Abstract
ZBTB48 (also known as TZAP) is a transcription factor that has previously been reported to bind to telomeres and act as a negative regulator of telomere length in human cell lines. To explore whether transcription factor activity and telomere length regulation are conserved at the organismal level in vertebrates, we generate a zbtb48−/− zebrafish line via CRISPR‒Cas genome editing. The zbtb48−/− mutants display no obvious physical or behavioral abnormalities in the first two generations. We find no statistically significant changes in telomere length in first-generation adults. However, for the gene regulatory aspect of Zbtb48, similar to that in human cancer cell lines, we observe downregulation of mtfp1 at both the mRNA and protein levels in the zbtb48−/− mutants. This suggests that mtfp1 is an evolutionarily conserved regulatory target of Zbtb48. Further investigation of the spatiotemporal expression of zbtb48 in previously published zebrafish data reveals low transcript expression in diverse tissues, except in germline stem cells and gametocytes of the gonads. Notably, Mtfp1 protein downregulation is detected in the ovaries of 40 dpf zbtb48−/− mutants and in the testes of both 40 dpf and 10.5-month-old zbtb48−/− mutants.
Similar content being viewed by others
Introduction
The zebrafish has emerged as an in vivo model for studying telomere regulation because its telomeres share several common properties with those of humans. Similar to those in humans, zebrafish telomeres consist of TTAGGG tandem repeats that typically range from 5 to 15 kilobases in length1. Despite the expression of telomerase in somatic tissue, telomeres in zebrafish shorten during aging2. In contrast to laboratory inbred mouse strains, zebrafish exhibit telomere deficiency phenotypes already in the first generation of tert−/− mutants that are correlated with significantly shorter telomeres compared to their wild-type counterparts1. The lifespan and telomere length of first-generation tert−/− mutants are reduced, and they experience premature aging with a rapid decline in fertility1,3. Critical short telomeres accumulate primarily in the gut4, and tissue-specific rescue of telomerase in the gut suppresses premature aging phenotypes and extends lifespan5. Second-generation tert−/− mutants do not survive to adulthood because the critical short telomeres activate p53-induced apoptosis at the embryonic stage3. Consequently, the tp53M214K mutation in tert−/− fish also results in partial lifespan extension6. In zebrafish, telomerase expression is further required for regeneration of the heart7 and for developmental hematopoiesis8. Notably, tert−/− zebrafish mutants exhibit phenotypes comparable to those of dyskeratosis congenita (DC), a bone marrow failure disorder that affects multiple parts of the human body. Overall, these findings make zebrafish a suitable model for studying telomere biology9.
In mammals, telomeres are bound by the shelterin complex, which consists of six core proteins (TRF1, TRF2, RAP1, POT1, TIN2, and TPP1), the CST complex (CTC1, STN1, TEN1) and other telomere-associated proteins10. These proteins have orthologs in zebrafish, and initial functional characterization has already been conducted.
Loss of Terfa (the TRF2 ortholog) in zebrafish is embryonic lethal, and heterozygous adult fish exhibit accelerated aging11. In addition to the induction of DNA damage, terfa−/− fish experience neurodevelopmental failure due to brain edema during embryo development based on transcriptional misregulation12. Generated pot1−/− and acd−/− (the TPP1 ortholog) lines die at the larval stage, with a few acd−/− mutant escapers showing premature aging13. Furthermore, acd morphants exhibit neural death, heart defects and extensive apoptosis during embryonic development14. In contrast, tinf2−/− (the TIN2 ortholog) and terf1−/− mutants seemed unaffected and developed normally into adults13.
Previously, a phylointeractomic study utilizing telomere pull-down assays coupled with mass spectrometry analysis identified TTAGGG-binding proteins in 16 vertebrate species, including zebrafish15. Apart from the core shelterin proteins, several recently characterized direct telomere-binding zinc finger proteins, such as ZBTB1016 and ZNF52417, bind to the telomeric TTAGGG repeat sequence. One of these zinc finger proteins, ZBTB48 (also known as TZAP or HKR3), showed conserved phylogenetic binding in 13 of the 16 species15. ZBTB48 is composed of an N-terminal BTB/POZ domain and a series of eleven adjacent C2H2-type zinc fingers at the C-terminus. Its human ortholog directly interacts with the TTAGGG telomere repeats via its eleventh zinc finger and adjacent C-terminal arm18. Telomere sequence recognition is exerted by a recurring RxxHxxR motif in the telomeric zinc finger proteins ZBTB10, ZNF524 and ZBTB4817,19.
Knockout of ZBTB48 in telomerase-positive human HeLa cancer cells and mouse embryonic stem cells results in telomere elongation20,21. Conversely, the overexpression of ZBTB48 in human U2OS cancer cells maintaining telomere length via the alternative lengthening of telomere (ALT) pathway results in telomere shortening21. This alteration was accompanied by the accumulation of extrachromosomal telomere repeat (ECTR) DNA; thus, it was proposed that ZBTB48 is involved in telomere trimming21. Further investigation revealed that ZBTB48 binds to telomeres in an open chromatin state induced by the absence of the ATRX/DAXX protein complex22. Although ZBTB48 has been extensively studied in cancer cell lines, alterations in this gene have been found in only 5% of tumors, and the prognosis varies depending on the tumor type23.
Apart from its telomeric functions, ZBTB48 can also regulate genes via direct binding to promoter regions of a selected subset of genes where it acts as a transcription activator. In human cancer cells, ZBTB48 knockout strongly downregulated mitochondrial fission process 1 (MTFP1), mirroring the mitochondrial matrix reorganization phenotype of MTFP1 depletion20. ZBTB48 was recently reported to regulate B-cell-specific CIITA expression in mice by acting as a pioneer transcription factor24.
To gain further insight into the telomeric and gene regulatory functions of ZBTB48 in an in vivo animal model, we established a zbtb48−/− zebrafish line.
Results
Identification of telomeric proteins in zebrafish by quantitative mass spectrometry
To recapitulate and expand on the previously reported putative telomere-binding proteins in zebrafish, we combined a telomere pull-down assay with label-free quantitative mass spectrometry (Fig. 1a)15. To this end, we concatenated chemically synthesized DNA oligonucleotides containing the telomeric TTAGGG sequence or a shuffled control sequence by in vitro ligation. The concatenated telomeric and control oligonucleotides were biotinylated in vitro, immobilized on paramagnetic streptavidin beads and incubated in quadruplicate with nuclear lysate from the zebrafish fin fibroblast BRF41 cell line. After several washes, the bound proteins were eluted from the beads, digested into tryptic peptides, and measured using a high-resolution mass spectrometer with subsequent label-free quantitative analysis based on MaxLFQ. In this experiment, we quantified 1,269 proteins, 46 of which were more than twofold enriched, with a p-value < 0.05 for the telomeric oligonucleotide (Fig. 1b; Supplementary Data 1). Among these proteins were all six orthologs of the core shelterin complex: Terfa, Terf1, Pot1, Tinf2, Terf2ip (also known as Rap1), and Acd (also known as Tpp1). By comparing our results to a previous screen covering 16 different vertebrates15, we successfully enriched all eight previously reported zebrafish candidates conserved among at least 5 species: the shelterin subunits Pot1, Terf2ip and Terfa, as well as the nuclear receptor Nr2c2, the transcription factor Runx2a, the zinc finger protein Zbtb7a, and the direct telomere-binding proteins Zbtb10 and Zbtb48 (Fig. 1c). Additionally, we also found Accs and Parp1, and zebrafish protein Zgc:171459 identified in the previous zebrafish experiment15. However, our current screen generated a more comprehensive list of candidates for zebrafish, as we listed here 9 additional proteins whose orthologs were already identified as TTAGGG binders in at least 5 other species in the previous phylointeractomics screen. These proteins include the Cbfb subunit of the heterodimeric Runx/Cbfβ complex, the shelterin-associated protein Dclre1b (also known as the 5’ exonuclease Apollo), the two direct telomere-binding proteins Hmbox1b and Znf827, the poly(C)-binding proteins Hnrnpk and Pcbp2, the nuclear apoptosis-inducing factor Naif1, the DNA helicase Recql, and the zinc-finger protein Vezf1a. Furthermore, we also identified two other proteins (i.e., Zbtb8a and its paralog Zbtb8b) that were more than 30-fold enriched and have not been reported in the previous study. Overall, of our 46 TTAGGG binders found in zebrafish, 20 overlap with the identified phylogenetic conserved candidates in the previous phylointeractomics study and thus constitute a solidified list of known and possible telomere-binding proteins.
a Schematic diagram showing an overview of the telomere pull-down experiment using nuclear lysate from the zebrafish fin fibroblast BRF41 cell line. b Volcano plot visualizing the results obtained by label-free quantification of the bound proteins. The pull-down experiment was conducted in quadruplicate using either concatenated telomeric TTAGGG or a control scrambled GTGAGT oligonucleotide as bait. The results were log-transformed and plotted on the x-axis as log2(fold change) and on the y-axis as -log10(p-value). The protein enrichment threshold was set at a fold change >2 and a p-value < 0.05 (Welch’s t test) with c = 0.05. Enriched proteins were annotated with their gene names and color-coded: shelterin subunits (magenta), proteins that were previously found in the study by Kappei et al. to be enriched in zebrafish (orange), and telomere-binding proteins previously identified as phylogenetically conserved in other species (green), which were also enriched in zebrafish (blue). c Comparison of the previous telomere zebrafish pull-down, the phylogenetic conserved binders and the candidates from the current study.
Generation of a zbtb48 knockout zebrafish line via CRISPR‒Cas
Among our highly enriched candidates was Zbtb48, which has recently been characterized as a direct telomere-binding protein in human HeLa and U2OS cells20,21. The zebrafish ortholog also features an N-terminal BTB domain and eleven C2H2-type zinc fingers (ZNFs) at the C-terminus (Fig. 2a). The amino acid sequence conservation of the BTB domain (aa 28–aa 93; 42% identity) is lower than that of the zinc finger region (aa 322–aa 637; 80% identity). Notably, the 10 amino acid residues involved in telomere binding in the region of zinc finger 11 and the adjacent C-terminal region are identical between human and zebrafish Zbtb4818.
a Protein sequence alignment of human ZBTB48 and zebrafish Zbtb48 showing greater homology in the telomere-binding zinc finger domain (80% identity; blue) than in the BTB domain (42% identity; beige). The ten amino acids highlighted in green that are involved in telomere binding activity in humans (Zhao et al.) are conserved in zebrafish. b The zebrafish zbtb48 gene consists of 11 exons (not drawn to scale). To create the zbtb48 CRISPR‒Cas knockout zebrafish line, a pair of guide RNAs was designed to target the second exon, resulting in a 110-base pair frameshift deletion with a premature stop codon. c Results of the genotyping of zbtb48 CRISPR‒Cas knockout zebrafish using agarose gel electrophoresis. d Comparison of zbtb48 mRNA levels between zbtb48−/− mutants and their wild-type counterparts via qRT‒PCR of 5 dpf larvae. The experiment was conducted in biological quadruplicate (25 pooled larvae), each with technical triplicates. Error bars represent the standard error of the mean (SEM), and p-values were calculated using two-tailed Welch’s t test. e mRNA sequencing tracks of the zbtb48 gene region in 5 dpf larvae of wild-type and zbtb48−/− mutant larvae. A magnified image of the boxed area is shown on the right. The CRISPR deletion is marked below the track. f Table showing the Zbtb48 peptides detected via mass spectrometry analysis of a telomere pulldown with lysates extracted from 5 dpf wild-type and zbtb48−/− larvae.
To investigate the role of Zbtb48 in zebrafish, we utilized a CRISPR-Cas9 system to generate a zbtb48 knockout zebrafish line. Using a pair of guide RNAs targeting the zbtb48 gene, we successfully deleted 110 base pairs from the second exon (Fig. 2b) and confirmed the deletion by PCR (Fig. 2c). We found a significant 4.6-fold increase (p = 0.004) in zbtb48 mRNA expression in the zbtb48−/− mutants sampled at 5 days post fertilization (dpf) (Fig. 2d). This observation was mirrored in the RNA-seq data of 5 dpf larvae (Fig. 2e), where the mRNA level increased 3.2-fold (p = 6.59e−08). Further examination of the deletion site in the RNA-seq tracks revealed a lack of sequencing reads in this region for the zbtb48−/− mutants (Fig. 2e, right). The deletion induced a frameshift mutation resulting in a 50 amino acid (5.5 kDa) long Zbtb48 mutant protein that lacked the ZNF domain essential for telomere binding. To confirm the loss of telomere binding of this truncated Zbtb48 protein, we performed a telomere pull-down assay coupled to mass spectrometric analysis with protein lysates from 5 dpf wild-type and zbtb48−/− larvae. Zbtb48 peptides were detected in the pull-down with lysates from the wild-type larvae but not in the pull-down with lysates from the zbtb48−/− larvae despite the heightened zbtb48 mRNA expression level (Fig. 2f). While no shelterin subunits were among the enriched proteins, Nr2f2 identified from the telomere pull-down with BRF41 lysate (Fig. 1b), was significantly enriched by 4.4-fold (p = 0.005) in zbtb48−/− mutants. (Supplementary Fig. 1 and Supplementary Data 1). In summary, we generated a zebrafish line with a telomere-binding defunct Zbtb48.
Breeding and investigation of telomere length in zbtb48 −/− mutants
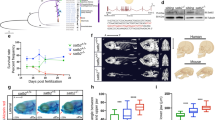
The knockout of some telomere-binding proteins was associated with a reduction in fitness or lethality in offspring. To examine the genotype distribution of our offspring bred from zbtb48+/− parents, 500 offspring from eight crosses were genotyped at various developmental stages. Generally, the distribution of all crosses followed a Mendelian inheritance pattern, with the exception of cross 6 (Fig. 3a). The results also suggest no significant reduction in the fitness of the zbtb48−/− mutant during development. Our first inbred generation of zbtb48−/− mutant fish was alive for two years before the study concluded. Unlike in tert−/− mutants1,4, we did not observe any obvious difference in lifespan between the zbtb48−/− mutants and their wild-type counterparts. Additionally, the zbtb48−/− mutants were fertile, and we successfully bred a second generation of zbtb48−/− mutants through in-crossing. This second generation of zbtb48−/− mutants was also viable and fertile even at an older age. A fertility test was performed on six zbtb48−/− mutant males and five wild-type fish at 1.5 years by crossing with wild-type females in a single mating. Despite the second-generation zbtb48−/− mutants (71%) showed a lower median of fertilized eggs as compared to their wild-type counterparts (96%), the difference was not statistically significant (p = 0.11) (Fig. 3b). Neither in the first nor second generation did we observe any physical or behavioral abnormalities in zbtb48−/− mutants. For example, we compared the appearance (Fig. 3c), weight (Fig. 3d), and length (Fig. 3e) of 10.5-month-old males of second-generation zbtb48−/− mutants and their wild-type specimens.
a Table showing the chi-square test results for 500 offspring from eight crosses between zbtb48+/− parents to determine the Mendelian genotype distribution. The test was performed on individual crosses (right) and overall as a group (bottom). For a test of significance at α = 0.05 with 2 degrees of freedom (df), the critical value of the chi-square (X²) is 5.99. b Boxplot showing the percentage of fertilized eggs from second-generation zbtb48−/− mutant males and their wild-type counterparts, both at 1.5-year-old, crossed with wild-type females. Each dot represents a mating setup (n = 6 for zbtb48−/− mutants and n = 5 for wild-type), indicating the percentage of fertilized eggs relative to the total number laid. P-values were calculated using Welch’s t-test. c Photograph of second-generation zbtb48−/− male zebrafish and their wild-type counterparts at 10.5 months of age. The scale bar equals 1 cm. Boxplot for weight (d) and size (e). Measurements were performed on four specimens, and p-values were calculated by Welch’s t test. f Bar plot of the mean telomere length of wild-type and zbtb48−/− mutant fish (n = 3 per genotype) quantified via telomere restriction fragment (TRF) analysis of the caudal fin collected from the first generation of 8-month-old males. Error bars represent the standard error of the mean (SEM), and p-value was determined by an unpaired two-tailed t test (Mann–Whitney).
Next, we quantified the telomere length of 8-month-old wild-type and zbtb48−/− males from the first incross by performing telomere restriction fragment (TRF) analysis on the collected caudal fins. We detected no change in the mean telomere length of the zbtb48−/− fins compared to that of the wild-type controls (p = 0.46) (Fig. 3f; Supplementary Fig. 2). As telomere length homeostasis was not affected in zbtb48−/− fish, we focused on the transcriptional activity of Zbtb48.
Transcriptomic and proteomic profiling
In addition to controlling telomere length, Zbtb48 has been reported to be a transcriptional regulator in human cells20. To investigate whether this function is conserved in zebrafish, we conducted transcriptome (Fig. 4a) and proteome (Fig. 4b, c) analyses of 5 dpf zbtb48−/− mutant and wild-type larvae.
a Volcano plot with quantitative comparison of RNA sequencing (RNA-seq) data from 5 dpf wild-type and zbtb48−/− mutant larvae (n = 4). The results were log-transformed and plotted on the x-axis as log2(fold change) and on the y-axis as -log10(p-value) (Welch’s t test). The transcript enrichment thresholds were set at a fold change > |2| and a p-value < 0.01. Genes annotated in this plot were also differentially expressed in the proteome analysis (b and c). Note: the p-value (4.14e−44) of zgc:153284 is beyond the upper limit of the y-axis. b, c Volcano plots for two biologically independent proteome analyses conducted on pooled 5 dpf wild-type and zbtb48−/− mutant larvae. The results were log-transformed and plotted on the x-axis as log2(fold change) and on the y-axis as -log10(p-value) (Welch’s t test). The protein enrichment threshold was set at a fold change > |2| and a p-value < 0.05. d Venn diagrams showing downregulated (left) and upregulated (right) transcripts or proteins in the zbtb48−/− mutants compared to the wild-type. e qRT‒PCR analysis of selected genes in 5 dpf larvae of wild-type and zbtb48−/− mutant zebrafish. The set includes differentially expressed genes from this study (left) and previously reported human target genes (Jahn et al.) (right). The experiment was conducted in biological quadruplicate (25 pooled larvae) with technical triplicates each. Error bars represent the standard error of the mean (SEM), and p-values were calculated using two-tailed Welch’s t test.
For RNA sequencing (RNA-seq) analysis (Fig. 4a; Supplementary Data 2), we collected four wild-type and zbtb48−/− siblings bred from zbtb48+/− parents through single mating as biological replicates. Among the 32,058 genes quantified by RNA-seq, only 25 genes exhibited downregulation, and 7 genes showed upregulation in the mutant (fold change > |2|, p-value < 0.01). In agreement with our qPCR data (Fig. 2d), among these upregulated genes was also zbtb48. Notably, zgc:153284, which is predicted to be involved in cell redox homeostasis, was the most strongly deregulated gene, with a 163-fold (p = 4.14e−44) increase in expression in the zbtb48−/− larvae.
We conducted the proteome experiment in two biological replicates with independently collected samples. Wild-type and zbtb48−/− larvae were bred from parents of the respective genotype, and they were pooled from a single mating. In the first replicate (Fig. 4b; Supplementary Data 3), a total of 8,075 proteins were quantified, with 80 downregulated and 109 upregulated (fold change > |2|, p-value < 0.05). In the second experiment from a new crossing (Fig. 4c; Supplementary Data 3), a total of 7601 proteins were quantified, 99 of which were downregulated and 152 of which were upregulated. Between the two replicates, we observed some variation, and only 11 proteins (Akr7a3, Cpa4, Ctrb.2, Cyp2aa7, Cyp2p8, Golgb1, Hdac10, Lmf2b, Mtfp1, Olfml3a and Zgc:111983) were downregulated, while 10 proteins (Coch, Col28a2a, Emc1, Myl7, Slc27a1a, Vtg2, Vtg3, Vtg5, Vtg7 and Zgc:153284) were upregulated in zbtb48−/− zebrafish according to both proteome analyses (Fig. 4d). To gain deeper insights into the dysregulated genes, gene ontology analysis of biological processes (GOBP) was performed on both transcriptomic and proteomic data. Although no significant downregulated GOBP terms were identified in the transcriptomic data, biological processes related to transition metal ion transport were consistently and significantly downregulated in both proteomic datasets. In contrast, the upregulated biological processes exhibited differences between the two gene regulatory layers. The transcriptomic data showed a broad range of processes, including translation and nucleotide metabolism. Meanwhile, the first proteomic replicate primarily highlighted processes related to proteolysis and apoptosis, while the second proteomic replicate featured macromolecule and transcription-related processes (Supplementary Fig. 3).
While no clear trend in dysregulated GOBP terms was observed, a small set of genes consistently exhibited significant changes in transcript and protein expression levels (Fig. 4d). This includes akr7a3, atox1, gbp3 and mov10a, which were downregulated, while zgc:153284 was upregulated. We validated the transcript downregulation of akr7a3 and atox1, as well as the upregulation of zgc:153284 at 5 dpf by qRT‒PCR (Fig. 4e). Additionally, consistent with the proteome data, we detected downregulation of the hdac10 and mtfp1 transcripts and upregulation of the col28a2a transcript in zbtb48−/− larvae, which were not significantly different at the transcript level according to the chosen RNA-seq cut-offs (Fig. 4e). These genes cover diverse biological aspects (Supplementary Data 3), but one of them, Mtfp1, is particularly interesting because its downregulation has also been reported in human ZBTB48 knockout cell lines20. We thus extended our investigation to other genes reported to be regulated by ZBTB48 in humans, namely, PXMP2, PP3CB, and VWA5A, by examining the transcription levels of their zebrafish orthologs via qRT‒PCR. There were no differences in the expression of pp3cb or vwa5a between zbtb48−/− and wild-type zebrafish larvae at 5 dpf, and only a slight 1.2-fold (p = 0.03) decrease in pxmp2 expression was detected (Fig. 4e). Therefore, strong mtfp1 downregulation upon zbtb48 knockout was the most pronounced similarity at the transcript and proteome levels between humans and zebrafish.
Spatiotemporal expression of zbtb48
Based on our proteome and RNA-seq data, Zbtb48 protein expression was low at 5 dpf because it was not detected in the proteome measurements and had a low number of transcript reads. Therefore, we assessed Zbtb48 expression based on publicly available data.
Using a developmental zebrafish study25, we detected high zbtb48 transcript levels in early development prior to zygotic genome activation, suggesting that zbtb48 mRNA is maternally loaded into the embryo (Fig. 5a). Transcript levels gradually decrease during development, and at 10–12 hpf, the liver and spinal cord show the highest zbtb48 transcription levels, followed by primordial germ cells (PGCs) at half of the level26,27 (Fig. 5b). By 5 dpf (or 120 hpf), the zbtb48 transcript became predominantly expressed in PGCs. Due to the strong expression of zbtb48 transcripts in larval PGCs, we anticipated similar high expression in adult gonads. Therefore, we extended our investigation with a previously published RNA-seq dataset of adult tissues28 and found prominent expression in the testis but 30-fold lower expression in the ovary (Fig. 5c). In addition to those in the gonads, low levels of the zbtb48 transcript were also detected in the skin, brain, fins and spleen.
a Developmental expression profile with zbtb48 mRNA highlighted (green thick line) in whole larvae replotted from previously published data (White et al.). Heatmap of zbtb48 expression in different tissues during the first five days of development (b) based on data from Farrell et al. and Sur et al. or in adult tissues (c) based on data from Jiang et al. d UMAP plots of single-cell RNA sequencing (scRNA-seq) of testes from 5- to 22-month-old fish. Figures were downloaded from the public website (Sposato et al.). e UMAP plots of scRNA-seq data from ovaries of 40 dpf fish. Figures were downloaded from a public website (Liu et al.). For d and (e), the top left panel shows the zbtb48 mRNA expression level, the top right panel shows ddx4 (also known as vasa, which is highly expressed in germ cells), the bottom left panel shows cdh17 (not expressed in germ cells), and the bottom right panel shows the cell population annotation.
We speculated that the zbtb48 transcript might be more highly expressed in highly proliferative tissue due to its involvement in telomere regulation. A single-cell RNA sequencing (scRNA-seq) study of zebrafish testes29 revealed the highest zbtb48 expression in spermatocytes (undergoing meiosis), followed by spermatogonia (stem cells) and then early round spermatids, irrespective of age (Fig. 5d). Notably, the expression of zbtb48 is high but restricted to a low number of cells, in contrast to cdh17, a gene expressed in the nephrons, liver and intestine30 that serves as a negative control, and ddx4 (also known as vasa), which is strongly expressed in the germline31,32. Consistently, published scRNA-seq data of 40 dpf old juvenile ovaries33 also revealed the highest zbtb48 expression in the germline stem cell population, atop meiotic cells and early oocytes I, compared to cdh17 as a negative control and ddx4 as a positive control (Fig. 5e). Overall, the data supported the expression of zbtb48 transcripts in the gonads, albeit at a small subset of cells and at lower levels than those of the germ cell marker ddx4.
We thus decided to focus our investigation on the juvenile gonads of 40 dpf fish. To easily distinguish the gonads, we crossed our zbtb48−/− zebrafish with a transgenic line expressing eGFP under the ddx4 promoter, also called vasa (Tg(vasa: eGFP))34. For this study, we used offspring from a single mating of Tg(vasa:eGFP);zbtb48+/− and collected ovaries from females and testes from male offspring for proteome analysis.
Among the 6015 proteins quantified via proteome analysis of the ovaries of zbtb48−/− versus wild-type zebrafish, 850 proteins were downregulated, and 38 proteins were upregulated in zbtb48−/− mutants (Fig. 6a; Supplementary Data 4). In contrast, using the same threshold (fold change > |2|, p-value < 0.05) among 5891 quantified proteins in the testis experiment, only 30 proteins were downregulated, and 22 proteins were upregulated in zbtb48−/− mutants (Fig. 6b; Supplementary Data 4). Notably, there was a stark difference in the number of downregulated proteins between the ovary and testis in 40 dpf zebrafish. Among the 11 proteins downregulated in the 5 dpf zbtb48−/− larvae, Golgb1 was also significantly downregulated in the testis, as were Akr7a3 and Mtfp1 in both the ovary and testis.
Volcano plots of the proteome analysis of ovaries from 40 dpf (a), testes from 40 dpf (b) and testes from 10.5-month-old (c) wild-type and zbtb48−/− mutant fish. Biological replicates of 40 dpf ovaries (n = 3) and 40 dpf and 10.5-month-old testes (n = 4) were measured for each genotype. The results were log-transformed and plotted on the x-axis as log2(fold change) and on the y-axis as -log10(p-value) (Welch’s t test). The protein enrichment thresholds were set at a fold change > |2| and a p-value < 0.05. Proteins common to Fig. 4d were annotated.
To better recapitulate the previously published scRNA-seq data for mature adult testes at 5–22 months of age29, we conducted another proteome analysis with 10.5-month-old second-generation zbtb48−/− mutants. Of the 5419 proteins quantified, 227 were downregulated and 9 were upregulated in the zbtb48−/− mutants. Thus, the number of downregulated proteins increased with age. Notably, among the downregulated proteins we could recapitulate Mtfp1 from previous 40 dpf ovary and testis experiments, with Mtfp1 showing the strongest downregulation among all measured proteins in 10.5-month-old testes (Fig. 6c; Supplementary Data 4). While mtfp1 was also downregulated at transcript level in the testes of 1-year-old first-generation zbtb48−/− fish (Supplementary Fig. 4a), unlike for the 5 dpf embryos, no significant increase in zbtb48 transcript levels was detected.
GOBP analysis of these proteome datasets did not reveal any consistent biological processes. Unlike the downregulation of transition metal ion transport at 5 dpf, the ovaries and testes of zbtb48−/− mutants at 40 dpf showed downregulation mainly related to carbohydrate metabolism, with no significant downregulation in the testes at 10.5 months. Instead, the 10.5-month-old testes exhibited upregulation of transcription-related processes, while no significant upregulation was observed in either gonad at 40 dpf (Supplementary Fig. 4b).
Discussion
We performed a telomere interactomics screen with lysates from a zebrafish cell line and extended the list of previously identified known and putative telomeric proteins. Among the 46 proteins that we found to be enriched with the TTAGGG oligonucleotide, 20 were already shown to recognize TTAGGG repeats in other vertebrate species15. These proteins include members of the shelterin complex and other previously reported direct telomere binding proteins, such as Hmbox135, Zbtb1016, Znf82736, and Zbtb4820,21. We decided to focus on Zbtb48, which has previously been reported to be a negative regulator of telomere length in human cancer cell lines and mouse stem cells20,21, by generating a zbtb48 knockout zebrafish line via CRISPR technology. However, we did not observe a telomere lengthening phenotype in our first-generation zbtb48−/− mutants. Thus, at present, we cannot comment on whether zbtb48−/− zebrafish require additional generations to exhibit telomere lengthening or whether Zbtb48 is not involved in telomere homeostasis in zebrafish. Additionally, there may be other factors that act redundantly with Zbtb48 to establish the telomere length mean in zebrafish. Therefore, a further long-term study of zbtb48−/− zebrafish mutants will be required to address this question.
Apart from its role as a telomere length regulator, ZBTB48 was also reported to be a transcription activator for a small group of genes20. Similarly, we observed gene regulatory changes in zbtb48−/− zebrafish in the first and second generations. In this study, we measured the transcriptome and proteome of 5 dpf zebrafish larvae as well as the proteome of ovaries from 40 dpf and testes from 40 dpf and 10.5-month-old zebrafish. Similar to the human ZBTB48 studies, in the zbtb48−/− mutants, we observed more downregulated genes (n = 25) than upregulated genes (n = 7) in our 5 dpf transcriptome, which is close to the low number of downregulated genes reported in U2OS (n = 23) and HeLa (n = 11) cells. Although this trend was not observed in the proteomic data collected at 5 dpf, a skewed distribution later manifested in the proteome of the gonads, with 40 dpf ovaries (850 downregulated, 38 upregulated) and 10.5-month-old testes (227 downregulated, 9 upregulated) showing a stronger trend than 40 dpf testes (30 downregulated, 22 upregulated).
The analyses revealed a small consistent group of genes that were repeatedly downregulated. One of the candidates is the nuclear-encoded mitochondrial protein mtfp1, which was previously reported to be downregulated in human ZBTB48 knockout cancer cell lines20. Its downregulation was detected via qRT‒PCR and proteomic analysis of 5 dpf zbtb48−/− larvae and adult testes, as well as in the proteomes of the juvenile ovaries and testes. Apart from mtfp1, we also found other candidates that were previously not reported in human cell lines. These genes are aldo-keto reductase akr7a3 and the guanylate binding protein gbp3 (involved in the innate immune response), which were consistently downregulated in the transcriptome and most of our proteome analyses. Conversely, Zbtb48 knockout increased the transcript levels of zgc:153284, which is involved in cell redox homeostasis, and consequently, protein upregulation was observed in the proteome at 5 dpf. Unlike the previously mentioned candidates, its upregulation was not detected in the gonad proteomes.
We also found that Zbtb48 is generally expressed at low levels in tissues because it was barely detected in our proteomes except for 40 dpf ovaries. The scRNA-seq expression data from various sources29,33 show that the zbtb48 transcript is expressed primarily in the stem cells and gametocytes of the gonads. Since germline stem cells are constantly dividing and rely on telomerase to replenish their telomere length to support ongoing cell division for reproduction, it is conceivable that high zbtb48 expression helps to reinforce the upper limit of telomere length.
Overall, we established a zbtb48−/− zebrafish line that can be used as an organismal model to further study the function of zbtb48 in gene regulation and telomere biology. Although the study of telomere homeostasis in this animal model requires further long-term investigations, we successfully recapitulated the previously reported zbtb48-mtfp1 regulatory loop in zebrafish. The finding that Mtfp1 is an evolutionarily conserved target of Zbtb48 may suggest a relationship between the role of Zbtb48 in telomere maintenance and mitochondrial regulation37,38,39. However, further investigations are needed to elucidate the function of Zbtb48 in zebrafish.
Methods
Zebrafish cell culture
The zebrafish-derived fibroblast line BRF41 (CVCL_4131) was cultured in Leibovitz’s L-15 medium (Gibco) supplemented with 15% fetal bovine serum (Gibco) and 1% penicillin‒streptomycin (Sigma). The cells were maintained at 33 °C in an incubator and split 1:5 by trypsinization every three to four days for passaging.
Preparing protein extracts from BRF41 cells
Nuclear protein extracts were prepared from BRF41 cells as previously described15. The cells were incubated in hypotonic buffer (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, and 10 mM KCl) on ice for 10 min and then supplemented with 0.2% Igepal CA630 (Sigma) and protease inhibitors before they were homogenized with 50 strokes with a douncer. Nuclei were pelleted and washed with 1X PBS. Nuclear proteins were extracted from the nuclei by incubating them in hypertonic buffer (420 mM NaCl, 20 mM HEPES (pH 7.9), 20% glycerol, 2 mM MgCl2, 0.2 mM EDTA, 0.1% Igepal CA630 (Sigma), 0.5 mM DTT and protease inhibitors) for 1 h at 4 °C on a rotating wheel. After centrifugation at maximum speed for 1 h at 4 °C, the supernatant containing the nuclear protein extract was collected. The protein concentrations were measured using a Bradford assay (Bio-Rad).
Telomere pull-down
Telomere pull-down was performed as described15. Briefly, oligonucleotide baits were biotinylated and immobilized on paramagnetic streptavidin beads (Dynabeads MyOne C1, Thermo Scientific). For each replicate, 450 µg of the beads were incubated with 400 µg of protein lysate, and all experiments were conducted with four replicates. Both telomere (TTAGGG) and scrambled (GTGAGT; control) baits were used in the BRF41 pull-down, while only telomere bait was used in the pull-down of wild-type versus zbtb48−/− mutant larvae. The incubation was carried out on a rotating wheel at 4 °C for 1 h, followed by three rounds of washes with PBB buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 0.5% Igepal CA630 (Sigma), freshly supplemented with 1 mM DTT and protease inhibitors). The samples were eluted with 20 μl of 0.1 M DTT and 1X NuPAGE™ LDS Sample Buffer (Thermo Scientific), followed by boiling at 70 °C for 10 min.
MS sample preparation
Lysates eluted from the telomere pull-down were separated on a 10% NuPAGE Bis-Tris precast gel (Thermo) at 180 V for 8 min. In-gel digestion was performed as previously described15. In brief, the samples were reduced with 10 mM DTT for 1 h at 56 °C, followed by alkylation with 55 mM iodoacetamide (Sigma) for 45 min in the dark. Tryptic digestion was carried out with 1 μg of MS-grade trypsin (Sigma) in 50 mM ammonium bicarbonate buffer (pH 8.0) at 37 °C overnight. Afterwards, the peptides were desalted and stored on activated C18 material (Empore) StageTips.
MS measurement and analysis (telomere pull-down)
Measurements of telomere pull-down lysates from BRF41 and 5 dpf larvae were conducted on an Exploris 480 (Thermo) coupled online to an Easy-nLC 1200 (Thermo) on an in-house packed C18 (Dr. Maisch GmbH) column. The data were acquired via data-dependent acquisition (DDA) with a top15 method. The BRF41 data were analyzed with MaxQuant40 version 1.6.10.43 using the UniProt zebrafish database (39,559 entries) with standard settings except label-free quantification (LFQ), and the match between run options were activated. The 5 dpf data were analyzed with MaxQuant (2.4.2.0) using a combined Swissprot/Trembl database (47,082 entries) with the standard settings except for peptides for quantification, which was switched to unique and LFQ quantitation activated. Protein quantification was performed as previously described41. In brief, contaminants, reverse database hits, protein groups identified only by site, and protein groups with fewer than two peptides (with at least one classified as unique) were filtered out from the MaxQuant proteinGroups.txt file. Missing values were imputed by shifting a beta distribution, based on the LFQ intensity values, to the limit of quantitation. Volcano plots were generated from Rstudio using ggplot242 and other packages. The threshold for protein enrichment was set at a fold change > |2| and a p-value < 0.05 (Welch’s t test).
Orthologous comparison
Amino acid sequences and domain annotations of human ZBTB48 (P10074) and zebrafish Zbtb48 (E7FDZ5) were obtained from UniProt and aligned online43. The percentage of domain identity was obtained through a BLAST search.
Zebrafish husbandry
The AB/Tübingen zebrafish strains were housed at the Institute of Molecular Biology in Mainz, and husbandry was carried out in accordance with the standards described44. The room and water temperature were maintained at 26–28 °C with a 14:10-h light:dark cycle. To induce embryo spawning, adult males and females were separated in a mating tank the evening prior to embryo spawning. The mating process was initiated by reuniting the fish the following morning. The embryos were collected in a 10-cm Petri dish filled with E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) and raised in a 28.5 °C incubator until 5 days post fertilization (dpf). Afterward, they were transferred to the aquarium facility. All experiments were approved and licensed by the local authorities of Rhineland-Palatinate and were conducted in accordance with European animal welfare law. We have complied with all relevant ethical regulations for animal use.
Creating the zbtb48 CRISPR-Cas9 knockout zebrafish line
A pair of guide RNAs (gRNAs; Supplementary Table 1) targeting the second exon of the zbtb48 gene (ENSDARG00000039263) were designed using CRISPRscan software (https://www.crisprscan.org). These gRNAs were cloned and inserted into the pDR274 vector and transcribed as previously described45. The injection mixture was prepared with 1.5 μl of each gRNA, 0.5 μl of NEB3.1, 0.5 μl of phenol red (Sigma) and 1 μl of EnGen® Spy Cas9 NLS (NEB). This mixture was injected into embryos at the one-cell stage, which were then raised to adulthood and outcrossed with wild-type fish. The strain is registered as ZDB-ALT-241104-4 at the zebrafish information network. The F1 offspring were screened for deletions in the zbtb48 locus using PCR. These F1 mutants were outcrossed with wild-type fish for another generation to create an F2 generation before in-crossed to generate zbtb48−/− mutants (F3). Additionally, the zbtb48−/− mutant fish line was crossed with the wild-type Tg(vasa:eGFP) reporter line34 to label germ cells with green fluorescence.
Genotyping of zebrafish specimens
The fish were either sacrificed on an ice bath or anesthetized with 0.02% tricaine (Sigma) for caudal fin clipping. The samples were lysed in lysis buffer (10 mM Tris-HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 0.45% Igepal CA630 (Sigma), 0.45% TWEEN® 20 (Sigma), 0.01% gelatin, and 100 μg*ml−1 proteinase K) at 60 °C for 1 h, followed by proteinase K deactivation at 90 °C for 15 min. The PCR mixture was prepared in a 10 μl reaction volume using Taq polymerase (IMB PPCF) and the primers listed in Supplementary Table 1. The amplification was performed as follows: denaturation at 95 °C for 5 min; 5 cycles of 95 °C for 15 s, 64 °C for 30 s, and 68 °C for 30 s; 5 cycles of 95 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s; and 27 cycles of 95 °C for 15 s, 55 °C for 30 s, 68 °C for 30 s, and a final extension at 68 °C for 5 min. The PCR products were resolved by 2% agarose gel electrophoresis. The genotype distribution of offspring bred from eight zbtb48+/− crosses was tested using the chi-square test. The test was performed on individual crosses and overall as a group. The outcome was determined by the X2 critical value of 0.05 significance level (α) and 2 degrees of freedom.
Fish fertility
Fertility test was conducted on second-generation males aged 1 year and 6 months (n = 5 for wild-type and n = 6 for zbtb48−/− mutants). The experiment was carried out in a single mating setup, using females from a single wild-type clutch that were aged 1 year 3 months. Adult males and females were separated in the mating tank the evening before embryo spawning and were reunited the following morning for mating. The eggs were collected within 30 min after laying, allowing for the removal of any defective eggs before counting the fertilized eggs around 3 h post-fertilization (hpf). The percentage of fertilized eggs (calculated as fertilized eggs divided by the total number of eggs laid) for each mating was represented in a boxplot. P-values were calculated using Welch’s t-test.
Telomere restriction fragment (TRF) analysis by Southern blot
Telomere length was measured on the caudal fins of three 8-month-old males of zbtb48−/− and the wild-type counterpart fish bred through group mating of zbtb48+/− parents. The TRF analysis was performed as previously described4. In brief, the fins were lysed overnight at 50 °C using lysis buffer (Fermentas) supplemented with 1 mg*ml−1 Proteinase K (Sigma) and 1:100 dilution RNase A (Sigma). Genomic DNA was extracted by equilibrated phenol-chloroform (Sigma) and chloroform-isoamyl alcohol extraction (Sigma). The same amount of genomic DNA was digested with RSAI and HINFI enzymes (NEB) for 12 h at 37 °C. Samples were electrophoresed on a 0.6% agarose gel, in 0.5% TBE buffer, at 4 °C for 17 h at 110 V. Gels were then processed for Southern blotting using a 1.6 kb telomere probe, (TTAGGG)n, labeled with [α-32P]-dCTP.
Quantitative real-time PCR
qRT‒PCR was performed on 5 dpf wild-type and zbtb48−/− mutant larvae bred from parents of the respective genotype through single mating. Testes were harvested from the first generation of zbtb48−/− mutants and their wild-type counterparts at 1-year-old. Each biological replicate consisted of a pool of 25 larvae or the gonad from a single individual. Samples were resuspended in 500 μl of TRIzol™ Reagent (Ambion) and sonicated by a Bioruptor Plus UCD-300I (Diagenode) at a low setting with three rounds of on for 30 s and off for 30 s, or until the samples were fully disintegrated, in a 4 °C water bath. RNA was purified using a Direct-zol RNA MicroPrep Kit (Zymo Research), and 1 μg of the extracted RNA was used for synthesizing complementary DNA (cDNA) with a First Strand cDNA Synthesis Kit (Thermo Fisher) according to the manufacturer’s instructions. qRT‒PCR was performed in technical triplicates, and each reaction consisted of a total volume of 10 μl: 5 μl of 2X Power SYBR Green PCR Master Mix (Applied Biosystems), 0.3 μl of each 10 μM primer, and 2 μl of 1:20 diluted cDNA. The primers used in this study are listed in Supplementary Table 1, and β-actin was used as a reference. The reactions were performed in a QuantStudio 5 Real-Time PCR system (Applied Biosystems) with the following amplification conditions: 50 °C for 2 min, 95 °C for 10 min, 40 cycles at 95 °C for 15 s, and 60 °C for 1 min, followed by 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s to determine the melting curve. The data were extracted from QuantStudioTM Design & Analysis software (Applied Biosystems) and processed with the 2 − ΔΔCt method46. Error bars represent the standard error of the mean (SEM), and p-values were calculated using two-tailed Welch’s t test.
Tissue harvesting
To obtain total protein lysates from 5 dpf wild-type and zbtb48−/− mutant larvae for the telomere pull-down experiment, approximately 200 larvae bred from parents of the respective genotype through single mating were pooled. The samples were sonicated in 2 ml of protein lysis buffer (25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1% Triton™ X-100 (Sigma), 1 mM DTT, and protease inhibitors) using Bioruptor Plus UCD-300I (Diagenode) with three rounds of on for 30 s and off for 30 s, or until the samples were fully disintegrated, in a 4 °C water bath. The lysates were centrifuged at maximum speed for 30 min at 4 °C, and the supernatant was collected. The protein concentrations were measured using a Bradford assay (Bio-Rad).
For proteome analysis, 5 dpf wild-type and zbtb48−/− mutant larvae were also bred from parents of the respective genotype through single mating and pooled. An additional deyolking step was performed by titrating the larvae in cold Ringer’s buffer (5 mM HEPES (pH 7.2), 116 mM NaCl, 2.9 mM KCl) supplemented with 0.3 mM PMSF and 1 mM EDTA (pH 7.0). To prepare the total protein lysates of larvae and adult testes from 10.5-month-old fish, samples were snap-frozen in liquid nitrogen and ground with micropestles before being resuspended in 50 μl of RIPA buffer. For the ovaries and testes harvested from 40 dpf juveniles, the fish were bred via single mating of Tg(vasa:eGFP);zbtb48+/− parents. The fish were genotyped upon harvesting the ovaries or testes. The samples were lysed through sonication in 25 μl of RIPA buffer with the settings mentioned above. All RIPA lysis buffer was centrifuged at maximum speed for 30 min at 4 °C to collect the supernatant. The protein concentrations were measured using a Bradford assay (Bio-Rad), and 50 μg of protein was prepared for each replicate with the addition of 0.1 M DTT and 1X NuPAGE™ LDS Sample Buffer (Invitrogen), followed by boiling at 70 °C for 10 min. For 5 dpf larvae, a 10% NuPAGE Bis-Tris gel was run at 180 V for 30 min, and each lane was cut into 3 fractions.
MS measurement and analysis (proteome)
Proteome analysis of 5 dpf larvae was conducted using an Exploris 480 (Thermo) coupled online to an in-house packed C18 (Dr. Maisch GmbH) column. For the ovaries and testes, proteomes were measured with a TimsTOF HT (Bruker) interfaced with a NanoElute2 HPLC system (Bruker). An Aurora Ultimate CSI 25 × 75 C18 UHPLC column (Ionopticks) was used at 50 °C. The peptides were eluted along a 95-min gradient from 4% to 32% acetonitrile (VWR) and measured with a DDA acquisition scheme. The measurement files were processed with MaxQuant (2.4.2.0) and a combined Swissprot/Trembl database (47,082 entries) with the standard settings except for peptides for quantification, which was switched to unique and LFQ quantitation activated. Protein analysis was performed as previously described41. In brief, contaminants, reverse database hits, protein groups identified only by site, and protein groups with fewer than two peptides (with at least one classified as unique) were filtered out from the MaxQuant proteinGroups.txt file. Missing values were imputed by shifting a beta distribution, based on the LFQ intensity values, to the limit of quantitation. Volcano plots were generated from Rstudio using ggplot242 and other packages. The threshold for protein enrichment was set at a fold change > |2| and a p-value < 0.05 (Welch’s t test).
Functional enrichment analysis
Genes were queried in the Gene Ontology database47 using the ClusterProfiler R package48. Terms found among the enriched genes were tested for over-representation with an adjusted p-value (false discovery rate) <0.05 (Fisher’s exact test) against terms found in the background (defined as all quantified genes in the comparison, whether enriched or not).
Next-generation mRNA sequencing and analysis
From a single mating of zbtb48+/− parents, 22 sibling larvae were picked at 5 dpf and processed individually. RNA and DNA were extracted using an AllPrep DNA/RNA/Protein Mini Kit (Qiagen) according to the manufacturer’s instructions. The larvae were sonicated in the provided RLT buffer using three rounds of Bioruptor Plus UCD-300I (Diagenode) for 30 s and off for 30 s, or until the samples were fully disintegrated, in a 4 °C water bath. Genotyping was performed on the extracted DNA, and subsequently, the RNA of four selected wild-type and zbtb48−/− mutant strains was subjected to mRNA sequencing.
NGS library preparation was performed with Illumina’s TruSeq Stranded mRNA LT Sample Prep Kit following the TruSeq Stranded mRNA Reference Guide (Oct. 2017) (Document # 1000000040498v00) using ¼ of the reagents. Libraries were prepared with a starting amount of 250 ng and amplified in 10 PCR cycles. Libraries were profiled on a high-sensitivity DNA chip on a 2100 Bioanalyzer (Agilent Technologies) and quantified using the Qubit dsDNA HS Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies). All 8 samples were pooled together with 20 samples from another project in equimolar ratio and sequenced on a NextSeq 500 High Output FC, SR for 1 × 75 cycles plus 2 × 8 cycles for the dual index read.
The library quality was assessed with FastQC (version 0.11.9) and FastQScreen (version 0.15.2) before alignment against the D. rerio genome assembly GRCz11 and its canonical gene annotations (Danio_rerio.GRCz11.100.chr.gtf) and its associated.GTF and.BED files annotations. Alignment was performed with STAR aligner49 version 2.7.3a (options: –runMode alignReads –outStd SAM –outSAMattributes Standard –outSJfilterReads Unique –outSAMunmapped Within –outReadsUnmapped None –outFilterMismatchNoverLmax 0.04 –outFilterMismatchNmax 999 –sjdbOverhang 75. Reads mapped to annotated features in the GTF file were counted with featureCounts50 version 1.6.2 using the featureCounts functionality (options: –donotsort -t exon). Coverage tracks were generated with deepTools51 version 3.1 (bamCoverage –binSize 1 –skipNonCoveredRegions –normalizeUsing CPM) and plotted using Gviz52 on an R framework. Finally, the overall quality of the reads and the alignment was assessed with MultiQC version 1.753.
Further filtering and exploratory analysis were performed in an R framework including ggplot242. Pairwise differential expression comparisons were performed with DESeq254. Gene expression in RPKM was used to filter out individuals with a replicate average lower than 0, thus considering them as non-expressed. Differentially expressed genes (DEGs) were selected with an adjusted p-value (false discovery rate) of <0.01, and a threshold of at least a 1 log2-fold-change difference between conditions was applied.
Analysis of published RNA-seq data
Metadata from RNA-seq analysis of embryo development25,26,27 and adult tissues28 were downloaded and processed in R.
The data and images of the scRNA measurements of 40 dpf ovaries33 and adult testes29 were obtained from the following websites: https://singlecell.broadinstitute.org/single_cell/study/SCP928/40dpf-ovary-all-cells and https://sposato.shinyapps.io/testis_shiny_app/.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The transcriptome data have been deposited in the SRA under accession number PRJNA1118296. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD052763. The data used in this paper and figures are available at Zenodo (https://doi.org/10.5281/zenodo.14793328).
References
Henriques, C. M., Carneiro, M. C., Tenente, I. M., Jacinto, A. & Ferreira, M. G. Telomerase is required for zebrafish lifespan. PLoS Genet. 9, e1003214 (2013).
Anchelin, M., Murcia, L., Alcaraz-Perez, F., Garcia-Navarro, E. M. & Cayuela, M. L. Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS One 6, e16955 (2011).
Anchelin, M. et al. Premature aging in telomerase-deficient zebrafish. Dis. Model Mech. 6, 1101–1112 (2013).
Carneiro, M. C. et al. Short Telomeres in Key Tissues Initiate Local and Systemic Aging in Zebrafish. PLoS Genet. 12, e1005798 (2016).
El Mai, M. et al. Gut-specific telomerase expression counteracts systemic aging in telomerase-deficient zebrafish. Nat. Aging 3, 567–584 (2023).
Serifoglu, N., Lopes-Bastos, B. & Ferreira, M. G. Lack of telomerase reduces cancer incidence and increases lifespan of zebrafish tp53(M214K) mutants. Sci. Rep. 14, 5382 (2024).
Bednarek, D. et al. Telomerase Is Essential for Zebrafish Heart Regeneration. Cell Rep. 12, 1691–1703 (2015).
Imamura, S. et al. A non-canonical function of zebrafish telomerase reverse transcriptase is required for developmental hematopoiesis. PLoS One 3, e3364 (2008).
Carneiro, M. C., de Castro, I. P. & Ferreira, M. G. Telomeres in aging and disease: lessons from zebrafish. Dis. Model Mech. 9, 737–748 (2016).
Lim, C. J. & Cech, T. R. Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat. Rev. Mol. Cell Biol. 22, 283–298 (2021).
Kishi, S. et al. The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet 4, e1000152 (2008).
Ying, Y. et al. The non-telomeric evolutionary trajectory of TRF2 in zebrafish reveals its specific roles in neurodevelopment and aging. Nucleic Acids Res. 50, 2081–2095 (2022).
Ma, J., Tang, D., Gao, P., Liang, S. & Zhang, R. Knockout of Shelterin subunit genes in zebrafish results in distinct outcomes. Biochem. Biophys. Res. Commun. 617, 22–29 (2022).
Xie, Y., Yang, D., He, Q. & Songyang, Z. Zebrafish as a model system to study the physiological function of telomeric protein TPP1. PLoS One 6, e16440 (2011).
Kappei, D. et al. Phylointeractomics reconstructs functional evolution of protein binding. Nat. Commun. 8, 14334 (2017).
Bluhm, A. et al. ZBTB10 binds the telomeric variant repeat TTGGGG and interacts with TRF2. Nucleic Acids Res. 47, 1896–1907 (2019).
Braun, H. et al. ZNF524 directly interacts with telomeric DNA and supports telomere integrity. Nat. Commun. 14, 8252 (2023).
Zhao, Y. et al. The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA. Cell Res. 28, 130–134 (2018).
Wang, S. et al. Structural insights into the recognition of telomeric variant repeat TTGGGG by broad-complex, tramtrack and bric-a-brac - zinc finger protein ZBTB10. J. Biol. Chem. 299, 102918 (2023).
Jahn, A. et al. ZBTB48 is both a vertebrate telomere-binding protein and a transcriptional activator. EMBO Rep. 18, 929–946 (2017).
Li, J. S. et al. TZAP: A telomere-associated protein involved in telomere length control. Science 355, 638–641 (2017).
Moreno, S. P. et al. TZAP overexpression induces telomere dysfunction and ALT-like activity in ATRX/DAXX-deficient cells. iScience 26, 106405 (2023).
Dos Santos, G. A. et al. Telomeric zinc-finger associated protein (TZAP) in cancer biology: friend or foe? Mol. Biol. Res. Commun. 10, 121–129 (2021).
Rane, G. et al. ZBTB48 is a priming factor regulating B-cell-specific CIITA expression. EMBO J. 43, 6236–6263 (2024).
White, R. J. et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 6, e30860 (2017).
Farrell, J. A. et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018).
Sur, A. et al. Single-cell analysis of shared signatures and transcriptional diversity during zebrafish development. Dev. Cell 58, 3028–3047.e3012 (2023).
Jiang, M. et al. Characterization of the Zebrafish Cell Landscape at Single-Cell Resolution. Front. Cell Dev. Biol. 9, 743421 (2021).
Sposato, A. L. Germ cell progression through zebrafish spermatogenesis declines with age. Development 151, dev204319 (2024).
Horsfield, J. et al. Cadherin-17 is required to maintain pronephric duct integrity during zebrafish development. Mech. Dev. 115, 15–26 (2002).
Hartung, O., Forbes, M. M. & Marlow, F. L. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 81, 946–961 (2014).
Krovel, A. V. & Olsen, L. C. Sexual dimorphic expression pattern of a splice variant of zebrafish vasa during gonadal development. Dev. Biol. 271, 190–197 (2004).
Liu, Y. et al. Single-cell transcriptome reveals insights into the development and function of the zebrafish ovary. Elife 11, e76014 (2022).
Krovel, A. V. & Olsen, L. C. Expression of a vas::EGFP transgene in primordial germ cells of the zebrafish. Mech. Dev. 116, 141–150 (2002).
Kappei, D. et al. HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment. EMBO J. 32, 1681–1701 (2013).
Yang, S. F. et al. ZNF827 is a single-stranded DNA binding protein that regulates the ATR-CHK1 DNA damage response pathway. Nat. Commun. 15, 2210 (2024).
Gao, X. et al. Telomeres and Mitochondrial Metabolism: Implications for Cellular Senescence and Age-related Diseases. Stem Cell Rev. Rep. 18, 2315–2327 (2022).
Sahin, E. et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365 (2011).
Zheng, Q., Huang, J. & Wang, G. Mitochondria, Telomeres and Telomerase Subunits. Front. Cell Dev. Biol. 7, 274 (2019).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Fradera-Sola, A., Nischwitz, E., Bayer, M. E., Luck, K. & Butter, F. RNA-dependent interactome allows network-based assignment of RNA-binding protein function. Nucleic Acids Res. 51, 5162–5176 (2023).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279 (2022).
Alestrom, P. et al. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 54, 213–224 (2020).
Hwang, W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Gene Ontology, C. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Hahne, F. & Ivanek, R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol. Biol. 1418, 335–351 (2016).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We are thankful to the IMB zebrafish facility, the IMB media lab, and the IMB protein production facility. Support from the IMB Genomics Core Facility and the use of its NextSeq500 (funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – INST 247/870-1 FUGG) is gratefully acknowledged. We thank Rachel Mullner for technical support. M.G.F. and N.S. were supported by Institut National du Cancer (INCa, PLBIO21-228) and La Ligue Contre le Cancer, France, and F.B. by the Deutsche Forschungsgemeinschaft Bu2996-1/3.
Author information
Authors and Affiliations
Contributions
S.Y.C.G. and F.B. conceived the idea and designed the experiments. S.Y.C.G., N.W. and N.S. performed experiments. A.F.-S. did bioinformatics analysis. M.G.F., R.K. and F.B. supervised the project. S.Y.C.G. and F.B. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Christina Karlsson Rosenthal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Goh, S.Y.C., Fradera-Sola, A., Wittkopp, N. et al. Zbtb48 is a regulator of Mtfp1 expression in zebrafish. Commun Biol 8, 277 (2025). https://doi.org/10.1038/s42003-025-07666-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07666-z








