing nanodiscs from the Biobeads, as described above, the samples were incubated with 0.5 CV of Ni2+-Sepharose resin. The resin was equilibrated with 10 CVs of ddH2O followed by 2 CVs of wash buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol plus 50 mM histidine). The nanodisc sample was incubated with the equilibrated resin at 4 °C for 1 h with nutation. The resin-nanodisc solution was then allowed to drain by gravity flow before washing with 10 CVs of wash buffer, followed by 4 CVs of elution buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl plus 5% (w/v) glycerol) without 3 C protease to remove excess histidine.
The column was closed before addition of 0.32 mL elution buffer (50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol, 0.04% (w/w) DDM plus 0.08 mL 3 C protease (final concentration 6 mg/mL)) and incubation for 2 h at 4 °C with nutation. The protein was then allowed to elute by gravity flow. Additional 0.25 mL elution fractions were collected by addition of elution buffer without 3 C protease in order to recover more protein. Fractions with the highest protein concentrations were pooled before centrifugation at 21,000 x g for 5 min at 4 °C. The eluted protein had a volume of ~0.7 mL and the concentration was ~0.2 mg/mL, using BCA assay. The purified protein was then flash frozen in liquid nitrogen before storage at -80 °C or used directly for activity assay or SEC analysis.
Reconstitution of CdaA in lipid vesicles
The extruder (Hamilton) was pre-washed using 1 mL 50 mM KPi pH 7.0 before all extrusion steps. Liposomes (20 mg/mL), composed of E. coli polar lipids/EggPC, were extruded 11 times through a 400 nm polycarbonate filter (Cytiva) to form large-unilamellar vesicles (LUVs). The LUVs were then diluted to 4 mg/mL in 50 mM KPi pH 7.0 plus 20% (w/v) glycerol and then titrated with aliquots of 10% (w/v) Triton X-100 at room temperature until an Rsaturation of 60% was reached. Destabilization of the liposomes was monitored by the optical density of the suspension at 540 nm. Purified CdaA was then mixed with the detergent-destabilized liposomes using a 1:100 (w/w) protein-to-lipid ratio by addition of 0.2 mL 1 mg/mL purified CdaA in 50 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 5% (w/v) glycerol plus 0.04% (w/w) to 5 mL of detergent-destabilized liposomes. The sample was then incubated for 15 min at room temperature with gentle agitation. To remove the detergents, 200 mg of Bio-beads SM-2 were then added per 5 mL of protein-liposome suspension, followed by incubation for 30 minutes at room temperature with gentle agitation. A further 200 mg of Bio-beads SM-2 was added to the protein-liposome suspension before incubation for 1 h at 4 °C with gentle agitation. Finally, a further 200 mg of Bio-beads SM-2 was added before incubation overnight at 4 °C with gentle agitation. In the morning, an additional 200 mg of Bio-beads SM-2 was added to the protein-liposome suspension before incubation for 2 h at 4 °C with gentle agitation. Bio-beads SM-2 were then removed by filtration over sintered glass in a disposable polyprep column and the proteoliposomes were collected in a 1.5 mL tube. Bio-beads SM-2 were washed using 50 mM KPi pH 7.0 to collect residual proteoliposomes and to dilute the glycerol concentration to 2% (w/v). The sample was then centrifuged for 45 min at 337,000 g 4 °C to pellet the proteoliposomes before resuspension to 20 mg/mL of lipid in buffer containing 50 mM HEPES-NaOH pH 8.0 plus 200 mM KCl. Proteoliposomes were then flash-frozen in liquid nitrogen in aliquots of 1 mL before slow thawing on ice. The freeze thaw process was repeated a minimum of three times before use of the sample in the activity assays.
In vitro activity assays
Activity of DDM-solubilised CdaA
The radiolabelled activity assays were performed with 1 µM of detergent-solubilised CdaA in buffer containing 50 mM HEPES-NaOH pH 8.0, 1 mM MnCl2, 200 mM NaCl, 100 µM ATP spiked with 3 µM 3H-ATP, in a final reaction volume of 200 µL. The reaction was carried out at 37 °C for 4 h. At the endpoint, the sample was incubated at 90 °C for 5 min to stop the reaction before proceeding to TLC analysis. Thin-layer chromatography was performed on a 5 × 10 cm silica 60 UV254 TLC plate (Macherey-Nagel). 2 µL of each sample was spotted onto a TLC plate and separation was performed for 1 h with running solvent (isopropanol:ammonia:water (11:7:2)). The plate was visualized by exposure to UV light at 254 nm before division into equal parts and addition of 2 mL liquid scintillation fluid (PerkinElmer Filter-Count). The radioactivity was measured using a PerkinElmer Tri-Carb 2800TR Liquid Scintillation Analyzer.
CdaA and CdaA-DAC activity assays
Unless stated otherwise, activity assays were performed using 16 µM of soluble CdaA-DAC and 400 µM ATP in 50 mM HEPES-NaOH pH 8.0 containing 1 mM MnCl2 plus 200 mM KCl in a final reaction volume of 0.2 mL or 0.3 mL. Activity assays using full-length membrane-bound CdaA were performed using 0.001 mM of CdaA-containing nanodiscs and 0.4 mM ATP in 50 mM HEPES-NaOH pH 8.0 containing 1 mM MnCl2 plus 200 mM KCl in a final reaction volume of 200 µL. The reactions were incubated at 37 °C for the time indicated in each figure. At the relevant time points, 20 µL samples were removed and incubated at 90 °C for 5 min to quench the reactions. Quenched samples were centrifuged at 16,000 x g for 5 min at room temperature before the supernatant was isolated and analysed via HPLC-UV.
HPLC was performed using two eluents: Eluent A, 5 mM ammonium acetate pH 5.0 and eluent B, 100% HPLC-grade acetonitrile. Eluent A was adjusted to pH 5.0 using acetic acid. Each sample was separated via HPLC using a single cycle of the following program: 1% acetonitrile for 0.5 min, a gradient from 1% to 10% acetonitrile over a 9.5 min period, followed by 10 min at 1% acetonitrile. Flow rate was set to 1 mL/min, oven temperature was 40 °C, and an injection volume of 10 µL was used. The samples were separated using a Nucleodur C18 pyramid column (Macherey-Nagel) with a length of 5 cm and particle size of 3 µM, equipped with a guard column.
A set of standard calibration samples with known nucleotide concentrations (0–1 mM) of cyclic di-AMP and ATP were run at the beginning of each experiment and were used to identify the peaks on the basis of the retention times of the nucleotides. The peak areas of samples with known concentrations were used to create a calibration curve. The calibration curve allowed us to determine and follow the synthesis of cyclic di-AMP over the course of the reaction.
In all experiments divalent metal cations were added to the final concentration of 1 mM and a pH of 8.0 unless stated otherwise. Experiments in which the pH was altered (Fig. 2D), the buffer was prepared using 25 mM HEPES-NaOH and 25 mM MES-NaOH containing 1 mM MnCl2 and 200 mM KCl. HEPES and MES have pKas of approximately 7.5 and 6.15, respectively. This ensured that the pH was stable in the range tested (pH 6.5–8.0).
Cryo-EM sample preparation, data acquisition and image processing
Preparation of cryo-EM samples of CdaA embedded in MSP1E3D1 lipid nanodiscs and data acquisition
Purified CdaA, reconstituted into MSP1E3D1 nanodiscs, was concentrated using a 100 kDa MWCO centrifugal concentrator (Millipore) to a final concentration prior to grid preparation. The final protein concentration was based on the respective yield of each purification, namely, 0.96 mg/ml – ligand free; 0.62 mg/ml – turnover condition (MnATP or MnAMP-PNP); 0.9 mg/ml – inhibited by ApCpp and 1 mg/ml - inhibited by GlmM. Prior to sample application, the concentrated protein was incubated for 30 minutes with the respective ligands. The final concentrations of each ligand were 10 mM ATP or AMP-PNP supplemented with 1 mM Mn−2, 10 mM MnAMP-PNP and 5 mM MnApCpp respectively (Supplementary Fig. 7). GlmM was additionally added to a 1:1 ratio substituted with 10 mM of MnATP. In sequence, a volume of 2.8 µL of each sample was then applied to Quantifoil Au R1.2/1.3 300-mesh grids, which were subsequently blotted for 3−5 s using a Vitrobot at 22 °C and 100% humidity. The grids were immediately flash frozen in a mixture of liquid ethane-propane cooled with liquid nitrogen. The grids were stored in liquid nitrogen until data acquisition.
Cryo-EM single particle datasets were acquired using a Talos Arctica microscope (Thermo Fisher Scientific) at 200 kV with a bioQuantum post-column energy filter (Gatan) in zero-loss mode (20 eV slit) and a 100 µm objective aperture. Movies were acquired in automated mode using SerialEM59,60 version 3.9.0beta7 with a K2 Summit detector (Gatan) in counting mode, with a pixel size of 1.022 Å (49.407 calibrated magnification) and a target defocus range of −0.5 to −2.5 µm. A 9 s exposure per target was captured in 60 frames with a total electron dose of 50.9 e- per Å2 for the ligand-free, MnATP and MnAMP-PNP datasets. The total electron dose of 53.6 e- per Å2 was used for the datasets of CdaA inhibited by GlmM and with MnApCpp respectively. An in-house-written script facilitated the screening and selection of optimal targets for acquisition based on ice thickness61, which was set between 20 and 40 nm. Data quality was monitored on-the-fly using FOCUS version 1.0.062.
Image processing of ligand-free CdaA embedded in MSP1E3D1 lipid nanodiscs
A dataset of 2234 dose-fractionated cryo-EM movies was recorded. Motion correction, frame dose weighting and CTF estimation was performed within FOCUS using MotionCor2 version 1.2.163 and CTFFIND4.1.1364, respectively. The processed movies were then visually inspected and poor-quality images were discarded on the basis of poor CTF estimation or the presence of contaminants. In total 1553 images remained. Particle picking was facilitated by the PhosaurusNet of crYOLO version 1.2.2 using the general model65. A total of 633,849 picked particles were imported into RELION version 4.0-beta-266 and extracted with a box size of 256 pixels. The extracted particles were subjected to two rounds of 2D classification using a particle mask diameter of 200 Å, followed by a 3D classification using the same particle mask diameter. The resulting 3D classes were inspected, and particles classified into robust classes were subjected to further 2D classifications in both RELION and CryoSPARC v467 (Supplementary Fig. 6).
Image processing of CdaA embedded in MSP1E3D1 lipid nanodiscs (MnAMP-PNP)
A total of 688 movies were collected from a single grid. Motion correction, frame dose weighting and CTF estimation was performed within CryoSPARCv467. Images that either showed contaminants or poor CTF estimation were discarded. In total 645 images remained for further processing. Particle picking was facilitated by the blob picker with a particle diameter range from 100 to 200 Å. A total of 359,459 particles were picked and extracted with a box size of 256 pixels. The extracted particles were then subjected a round of 2D classification using settings as shown in Supplementary Fig. 7A.
Image processing of CdaA embedded in MSP1E3D1 lipid nanodiscs (MnATP)
A total of 1435 movies were collected from a single grid. Motion correction, frame dose weighting and CTF estimation was performed within FOCUS using MotionCor2 version 1.2.163 and CTFFIND4.1.1364, respectively. Images that either showed contaminants or poor CTF estimation were discarded. In total 949 images remained for further processing. Particle picking was facilitated by the PhosaurusNet of crYOLO version 1.2.2 using the general model65. A total of 182,288 particles were picked and subsequently imported into RELION version 4.0-beta-266 and extracted with a box size of 256 pixels. The extracted particles were then subjected to three consecutive rounds of 2D classification using a particle mask diameter of 200 Å while limiting the resolution E-step to 7 Å (Supplementary Fig. 7B).
Image processing of CdaA embedded in MSP1E3D1 lipid nanodiscs, inhibited by MnApCpp
A total of 5178 movies were collected from a single grid. Motion correction, frame dose weighting and CTF estimation was performed within FOCUS using MotionCor2 version 1.2.163 and CTFFIND4.1.1364, respectively. Images with extensive contamination or poor CTF estimation were discarded. In total 4497 images remained for further processing. Particle picking was facilitated by the PhosaurusNet of crYOLO version 1.2.2 using the general model65. A total of 1,543,977 particles were picked and subsequently imported into RELION version 4.0-beta-266 and extracted with a box size of 256 pixels. The extracted particles were then imported to CryoSPARC v467 and subjected to 2D classification using settings as shown in (Supplementary Fig. 7D). The selected particles (652,535) where classified in 3D using ab-initio and subsequently hetero refinement with K = 3 and default settings.
Image processing of CdaA embedded in MSP1E3D1 lipid nanodiscs, inhibited by GlmM
A total of 5150 movies were collected from a single grid. Motion correction, frame dose weighting and CTF estimation was performed within FOCUS using MotionCor2 version 1.2.163 and CTFFIND4.1.1364, respectively. Images with extensive contamination or poor CTF estimation were discarded. In total 4,132 images remained for further processing. The detailed processing workflow can be found in Supplementary Fig. 8. In brief, particle picking was facilitated by the PhosaurusNet of crYOLO version 1.8.4 using the general model65. A total of 1,908,105 particles were picked and extracted with a box size of 256 Å in Relion 5.0. The extracted particles were imported to CryoSPARC v4 and subjected to a round of 2D classification to remove bad quality and false positive particles. The remaining 1,007,405 particles were classified in 3D by a consecutive round of ab-initio and subsequently Hetero refinement using 3 classes. The class corresponding to the DAC-GlmM complex (404,283 particles) was further cleaned using 2D classification with default parameters. The selected 230,609 particles were subjected to an additional round of ab-initio/hetero refinement. The most homogeneous class (133,430 particles) was refined via non-uniform refinement to 6.6 Å resolution. The homogeneous particle stack was then imported to Relion 5.0 for further processing. An initial masked 3D refinement without utilizing the blush algorithm resulted in a reconstruction of 7.5 Å. Subsequently the particles were subjected to three consecutive rounds of CTF refinements correcting magnification anisotropy, per particles defocus and per micrograph astigmatism. The refinements after each round of CTF refinement were done using the blush algorithm. Ultimately the particles converged to a final reconstruction of 4.84 Å global resolution after post processing. The data collection and processing parameters are summarized in Supplementary Table 4. The AF3 model was fitted in the cryo-EM density in ChimeraX. Due to the limited global resolution of the cryo-EM map all sidechains were deleted and the model was subsequently iteratively refined in real-space against the post-processed map in PHENIX and manually improved in COOT. The domain 4 of each GlmM protomer (residues 367-452) and the ligand in one of the DAC-active sites were not modelled due to poor or absent densities. Cryo-EM map, model visualization and Figure preparation were done in ChimeraX v1.8.
Alphafold structure prediction
The full-length cdaA sequence (llmg_0448) was used as the input for the prediction of the tetrameric complex of CdaA using the Alphafold 2 multimer13 in ColabFold v1.5.568. Visualisation of the PAE scores was done using the PAE Viewer webserver69. Alphafold 3 server (https://www.alphafoldserver.com)12 was used to predict the ligand bound state in Fig. 3B.
Statistics and Reproducibility
All statistical analysis and graph preparation for the biochemical characterization of CdaA and CdaA-DAC was performed using Prism 5 (GraphPad Inc.). All equations and model fitting used in the analysis were standard for Prism 5 software. Where appropriate, the number and type of replicates for each experiment are defined in the appropriate figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and are available upon request. The SDS gels presented in Fig. 1 are included uncropped and unedited as Supplementary Fig. 2. Raw cryoEM movies are currently being deposited in the Electron Microscopy Public Image Archive (EMPIAR) and the respective accession numbers updated later. The cryo-EM density map, half maps, and mask have been deposited in the Electron Microscopy Data Bank (EMDB). The atomic model of CdaA-DAC in complex with GlmM is available through the Protein Data Bank (PDB). The respective accession codes are: EMDB-51096 (https://www.ebi.ac.uk/emdb/EMD-51096) and PDB-9G69 (PDB https://doi.org/10.2210/pdb9G69/pdb).
References
Stülke, J. & Krüger, L. Cyclic di-AMP Signaling in Bacteria. Annu. Rev. Microbiol. 74, annurev-micro-020518-115943 (2020).
He, J., Yin, W., Galperin, M. Y. & Chou, S. H. Cyclic di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 48, 2807–2829 (2020).
Gibhardt, J. et al. An extracytoplasmic protein and a moonlighting enzyme modulate synthesis of c-di-AMP in Listeria monocytogenes. Environ. Microbiol. 22, 2771–2791 (2020).
Rosenberg, J. et al. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J. Biol. Chem. 290, 6596–6606 (2015).
Heidemann, J. L., Neumann, P., Dickmanns, A. & Ficner, R. Crystal structures of the c-di-AMP-synthesizing enzyme CdaA. J. Biol. Chem. 294, 10463–10470 (2019).
Gundlach, J. et al. An essential poison: Synthesis and degradation of cyclic Di-AMP in Bacillus subtilis. J. Bacteriol. 197, 3265–3274 (2015).
Pathania, M. et al. Structural basis for the inhibition of the Bacillus subtilis c-di-AMP cyclase CdaA by the phosphoglucomutase GlmM. J. Biol. Chem. 297, 101317 (2021).
Tosi, T. et al. Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM. PLoS Pathog. 15, 1–28 (2019).
Zhu, Y. et al. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol. Microbiol. 99, 1015–1027 (2016).
Müller, M., Deimling, T., Hopfner, K. P. & Witte, G. Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3′-dATP. Biochem. J. 469, 367–374 (2015).
Bai, Y. et al. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-amp. PLoS One 7, 1–10 (2012).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer https://doi.org/10.1101/2021.10.04.463034. (2022).
Ayoub, N., Roth, P., Ucurum, Z., Fotiadis, D. & Hirschi, S. Structural and biochemical insights into His-tag-induced higher-order oligomerization of membrane proteins by cryo-EM and size exclusion chromatography. J. Struct. Biol. 215, 107924 (2023).
Bailoni, E. et al. Minimal Out-of-Equilibrium Metabolism for Synthetic Cells: A Membrane Perspective. ACS Synth. Biol. https://doi.org/10.1021/acssynbio.3c00062 (2023).
de Azambuja, F., Steens, N. & Parac-Vogt, T. N. Kinetic and interaction studies of adenosine-5′-triphosphate (ATP) hydrolysis with polyoxovanadates. Metals (Basel) 11, 1678 (2021).
Stockbridge, R. B. & Wolfenden, R. The Intrinsic reactivity of ATP and the catalytic proficiencies of kinases acting on glucose, N-acetylgalactosamine, and homoserine. A thermodynamic analysis. J. Biol. Chem. 284, 22747–22757 (2009).
Gupta, R. K., Gupta, P., Yushok, W. D. & Rose, Z. B. Measurement of the dissociation constant of MgATP at physiological nucleotide levels by a combination of 31P NMR and optical absorbance spectroscopy. Biochem. Biophys. Res Commun. 117, 210–216 (1983).
Pecoraro, V. L., Hermes, J. D. & Cleland, W. W. Stability constants of magnesium and cadmium complexes of adenine nucleotides and thionucleotides and rate constants for formation and dissociation of magnesium-ATP and magnesium-ADP. Biochemistry 23, 5262–5271 (1984).
Wilson, J. E. & Chin, A. Chelation of divalent cations by ATP, studied by titration calorimetry. Anal. Biochem 193, 16–19 (1991).
Zea, C. J., Camci-Unal, G. & Pohl, N. L. Thermodynamics of binding of divalent magnesium and manganese to uridine phosphates: Implications for diabetes-related hypomagnesaemia and carbohydrate biocatalysis. Chem. Cent. J. 2, 1–7 (2008).
Yaginuma, H. et al. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci. Rep. 4, 6522 (2014).
Groisman, E. A. et al. Bacterial Mg 2+ Homeostasis, Transport, and Virulence. Annu Rev. Genet 47, 625–646 (2013).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Hao, X. et al. Structure and function of diadenylate cyclase DacM from Mycoplasma ovipneumoniae. bioRxiv 2022.03.11.483894; 03.11.483894 https://doi.org/10.1101/2022 (2022).
Whiteley, A. T. et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199 (2019).
Wentinck, K., Gogou, C. & Meijer, D. H. Putting on molecular weight: Enabling cryo-EM structure determination of sub-100-kDa proteins. Curr. Res. Struct. Biol. 4, 332–337 (2022).
Biemans-Oldehinkel, E., Mahmood, N. A. B. N. & Poolman, B. A sensor for intracellular ionic strength. Proc. Natl Acad. Sci. USA 103, 10624–10629 (2006).
van der Heide, T., Stuart, M. C. A. & Poolman, B. On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J. 20, 7022–7032 (2001).
Sikkema, H. R. et al. Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA. Sci. Adv. 6, eabd7697 (2020).
Grasberger, B., Minton, A. P., DeLisi, C. & Metzger, H. Interaction between proteins localized in membranes. Proc. Natl Acad. Sci. USA 83, 6258–6262 (1986).
Bowman, L., Zeden, M. S., Schuster, C. F., Kaever, V. & Gründling, A. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureu. J. Biol. Chem. 291, 26970–26986 (2016).
Rørvik, G. H. et al. Cyclic Di-adenosine Monophosphate Regulates Metabolism and Growth in the Oral Commensal Streptococcus mitis. Microorganisms 8, 1269 (2020).
Rao, F. et al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285, 473–482 (2010).
Rallu, F., Gruss, A., Ehrlich, S. D. & Maguin, E. Acid- and multistress-resistant mutants of Lactococcus lactis: Identification of intracellular stress signals. Mol. Microbiol 35, 517–528 (2000).
Zhu, Z. et al. Enhanced acid-stress tolerance in Lactococcus lactis NZ9000 by overexpression of ABC transporters. Micro. Cell Fact. 18, 1–14 (2019).
Witte, C. E. et al. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 4, 1–10 (2013).
Zarrella, T. M., Yang, J., Metzger, D. W. & Bai, G. Bacterial Second Messenger Cyclic di-AMP Modulates the Competence State in Streptococcus pneumoniae. J. Bacteriol. 202, 1–17 (2020).
Zarrella, T. M., Metzger, D. W. & Bai, G. Stress suppressor screening leads to detection of regulation of cyclic di-AMP homeostasis by a Trk family effector protein in Streptococcus pneumoniae. J. Bacteriol. 200, 1–15 (2018).
Zarrella, T. M. & Bai, G. The many roles of the bacterial second messenger cyclic di-AMP in adapting to stress cues. J. Bacteriol. https://doi.org/10.1128/jb.00348-20 (2020).
Juttukonda, L. J. & Skaar, E. P. Manganese homeostasis and utilization in pathogenic bacteria. Mol. Microbiol. 97, 216–228 (2015).
Bosma, E. F., Rau, M. H., Van Gijtenbeek, L. A. & Siedler, S. Regulation and distinct physiological roles of manganese in bacteria. FEMS Microbiol. Rev. 45, 1–16 (2021).
Archibald, F. S. & Duong, M. N. Manganese acquisition by Lactobacillus plantarum. J. Bacteriol. 158, 1–8 (1984).
Luo, Y. & Helmann, J. D. Analysis of the role of Bacillus subtilis σ M in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol 83, 623–639 (2012).
Mehne, F. M. P. et al. Cyclic Di-AMP homeostasis in bacillus subtilis: Both lack and high level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 288, 2004–2017 (2013).
Wendel, B. M. et al. A Central Role for Magnesium Homeostasis during Adaptation to Osmotic Stress. mBio 13, e0009222 (2022).
Gundlach, J. et al. Sustained sensing in potassium homeostasis: Cyclic di-AMP controls potassium uptake by KimA at the levels of expression and activity. J. Biol. Chem. 294, 9605–9614 (2019).
Schuster, C. F. et al. The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus. Sci. Signal 9, 1–31 (2016).
Pham, H. T. et al. Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLoS Genet 14, 1–23 (2018).
Shabala, L. et al. Ion transport and osmotic adjustment in Escherichia coli in response to ionic and non-ionic osmotica. Environ. Microbiol 11, 137–148 (2009).
Whatmore, A. M., Chudek, J. A. & Reed, R. H. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol 136, 2527–2535 (1990).
Ochrombel, I., Ott, L., Krämer, R., Burkovski, A. & Marin, K. Impact of improved potassium accumulation on pH homeostasis, membrane potential adjustment and survival of Corynebacterium glutamicum. Biochimica et. Biophysica Acta (BBA) - Bioenerg. 1807, 444–450 (2011).
Goel, A., Santos, F., de Vos, W. M., Teusink, B. & Molenaar, D. Standardized assay medium to measure Lactococcus lactis enzyme activities while mimicking intracellular conditions. Appl Environ. Microbiol 78, 134–143 (2012).
Karasawa, A. et al. Physicochemical factors controlling the activity and energy coupling of an ionic strength-gated ATP-binding cassette (ABC) transporter. J. Biol. Chem. 288, 29862–29871 (2013).
Geertsma, E. R. & Dutzler, R. A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry 50, 3272–3278 (2011).
Geertsma, E. R. & Poolman, B. High-throughput cloning and expression in recalcitrant bacteria. Nat. Methods 4, 705–707 (2007).
Solem, C., Defoor, E., Jensen, P. R. & Martinussen, J. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl Environ. Microbiol 74, 4772–4775 (2008).
Slotboom, D. J., Duurkens, R. H., Olieman, K. & Erkens, G. B. Static light scattering to characterize membrane proteins in detergent solution. Methods 46, 73–82 (2008).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Schorb, M., Haberbosch, I., Hagen, W. J. H., Schwab, Y. & Mastronarde, D. N. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019).
Rheinberger, J., Oostergetel, G., Resch, G. P. & Paulino, C. Optimized cryo-EM data-Acquisition workflow by sample-Thickness determination. Acta Crystallogr D. Struct. Biol. 77, 565–571 (2021).
Biyani, N. et al. Focus: The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017).
Zheng, S. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 https://doi.org/10.1038/s42003-019-0437-z (2019).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochemical J. 478, 4169–4185 (2021).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Elfmann, C. & Stülke, J. PAE viewer: A webserver for the interactive visualization of the predicted aligned error for multimer structure predictions and crosslinks. Nucleic Acids Res 51, W404–W410 (2023).
Acknowledgements
Thanks to Prof. Dr. Dirk Slotboom for assistance with the SEC-MALLS and data analysis. Jan Rheinberger for his support and consultation in the cryo-EM data acquisition and image processing. Michiel Punter for the IT support and maintenance of the image processing cluster. This work was financed by the NWO Gravitation Program BaSyC and NWO OCENW.KLEIN.526.
Author information
Authors and Affiliations
Contributions
A.J.F., P.D., H.L. and B.P. designed the studies. A.J.F. designed and performed cloning. A.J.F., Jt.K., and H.L. performed protein purification, S.E.C. and activity assays. P.D. performed grid preparation, cryo-electron microscopy and data processing/analysis. A.J.F. and D.J.S. performed SEC-MALLS. GKS-W performed part of the gene cloning. P.D. and B.P. wrote the manuscript. B.P. and C.P. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Jorg Stulke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Dario Ummarino and Huan Bao. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Foster, A.J., Li, H., Drougkas, P. et al. Membrane-embedded CdaA is required for efficient synthesis of second messenger cyclic di-AMP. Commun Biol 7, 1710 (2024). https://doi.org/10.1038/s42003-024-07420-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07420-x