Abstract
The human relaxin family peptide receptors RXFP3 and RXFP4 play important physiological roles through interactions with endogenous hormones, relaxin-3 and insulin-like peptide 5 (INSL5). They are implicated in certain neurological and metabolic disorders. While INSL5 only activates RXFP4, relaxin-3 is recognized by both receptors. Here, we report the cryo-electron microscopy structures of RXFP3–Gi complexes bound by relaxin-3 or a small-molecule dual agonist (compound 4), and relaxin-3 in complex with RXFP4–Gi, with global resolutions of 2.91 Å, 2.95 Å, and 3.10 Å, respectively. It is found that relaxin-3 adopts a conserved binding conformation within the transmembrane domain (TMD) bundle of RXFP3 and RXFP4, where the C-terminal tip residues of its B chain, R26 and W27, make extensive contacts with conserved receptor residues, thereby activating RXFP3 and RXFP4. Compound 4 mimics these key interactions by binding to both receptors. In contrast, the C-terminal residues composition and TMD-binding angle of INSL5 in RXFP4 differ significantly from that of relaxin-3, ensuring its selectivity for RXFP4. These findings deepen our understanding about the structural basis of ligand recognition and selectivity in this G protein-coupled receptor subfamily.
Similar content being viewed by others
Introduction
Relaxin/insulin-like family peptide receptor 3 (RXFP3), also known as GPCR135 and somatostatin- and angiotensin-like peptide receptor (SALPR), is a typical class A neuropeptide-like G protein-coupled receptor (GPCR) that exhibits a high sequence similarity with RXFP41,2. Relaxin-3, a member of the relaxin family of peptides consisting of two chains and three disulfide bonds, serves as the endogenous ligand for RXFP3, as supported by its expression and binding properties3. It activates RXFP3 by engaging pertussis toxin (PTX)-sensitive Gαi/o proteins and inhibits cAMP production2. This ligand-receptor pair is predominantly expressed in the brain, particularly in the paraventricular nucleus4, and is implicated in feeding5,6, stress responses7,8,9, arousal10,11, cognition12,13, and alcohol addiction14,15,16. As a result, relaxin-3 and RXFP3 have emerged as targets for treating various central nervous system (CNS) diseases.
Despite their therapeutic potential, there are currently no ideal ligands available for this receptor. While relaxin-3 and its peptidic analogs were proven challenging to synthesize, several small-molecule modulators have been identified in recent years17,18,19. However, all of them exhibit cross-reactivity with RXFP4 or RXFP1 in vitro. The lack of structural information has hindered our understanding of the mechanism underlying ligand recognition and subtype selectivity between RXFP3 and RXFP4. Therefore, we determined the cryogenic electron microscopy (cryo-EM) structures of the full-length, Gi-coupled human RXFP3 and RXFP4 bound by relaxin-3, as well as Gi-coupled human RXFP3 in complex with compound 4 (a non-selective small-molecule dual agonist), with global resolutions of 2.91 Å, 2.95 Å, and 3.10 Å, respectively. Together with mutagenesis and functional analyses, we elucidated a common recognition mechanism for RXFP3 and RXFP4 shared by relaxin-3 and compound 4. We further clarified the differences in receptor selectivity exhibited by relaxin-3 and insulin-like peptide 5 (INSL5). Our study provides useful information for the rational design of novel RXFP3 and RXFP4 agonists either as research probes or as drug candidates.
Results
Overall structure
To stabilize the RXFP3 and RXFP4 complexes for structure determination, we employed multiple strategies to overcome challenges as previously reported20, including a haemagglutinin (HA) signal peptide addition, N-terminal cytochrome b562RIL (BRIL) insertion, and the NanoBiT tethering strategy (Supplementary Fig. 1a, b). The activity of these modifications was verified by cAMP accumulation assay (Supplementary Fig. 1c–h, Supplementary Tables 1–3 and Supplementary Data 1). The complexes were then purified, resolved as monodispersed peaks on size-exclusion chromatography (SEC), and confirmed by SDS gel to ascertain all the expected components (Supplementary Fig. 1c and Supplementary Data 1). After sample preparation, cryo-EM data collection and analysis, three-dimensional (3D) consensus density maps were reconstructed with global resolutions of 2.91 Å, 2.95 Å, and 3.10 Å (Supplementary Fig. 2) for the relaxin-3–RXFP3–Gi, relaxin-3–RXFP4–Gi and compound 4–RXFP3–Gi, respectively (Fig. 1, Supplementary Figs. 1–5 and Table 1). The clarity of the cryo-EM density maps allowed for the near-atomic level models of most regions of the complexes, encompassing ligands, the receptors, the Gαβγ heterotrimer, and scFv16 (Fig. 1e–j; Supplementary Fig. 3). In terms of receptor composition in the final structure models, RXFP3 spans residues 77–409 except for the intracellular loop 2 (ICL2) between T193–C210 and ICL3 between A300–G310, and RXFP4 covers residues 35–325 except for the ICL1 between N66 to P72. Regarding relaxin-3, G6B–W27B (B indicates that the residue belongs to the B chain of relaxin-3) in the relaxin-3‒RXFP3‒Gi complexes, and L9B–W27B in the relaxin-3‒RXFP4‒Gi complexes are well-defined due to the relatively strong cryo-EM density (Fig. 1e, f, h, i; Supplementary Fig. 3). In contrast, the A chain of relaxin-3 is only visible at one disulfide bond (C24A–C22B) when bound to RXFP4 and includes residues S16A to C24A in the case of RXFP3, while the remainder of the structure appears more dynamic, resulting in insufficient density for constructing a complete 3D model. To ensure structural validity, we retained the A-chain backbone coordinates but omitted non-disulfide-bonded side-chains. These observations regarding relaxin-3 are consistent with previous structure-activity relationship (SAR) studies demonstrating that its B chain, excluding the N-terminal seven residues, is the sole determinant for binding and activating RXFP3 and RXFP421.
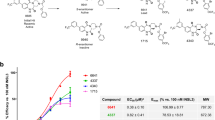
a Activity of INSL5 at RXFP4 and cross-reactivity of compound 4 and relaxin-3 at RXFP3 and RXFP4, with line thickness indicating efficiency. b Sequence and conformational comparison of relaxin-3 and INSL5. c, d cAMP signaling induced by different ligands in cells expressing wild-type RXFP3 and RXFP4. Data shown are means ± S.E.M. of three independent experiments (n = 3) performed in quadruplicate. e–j Cryo-EM density maps (e–g) and cartoon representation (h–j) of the relaxin-3−RXFP3−Gi complex (e, h), relaxin-3−RXFP4−Gi complex (f, i), and compound 4−RXFP3−Gi complex (h, j). Atomic models and EM densities of the three ligands are shown as sticks and surfaces, respectively. Compound 4 is shown in peru, the A chain of relaxin-3 is shown in light steel blue or light coral, and the B chain is in medium purple or violet in relaxin-3-bound RXFP3 or RXFP4, respectively. Compound 4 and relaxin-3-bound RXFP3 are shown in pale violet red and rosy brown, respectively, while relaxin-3-bound RXFP4 is shown in pale blue. Gαi in cornflower blue, Gβ in dark salmon, Gγ in plum, and scFv16 in dark gray.
The overall structures of the relaxin-3-bound RXFP3 and RXFP4 exhibit a similar conformation, with a Cα root mean squared deviation (RMSD) of 0.84 Å. Both structures mirror the previously reported INSL5–RXFP4 complex structure20 with Cα RMSD values of 1.01 Å and 0.84 Å, respectively. When compared to the representative inactive conformation of the β2-adrenergic receptor22, both relaxin-3-bound RXFP3 and RXFP4, as well as compound 4-bound RXFP3, display a distinctive outward movement of the intracellular half of transmembrane helix 6 (TM6), indicative of a fully active state (Supplementary Fig. 4). By adopting a similar binding mode in both receptors, relaxin-3 penetrates the transmembrane domain (TMD) core via its C-terminal segments, which include a short α-helix (from R12B to C22B) and a loop (from G23B to W27B) of the B chain. Notably, the two C-terminal residues R26B and W27B extend deeply to the orthosteric pocket (Fig. 2a, b), a conformation distinct from the upward-folded orientation reported in the NMR structure23 (Supplementary Fig. 6). These observations demonstrate that receptor binding and interactions significantly influence relaxin-3 conformational dynamics, particularly altering the spatial positioning of critical residues (e.g., R26B and W27B) that drive receptor activation. The A chain of relaxin-3 maintains the structural integrity of the B chain through two interchain disulfide bonds (C11A‒C10B and C24A‒C22B), facilitating stable receptor-ligand interactions within the binding pocket. In comparison, compound 4 also inserts deeply into the orthosteric pocket by occupying the positions of R26B and W27B of relaxin-3 without interacting with the extracellular loops (ECLs) (Fig. 3a). To accommodate the binding of these ligands, ECLs 1–3 and the extracellular halves of TM1, TM4 and TM7 display notable structural variations between RXFP3 and RXFP4. Importantly, the ECL2 of RXFP3 is longer than that of RXFP4, displaying distinct and ligand-specific structural features in addition to the conserved β-hairpin found in the ECL2 of both receptors (Fig. 2a).
a Superimposition of relaxin-3-bound RXFP3 and RXFP4 structures. b Cross-section view of the relaxin-3 binding pocket in RXFP3 and RXFP4. c Comparison of relaxin-3 interaction with RXFP3 and RXFP4. Close-up views of these interactions are shown with polar contacts depicted as black dashed lines. Receptor residues are labeled with Ballesteros–Weinstein numbering and colored by interaction type: red (salt bridge), yellow (hydrogen bond), green (stacking), and gray (hydrophobic interactions). d, e Effects of RXFP3 (d) and RXFP4 mutants (e) on relaxin-3 activity. Data are means ± S.E.M. of at least three independent experiments shown in parentheses.
a The EM density of compound 4 in the RXFP3 binding pocket. b Detailed interaction between compound 4 (peru) and residues in RXFP3 (pale violet red). c Effects of receptor mutation on compound 4-induced cAMP accumulation. Data are as means ± S.E.M. of at least three independent experiments shown in parentheses. d, e Structural superposition of compound 4-bound RXFP3 (pale violet red) and RXFP4 (dark sea green). f Schematic diagram of the interaction between compound 4 and the receptors. Different interaction types are represented by assorted colors, with salt bridges in red, hydrogen bonds in yellow, π–π stacking interactions in green, and hydrophobic interactions in gray. Different residues in the homologous binding pocket sites of RXFP3 and RXFP4 are highlighted with a pink background. g Schematic representation of the interactions between the indole ring substituents and different residue types in RXFP3 and RXFP4. h Effects of mutation at conserved sites in RXFP3 and RXFP4 on the activity of compound 4. Data are means ± S.E.M. of at least three independent experiments shown in parentheses.
Dual agonism of relaxin-3
The cryo-EM structures of relaxin-3–RXFP3 and relaxin-3–RXFP4 reveal that non-selective binding of relaxin-3 is achieved via interactions with ECLs 1‒3 and the extracellular halves of TMs 2, 3, and 5‒7 (Fig. 2a–c). Specifically, the short α-helix (from R12B to C22B) of the B chain penetrates the receptor TMD core at a consistent angle, establishing notable contacts with ECL2 and the extracellular tips of TMs 2 and 5–7 (Fig. 2a). Overlap is also observed at the C-terminal residue W27B, located at the bottom of the orthosteric pocket. Notably, the loop connecting the α-helix to W27B exhibits receptor-specific orientation: residues G24B-S25B are positioned closer to ECL2 and TM2 in RXFP3, while they are nearer to TM7 in RXFP4, thereby maximizing interactions with these subtype-specific residues (Fig. 2c).
W27B forms the most extensive interactions to stabilize peptide binding (Fig. 2b, c). In RXFP3, the indole moiety of W27B likely makes a water-mediated hydrogen bond with T1623.32 (superscripts denote Ballesteros–Weinstein numbering24) in a manner similar to that of the INSL5–RXFP4 complex20, and engages in π–π stacking with W1382.60. Additionally, multiple aromatic residues, including F1312.53, W3396.48, F3717.35, and H3797.43, synergistically stabilize the orthosteric pocket through stacking interactions. These interactions are also observed in RXFP4 due to residue conservation at these equivalent sites (Fig. 2c). A slight difference is noted for hydrophobic contacts, where RXFP3 residues F1312.53 and M1663.36 provide stronger interactions compared to RXFP4 residues L902.53 and V1253.36. Additionally, the carboxylic acid group of W27B points toward TM5 with the formation of one hydrogen bond (via H2685.39 in RXFP3 and Q2055.39 in RXFP4) and one salt bridge (via K2715.42 in RXFP3 and R2085.42 in RXFP4) (Fig. 2c, Supplementary Table 1). These observations are supported by mutagenesis studies, where the relaxin-3-induced cAMP responses in RXFP3 were markedly diminished in mutants F1312.53A, W1382.60A, T1622.60A, F3717.35A and H3797.43A by 6-fold, 117-fold, 28-fold, 32-fold and 5-fold, respectively (Fig. 2d, Supplementary Table 4 and Supplementary Data 1). Similarly, the action of relaxin-3 was completely abolished in RXFP4 mutant T1213.32A, and profoundly reduced in W972.60A, R2085.42A, and H2997.43A by 537-fold, 129-fold, and 62-fold, respectively (Fig. 2e, Supplementary Table 4 and Supplementary Data 1). Another pivotal residue at the C terminus of the B chain is R26B, which forms a salt bridge with TM2 (via E1002.63) in RXFP4, but two hydrogen bonds with TM7 (via Q3677.31) in RXFP3. Additionally, E1412.63 in RXFP3 establishes a hydrogen bond with S25B (Fig. 2c). Consistently, mutations E1412.63A in RXFP3 and E1002.63A in RXFP4 both abolished the ability of relaxin-3 to activate the receptors (Fig. 2d, e, Supplementary Table 4 and Supplementary Data 1), in line with the reduced potencies of R26BE25 in relaxin-3.
The α-helix (from R12B to C22B) of relaxin-3 engages significant polar and hydrophobic interactions with the upper half of the ligand-binding pocket. Specifically, R12B and R16B participate in a polar network involving RXFP3 residues D145ECL1 and E244ECL2, or RXFP4 residue D104ECL1 (Fig. 2c). This observation is in line with the reduced potencies of relaxin-3-induced cAMP response for the relaxin-3 mutants R12BE and R16BE as well as the RXFP4 mutant D104ECL1R25. Regarding nonpolar interaction, relaxin-3 contributes substantial hydrophobic contacts with RXFP3 residues including F146ECL1, V241ECL2, M242ECL2, L246ECL2, L24845.51, W2635.34, Y3637.27, F3647.28 and V3687.32 (via I15B, V18B, I19B, F20B, T21B and C22B), or RXFP4 residues F105ECL1, V185ECL2, C186ECL2, V188ECL2, L190ECL2, L19245.51, L2015.35, W279ECL3, F2837.27, Y2847.28 and T2887.32 (via I15B, A17B, V18B, I19B, F20B and C22B) (Fig. 2c). Disruption of them by RXFP3 mutations L246ECL2A, L24845.51A and L246ECL2D reduced relaxin-3 potency by 38-fold, 66-fold and 537-fold, respectively, while RXFP4 mutations L190ECL2A and L19245.51A decreased its potency by 14-fold and 58-fold, respectively (Fig. 2d, e, Supplementary Table 4 and Supplementary Data 1).
Peptidomimetics of compound 4
Compound 4 exhibits clear electron density at the orthosteric site of RXFP3, adopting a “C”-shaped conformation, with its indole core inserting deeply into the binding pocket and its chlorophenyl moiety extending outward (Fig. 3a, b). The indole core engages in a series of polar and hydrophobic interactions with the surrounding RXFP3 residues, including hydrogen bonds (via S1593.29, T1623.32, S1633.33 and K2715.42), π–π stacking (via W1382.60), cation-π interaction (via K2715.42), and hydrophobic interactions with F1312.53, F3717.35, V3757.39 and H3797.43. The positively charged guanidine moiety in the central region forms a salt bridge with the negatively charged side chain of E1412.63. The chlorophenyl moiety extends outward to cover the orthosteric site composed of F146ECL1 and F3717.35, as well as forms hydrophobic interactions with L246ECL2 and L24845.51 (Supplementary Table 2). Mutagenesis and functional studies support these observations: compound 4 is completely inactive on the W1382.60A and E1412.63A mutants. Significant impairment in cAMP responses was observed in mutants T1623.32A, F146ECL1A, and H3797.43A: EC50 values were negatively impacted by 200-fold, 13-fold, and 7-fold, respectively, while a 40% decrease in Emax was noted in mutants K2715.42A, F146ECL1A, and H3797.43A (Fig. 3c, Supplementary Table 5, and Supplementary Data 1). Removal of the hydroxyl group at indole 5 position or substitution of the ethyl group with a smaller methyl group at indole 7 position weakened hydrophobic interactions with TM7 residues and polar interactions with TM3 residues, consequently affecting the agonistic potencies of compound 4 as reported previously17,19.
Because of the high sequence identity of the ligand-binding pocket between RXFP3 and RXFP4, the development of subtype-selective ligands has been challenging. The structures of compound 4-bound RXFP3 and RXFP4 were superimposed using the Cα atoms of the receptors, revealing commonalities in the molecular mechanism by which compound 4 exerts its peptidomimetic property, particularly in the relative positions and interactions of W2.60 and E2.63. Since compound 4 is positioned slightly deeper in the RXFP3 binding pocket than in RXFP4, the orientations of the chlorophenyl moiety and the indole core show conformational variations. This results in weaker π–π interactions between the indole core and RXFP3 residue W1382.60 compared to RXFP4 residue W972.60, as well as a larger distance between the guanidine moiety and RXFP3 residue E1412.63 (Fig. 3d, e). These observations partially explain the differential activity of compound 4 manifested in the two receptor subtypes (the agonistic activity of compound 4 at RXFP4 is approximately 40-fold stronger than at RXFP3, with EC50 values of 2 nM and 82 nM, respectively)19. Furthermore, such a differentiated behavior led to changes in the orientation of hydroxyl and ethyl in the indole ring, which conveniently interact with residues involved in subtype selectivity, including S1593.29 (RXFP3)/L1183.29 (RXFP4), S1633.33 (RXFP3)/V1223.33 (RXFP4), and V3757.39 (RXFP3)/T2957.39 (RXFP4) (Fig. 3e–g). To further explore the determinants of subtype selectivity, we conducted a comparative analysis of the effects of amino acid switch and alanine mutation in the equivalent positions between RXFP3 and RXFP4 around the ligand-binding pocket (Fig. 3h and Supplementary Data 1). It was found that the former significantly alters the activity of compound 4 at RXFP3. This may result from disrupting the corresponding polar interactions (S1593.29 and S1633.33 with hydroxy) and hydrophobic interactions (V3757.39 with ethyl) due to an amino acid switch in RXFP3. In RXFP4, hydrophobic interactions between the indole ring of compound 4 and TM3 residues (L1183.29, T1213.32, and V1223.33) are critical for agonistic efficacy, as demonstrated in our prior studies20. The mutant L1183.29D completely abolished compound 4 activity, while L1183.29A substitution enhanced compound 4 potency (Supplementary Table 5), underscoring the critical role of hydrophobic complementarity at this locus.
Receptor selectivity
Despite sharing the same structural scaffold (three α-helices constrained by one intra- and two interchain disulfide bonds), along with the conserved C-terminal residues (R26BW27B in relaxin-3 and R23BW24B in INSL5) pivotal for receptor activation, relaxin-3 and INSL5 present distinct receptor selectivity between RXFP3 and RXFP4 (Fig. 1a). The former demonstrates a dual agonistic profile across both two subtypes (pEC50 = 8.86 ± 0.07 in RXFP3 and pEC50 = 7.56 ± 0.08 in RXFP4, Fig. 1c, d and Supplementary Data 1), while the latter selectively activates RXFP4 (pEC50 = 8.61 ± 0.07, Fig. 1c and Supplementary Data 1). Although both peptides are anchored in the orthosteric binding pocket of RXFP4 bordered by ECLs as well as TMs 2, 3, and 5–7, common and deviated conformational features are noticeable (Fig. 4a), shedding light on a better understanding of subtype selectivity from ligand and receptor perspectives.
a Structural superposition of relaxin-3- or INSL5-bound RXFP4, shown in side-view (left) and top view (right), highlighting differences in binding modes. b–f Close-up views of the interactions between the B chain of peptides and the key residues in RXFP4. g, h Effects of RXFP4 mutations on relaxin-3 and INSL5 induced cAMP accumulation. Data are as means ± S.E.M. of at least three independent experiments shown within bars. N.A. not active. N.D. not determined.
The C-terminal tip residue (W27B in relaxin-3 and W24B in INSL5) is the most prominent overlay deeply buried in the bottom of the orthosteric pockets (Fig. 4a). The indole moiety forms a hydrogen bond with T1213.32, engages in cation-π stacking with R2085.42, π–π stacking with W972.60, F2917.35 and H2997.43, alongside the hydrophobic interactions with L1183.29, V1223.32 and V1253.35. Meanwhile, the free carboxylic acid of the peptide C terminus establishes polar contacts with Q2055.39 and R2085.42, and the penultimate arginine (R26B in relaxin-3 and R23B in INSL5) makes a salt bridge with E1002.63. These conserved ligand-RXFP4 interactions between relaxin-3 and INSL5 indicate a common signaling initiation mechanism between these two peptides (Fig. 4b–d, g, h). Besides, several ligand-specific interactions such as R26B‒Y461.39, R26B‒S1012.64 and S25B‒Q2877.31 were found for relaxin-3, but not INSL5 (Fig. 4d).
Interestingly, the B chain of INSL5 features a rigid conformation at its C terminus due to an extended α-helix. In contrast, relaxin-3, which possesses two adjacent glycine residues (G23B and G24B), adopts more flexible loop conformations at its C terminus to recognize both RXFP3 and RXFP4 (Fig. 4a, Fig. 1b). Correspondingly, TM1, TM7 and ECLs of RXFP4 exhibit structural variations to accommodate the different binding modes of these two peptides (Fig. 4a). The presence of R12B and R16B in relaxin-3 enables stronger polar interactions with D104ECL1, while the equivalent residues (R13B and Y17B) in INSL5 form one salt bridge with D104ECL1 and engage in π–π stacking with F105ECL1, respectively (Fig. 4e). Furthermore, F14B, I15B, A17B, V18B, I19B and F20B in relaxin-3 promote the inward shift of F105ECL1, W279ECL3 and F2837.27 via stacking and hydrophobic contacts (Fig. 4f). These interactions collectively contribute to the constriction of the ECLs of relaxin-3 binding pocket, as opposed to INSL5. Notably, the ECLs are the regions showing significant sequence and conformational diversity between RXFP3 and RXFP4, influencing the binding properties of different ligands. Specific interactions, such as R12B‒E244ECL2, along with the hydrophobic contacts within C22B, T21B, and W2635.34, were observed in the relaxin-3‒RXFP3 complexes, but were absent in the INSL5‒RXFP4 complex due to variations in the corresponding receptor residues. Our mutagenesis experiments support these observations. Specifically, mutations E1002.63A, T1213.32A, and R2085.42A completely abolished the activity of both peptides, while W972.60A reduced the potency of relaxin-3 and INSL5 by 537-fold and 20-fold, respectively, and Q2055.39A decreased their potency by 9.6-fold and 5-fold, respectively (Fig. 4g, h, and Supplementary Data 1).
Gi coupling
The conformations of RXFP3 intracellular side in active state resembled those of RXFP4, characteristic of an outward movement of the cytoplasmic half of TM6 (Fig. 5a). The GαiH5 helix is anchored in an intracellular cavity formed by TM2, TM3, TM5, TM6, TM7, and ICLs, and is stabilized through extensive hydrophobic and polar interactions. Using the receptor Cα atoms as an alignment standard, the RMSD between RXFP3 and RXFP4 in compound 4 activated state is 0.93 Å, with a displacement of 7.85 Å in the GαN (measured at the Cα atom of S6G.HN.30). Hydrophobic residues at the carboxy terminal of GαiH5 helix (I345G.H5.16, L349G.H5.20, C352G.H5.23, L354G.H5.25 and F355G.H5.26) form numerous hydrophobic interactions with residues in TM3, TM5, and TM6, which are largely conserved across both complexes. Notable differences were mainly present in G protein interactions with ICL2 and ICL3 across the two receptors. In the RXFP3 complexes, regions of ICL2 and ICL3 lacked electron density, likely due to their disordered structure and absence of interaction with Gαi. In contrast, RXFP4 established a polar interaction network with Gαi through three arginine residues (R234ICL3, R236ICL3, and R237ICL3), but only one hydrogen bond (R312ICL3−T322G.S6.3) was identified in the RXFP3−Gi complexes. The salt bridge, formed between R192ICL2 and D194G.s2s3.2, along with the extensive hydrophobic contacts (A187ICL2, L188ICL2, H191ICL2, V1843.54, A1853.55, and F2935.64 via I344G.H5.16, I345G.H5.16, and L349G.H5.20 in GαiH5 helix), suggests a stronger interaction between Gαi subunit and ICL2 of RXFP3.
a Structural superposition of the RXFP3−Gi and RXFP4−Gi complexes (PDB: 7YK6) in different views. b Interaction between RXFP3 (pale violet red) and GαiH5 helix (cornflower blue). c Conformational differences in intracellular loops 2 (ICL2) and 3 (ICL3), and the interactions between RXFP3 ICL2/ICL3 and GαiH5. Polar interactions are represented by black dashed lines. d Sequence alignment of ICL2 and ICL3 between RXFP3 and RXFP4.
Discussion
Given the high sequence homology of the ligand-binding pocket between RXFP3 and RXFP4, the development of receptor subtype-selective modulators faces significant challenges. In this study, we present the cryo-EM structures of RXFP3–Gi bound by relaxin-3 or a small-molecule RXFP3/RXFP4 dual agonist compound 4, and RXFP4–Gi bound by relaxin-3. Combined with mutagenesis studies and SAR analysis21, we delineate the molecular mechanisms underlying the dual agonism manifested relaxin-3 and compound 4 in conjunction with receptor subtype selectivity of INSL5, thereby expanding our understanding of ligand-receptor interaction in this important class A GPCR subfamily.
The phenomenon of combinatorial agonism among endogenous peptides at their cognate receptors is well-documented, with bound peptides consistently adopting nearly identical binding conformations and recognition patterns. For instance, α-melanocyte-stimulating hormone (α-MSH), a non-selective full agonist of melanocortin 1 receptor (MC1R), MC2R, MC4R and MC5R, shows a significant overlap, particularly in the middle region (H6‒F7‒R8‒W9), where it interacts with conserved MCR residues in the lower half of the TMD pocket26. Similar observations have been reported for pasireotide in the somatostatin receptor 1 (SSTR1) and SSTR3 binding pockets27. In contrast, while the short α-helix in the middle region of the relaxin-3 B chain anchors in the upper half of the orthosteric pocket bordered by ECLs and TM7 at a consistent angle, the C-terminal segment of which adopts distinct loop conformations in the two receptors, culminating in the overlap of the tip residue, W27B, which is deeply buried in the TMD. Notably, R26B and W27B are critical for receptor activation, recognized by RXFP3/RXFP4 conserved residues including W1382.60 (RXFP3)/W972.60 (RXFP4), E1412.63 (RXFP3)/E1002.63 (RXFP4), T1623.32 (RXFP3)/T1213.32 (RXFP4) and H3797.43 (RXFP3)/H2997.43 (RXFP4) to achieve dual agonism. Meanwhile, non-conserved residues in the ligand-binding pocket, such as S1593.29 (RXFP3)/L1183.29 (RXFP4), S1633.33 (RXFP3)/V1223.33 (RXFP4), and V3757.39 (RXFP3)/T2957.39 (RXFP4), introduce variations in the fine-tuning of specific interactions within the two binding pockets. These variations make the flexible C terminus adopt distinct conformations to ensure the correct positioning of W27B and R26B for receptor activation.
Similarly, compound 4-bound RXFP3 and RXFP4 share a conserved activation mechanism, involving the formation of salt bridges between a centrally located guanidine moiety and E1412.63 (RXFP3) or E1002.63 (RXFP4). However, the central urea moiety in RXFP4-specific agonist DC59105320 failed to establish strong polar interactions with E1002.63 (RXFP4). Although this deficiency partially affected the activity of DC591053, it is compensated by other interactions, including occupancy of an additional binding pocket by the piperidine ring, formation of two moderately strong hydrogen bonds with Q2055.39 (RXFP4) and R2085.42 (RXFP4), multiple π–π stacking interactions with F105ECL1 (RXFP4) and F2917.35 (RXFP4), the cation-π stacking with R194ECL2 (RXFP4), as well as hydrophobic interactions with L190ECL2 (RXFP4), L19245.51 (RXFP4) and P2927.36 (RXFP4) facilitated by the tetrahydroisoquinoline group. This “avoidance” of the core activation mechanism and “compensation” by other interactions allow DC591053 to achieve both specificity and activity for RXFP4. Considering the larger interface between endogenous peptides and the receptors, as well as both common and distinct recognition modes of RXFP3 and RXFP4, it might be possible to design specific modulators by deliberately avoiding common features and leveraging their respective specificities.
Relaxin-3 and INSL5 share a common structural scaffold and high sequence identity, particularly in the C terminus of the B chain, where identical residues (RW) are crucial for receptor activation. Despite these similarities, they exhibit distinct receptor preferences: INSL5 specifically activates RXFP4, while it acts as a weak antagonist for RXFP3, though the underlying mechanism remains unclear. In relaxin-3, positions 23B and 24B are typically occupied by two small and flexible glycine residues, providing a greater degree of freedom28. In contrast, the corresponding positions (20B and 21B) of INSL5 are usually occupied by larger alanine and serine residues, leading to a more rigid, extended C-terminal α-helix. In terms of ligand-binding pocket, different shapes and compositions of RXFP3 and RXFP4 may require distinct ligand conformations. The rigid C terminus of the INSL5 B chain likely prevents it from adopting the necessary conformation to engage RXFP3 effectively, potentially causing mismatches at positions R23B and/or W24B in the ligand-binding pocket. This is supported by experimental findings where substituting A20B and S21B with glycine in INSL5 conferred flexibility, allowing it to activate RXFP3, albeit with a slight reduction in RXFP4 activity28. Furthermore, the extracellular interactions between the A chain of INSL5 and the ECLs of RXFP4, absent in relaxin-3, suggest a unique role for the A chain of INSL5. This aligns with previous studies showing that deletion of the A chain abolished INSL5 binding and activity29, while the B chain of relaxin-3 functions as a weak agonist30. These results underscore the distinct binding dynamics of relaxin-3 and INSL5 to RXFP3/RXFP4.
In conclusion, our work offers a comprehensive understanding of ligand recognition and the activation process of RXFP3 and RXFP4. The findings may pave the way for developing specific and effective modulators useful to treat certain neuroglial and metabolic diseases.
Methods
Construct
Full-length human RXFP3 (NCBI Reference Sequence: NM_016568.3) and RXFP4 (NCBI Reference Sequence: NM_181885.3) were cloned into a modified pFastBac vector (Invitrogen, Carlsbad, CA, USA). Modification of RXFP3 was made in a way identical to that of RXFP4 described previously20. Briefly, the haemagglutinin (HA) signal peptide, followed by a 10× histidine tag and BRIL, were inserted at the N terminus, while the LgBiT subunit (Promega, Madison, WI, USA) was fused at the C terminus connected by a 15-amino acid polypeptide linker (Supplementary Fig. 1a, b). These modifications had a negligible effect on the signaling properties of RXFP3 and RXFP4 as verified by cAMP accumulation assays (Supplementary Fig. 1c–e and Supplementary Data 1). A dominant-negative human Gαi2 (DNGi2) with 4 mutations (S47N, G204A, E246A, and A327S) was generated to limit G protein dissociation. Human Gβ1 was cloned with a C-terminal HiBiT (peptide 86, Promega) connected with a 15-amino acid polypeptide linker to help stabilize the complex. The engineered human Gαi2, Gβ1, bovine Gγ2, and scFv16 were individually cloned into the pFastBac vector (Invitrogen), respectively. To conduct functional assays, RXFP4 and RXFP3 were cloned into pCMV6 constructs (OriGene Technologies, Maryland, MD, USA). Mutant receptors were generated by site-directed mutagenesis using the wild-type (WT) constructs as templates, with primers designed in QuickChange Primer Design [QuickChange Primer Design (http://agilent.com.cn)] and made by Phanta Max Master (Vazyme, Nanjing, China). An N-terminal Flag tag was added to both WT and mutant receptors for surface expression measurement. Sequences of all primers used in this study were provided in Supplementary Data 2, and all constructed entities were verified by DNA sequencing.
Peptide
The recombinant human relaxin-3 and INSL5 used in this study were synthesized and processed by Genova (Changzhou, China). The bioactivity was verified in Chinese hamster ovary (CHO-K1) cells stably transfected with RXFP3 and RXFP4. As shown in Supplementary Fig. 5 and Supplementary Data 1, the recombinant human relaxin-3 was able to inhibit forskolin-induced cAMP responses in stably transfected CHO-K1 cells (pEC50 = 8.86 ± 0.07 in RXFP3 and pEC50 = 7.76 ± 0.11 in RXFP4, from three independent experiments) compared to the native relaxin-3 standard (pEC50 = 8.70 ± 0.07 in RXFP3 and pEC50 = 7.66 ± 0.13 in RXFP4, Phoenix Pharmaceuticals, CA, USA). The bioactivity of INSL5 was reported in our previous work20.
ScFv16 expression and purification
ScFv16 was modified with an N-terminal GP67 signaling peptide, and a C-terminal 8× histidine tag was cloned into the pFastBac vector and expressed in High Five™ insect cells (Invitrogen) as a secreted protein purified by Ni-sepharose chromatography column31. The HiLoad 16/600 Superdex 75 column (GE HealthCare, Little Chalfont, Buckinghamshire, UK) was used to separate the monomeric fractions of scFv16 with running buffer containing 20 mM HEPES and 100 mM NaCl (pH 7.4). The purified scFv16 was concentrated to 1.5 mg/mL and flash frozen in liquid nitrogen with 10% glycerol and stored in −80 °C until use.
Expression and purification of the RXFP3–Gi and RXFP4–Gi complexes
Recombinant viruses of RXFP3, RXFP4, Gαi2, Gβ1, and Gγ2 were prepared using the Bac-to-Bac baculovirus expression system (Invitrogen) in Spodoptera frugiperda (Sf9) insect cells (Invitrogen). High Five™ insect cells were grown to a density of 3.2 × 106 cells per mL and infected with receptor, Gαi2, Gβ1 and Gγ2 P1 viral stocks at a ratio of 6:1:1:1. After 48 h incubation at 27 °C, the cells were harvested by centrifugation at 813 × g for 20 min and stored at −80 °C until use.
The pellets (from 2 L cell suspension) expressing the RXFP4–Gi or RXFP3–Gi complex were resuspended in a buffer containing 20 mM HEPES, 100 mM NaCl, and 100 μM TCEP, pH 7.4, supplemented with 10% (v/v) glycerol and EDTA-free protease inhibitor mixture (Biomake, Houston, TX, USA). The suspension was transferred to a pre-chilled Dounce homogenizer and underwent 50–60 cycles of mechanical disruption at constant speed under ice-cold conditions, ensuring complete cell lysis while maintaining sample viability and structural integrity, and the membrane was collected at 65,000 × g for 30 min. The formation of complex was initiated by addition of 10 mM MgCl2, 1 mM MnCl2, 5 mM CaCl2, 25 mU/mL apyrase (New England Biolabs, Beverly, MA, USA), 15 μg/mL scFv16, ligands (20 μM relaxin-3 or 50 μM compound 4), 100 μM TCEP and 100 U salt active nuclease (Sigma-Aldrich, St. Louis, MO, USA) supplemented with protease inhibitor cocktail. After 1.5 h incubation at room temperature (RT), the complex was extracted from the membranes with 0.5% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace, Maumee, OH, USA) and 0.1% (w/v) cholesterol hemisuccinate (CHS, Anatrace) with additional protease inhibitor cocktail and ligands for 3 h at 4 °C. The supernatant was isolated by centrifugation at 65,000 × g for 1 h and incubated with Ni-NTA beads (GE HealthCare) for 1.5 h at 4 °C. One milliliter of settled bed volume of chromatographic media was employed for 2 L cell suspension, which was standardized to one column volume. The resin was washed with five column volumes of buffer A [20 mM HEPES, 100 mM NaCl, 5 mM MgCl2, 1 mM MnCl2 100 μM TCEP, ligands (4 μM relaxin-3 or 10 μM compound 4), 0.1% (w/v) LMNG, 0.02% (w/v) CHS and 30 mM imidazole, pH 7.4], followed by washing with twenty-five column volumes of buffer B [essentially the same as buffer A with decreased concentrations of detergents 0.03% (w/v) LMNG, 0.01% (w/v) GDN and 0.006% (w/v) CHS containing 60 mM imidazole, pH 7.4]. The protein was eluted with five column volumes of buffer C (buffer B with 300 mM imidazole, pH 7.4). The complexes were then concentrated using a 100-kD Amicon Ultra centrifugal filter (Merck Millipore, Billerica, MA, USA) and subjected to a Superdex 200 10/300 GL column (GE HealthCare) with running buffer containing 20 mM HEPES, 100 mM NaCl, 100 μM TCEP, ligands (4 μM relaxin-3 or 10 μM compound 4), 0.00075% (w/v) LMNG, 0.00025% (w/v) GDN, and 0.00015% (w/v) CHS, pH 7.4. The monomeric peak fractions were combined and concentrated to 8–10 mg/mL for relaxin-3–RXFP3–Gi and relaxin-3–RXFP4–Gi complexes, and to 3–5 mg/mL for compound 4–RXFP3–Gi complexes.
Cryo-EM data acquisition
The purified complex samples (3 μL) were applied to glow-discharged holey grids (Quantifoil R1.2/1.3, Au 300 mesh) and subsequently vitrified using a Vitrobot Mark IV (ThermoFisher, Waltham, MA, USA) set at 100% humidity and 4 °C. For relaxin-3–RXFP3–Gi and relaxin-3–RXFP4–Gi complexes, the optimal vitrification conditions were achieved with a 3 N blot force applied for 1 s followed by 5 s waiting time, while compound 4–RXFP3–Gi complexes required adjusted parameters of 2 N blot force sustained for 3 s. Cryo-EM images were acquired on a Titan Krios microscope (FEI, Oregon, OR, USA) equipped with a Gatan energy filter, a K3 direct electron detector, and serial EM3.732. For the relaxin-3–RXFP3–Gi and relaxin-3–RXFP4–Gi complexes, the microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. Totally, 6172 movies of the relaxin-3–RXFP3–Gi complexes and 5354 movies of the relaxin-3–RXFP4–Gi complexes were obtained with a defocus range of −1.2 to −2.2 μm, respectively. An accumulated dose of 80 electrons per Å2 was fractionated into a movie stack of 36 frames. For compound 4–RXFP3–Gi complexes, the microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 60,680× in counting mode, corresponding to a pixel size of 0.824 Å. 13,698 movies of the compound 4–RXFP3–Gi were obtained with a defocus range of −1.0 to −2.0 μm. An accumulated dose of 70 electrons per Å2 was fractionated into a movie stack of 36 frames.
Cryo-EM data processing
Dose-fractionated image stacks were subjected to beam-induced motion correction using MotionCor2.133. A sum of all frames, filtered according to the exposure dose, in each image stack was used for further processing. Contrast transfer function parameters for each micrograph were determined by Gctf v1.0634. Particle selection, 2D and 3D classifications were performed using cryoSPARC v3.2.035.
For the relaxin-3–RXFP3–Gi complex, auto-picking yielded 6,040,397 particle projections that were subjected to two rounds of reference-free 2D classification to discard false-positive particles or particles categorized in poorly defined classes, producing 1,861,948 particle projections for further processing. This subset of particle projections was subjected to a round of maximum-likelihood-based 3D classification with a pixel size of 2.142 Å, resulting in one well-defined subset with 758,481 projections. Further 3D classification with a mask on the receptor and ligand sequentially, and finally produced one good subset accounting for 364,523 particles, which were then subjected to 3D refinement and Bayesian polishing with a pixel size of 1.071 Å. After the last round of refinement, the final map has an indicated global resolution of 2.91 Å at a Fourier shell correlation (FSC) of 0.143. It was subsequently optimized using DeepEMhancer36 before model building. Local resolution was determined using the Bsoft package (v2.0.3) with half maps as input maps.
For the relaxin-3–RXFP4–Gi complex, auto-picking yielded 5,472,514 particle projections that were subjected to two rounds of reference-free 2D classification to discard false-positive particles or particles categorized in poorly defined classes, producing 1,832,828 particle projections for further processing. This subset of particle projections was subjected to a round of maximum-likelihood-based 3D classification with a pixel size of 2.142 Å, resulting in one well-defined subset with 937,571 projections. Further 3D classification with a mask on the receptor and ligand sequentially, and finally produced one good subset accounting for 339,434 particles, which were then subjected to 3D refinement and Bayesian polishing with a pixel size of 1.071 Å. The map with an indicated global resolution of 2.95 Å at a FSC of 0.143 was generated from the final 3D refinement. Local resolution was determined using the Bsoft package (v2.0.3) with half maps as input maps.
For the compound 4–RXFP3–Gi complex, auto-picking yielded 10,827,690 particle projections that were subjected to two rounds of reference-free 2D classification to discard false-positive particles or particles categorized in poorly defined classes, producing 1,959,458 particle projections for further processing. This subset of particle projections was subjected to a round of maximum-likelihood-based 3D classification with a pixel size of 2.142 Å, resulting in one well-defined subset with 818,270 projections. Further 3D classification with a mask on the receptor produced one good subset accounting for 407,171 particles, which were then subjected to 3D refinement and Bayesian polishing with a pixel size of 1.071 Å. After the last round of refinement, the final map has an indicated global resolution of 3.10 Å at a FSC of 0.143. Local resolution was determined using the Bsoft package (v2.0.3) with half maps as input maps.
Model building and refinement
According to the expected quality of the resulting models using SWISS-MODEL (https://swissmodel.expasy.org/interactive) with the quality estimated by Global Model Quality Estimate (GMQE)37, the cryo-EM structure of compound 4–RXFP4–Gi complex (PDB code: 7YK6)20 was used as the initial model of RXFP3, while the other parts were used to generate an initial model of G proteins and scFv16. Ligand coordinates and geometry restraints were generated using electronic Ligand Builder and Optimization Workbench (eLBOW)38 and fitted to the cryo-EM density by LigandFit GUI39 in PHENIX v1.1840. The model was docked into the EM density maps using UCSF Chimera v1.13.141, followed by iterative manual adjustment and rebuilding in COOT 0.9.4.142. Real space refinement was performed using PHENIX v1.1840,43. The model statistics were validated using the module comprehensive validation (cryo-EM) in PHENIX v1.1843. Structural figures were prepared in UCSF Chimera v1.13.141, UCSF ChimeraX v1.044, and PyMOL v.2.1 (https://pymol.org/2/). The refinement statistics are provided in Table 1.
Cell culture and transfection
CHO-K1 (ATCC, Virginia, VA, USA) cells stably expressing human RXFP3 (hRXFP3-CHO) or RXFP4 (hRXFP4-CHO) were maintained in DMEM/F12 (ThermoFisher) supplemented with 10% (v/v) fetal bovine serum (FBS) and 2 mM L-glutamine. Human embryonic kidney 293T cells containing SV40 large T-antigen (HEK293T, ATCC) were maintained in DMEM (ThermoFisher) supplemented with 10% (v/v) FBS, 1 mM sodium pyruvate (ThermoFisher), 100 units/mL penicillin, and 100 μg/mL streptomycin at 37 °C in 5% CO2. For cAMP assays, HEK293T cells were seeded onto 6-well cell culture plates at a density of 7 × 105 cells per well. After overnight incubation, cells were transfected with WT or mutant receptors using Lipofectamine 3000 transfection reagent (Invitrogen). Following 24 h of culturing, the transfected cells were ready for detection.
cAMP accumulation assay
Inhibition of forskolin-induced cAMP accumulation by compound 4 and relaxin-3 was measured by a LANCE Ultra cAMP kit (PerkinElmer, Waltham, MA, USA). Ligands were verified for their bioactivity in the beginning in hRXFP4-CHO, which were ready for use after 24 h of culturing. For assaying mutants, HEK293T cells were used 24 h post-transfection. Cells were digested with 0.02% (w/v) EDTA and seeded onto 384-well microtiter plates at a density of 8 × 105 cells/mL in cAMP stimulation buffer [HBSS supplemented with 5 mM HEPES, 0.1% (w/v) bovine serum albumin (BSA; ABCone, East Hame, London, UK) and 0.5 mM 3-isobutyl-1-methylxanthine]. The cells were stimulated with different concentrations of ligands plus 4 μM forskolin in RXFP3 and 1.5 μM forskolin in RXFP4. After 40 min incubation at RT, the Eu-cAMP tracer and ULight-anti-cAMP working solution were added to the plates separately to terminate the reaction, followed by 60 min additional incubation. The time-resolved fluorescence resonance energy transfer (TR-FRET) signals were detected by an EnVision multilabel plate reader (PerkinElmer) with the emission window ratio of 665 nm over 620 nm under 320 nm excitation. Data were normalized to the maximal response of WT receptors.
Receptor surface expression
Cell membrane expression was determined by flow cytometry to detect the N-terminal Flag tag on the WT and mutant receptor constructs transiently expressed in HEK293T cells. Briefly, approximately 2 × 105 cells were blocked with PBS containing 5% BSA (w/v) at RT for 15 min, and then incubated with 1:300 anti-Flag primary antibody (diluted with PBS containing 5% BSA, Sigma-Aldrich, Cat# F3165, purified IgG1 subclass) at RT for 60 min. The cells were then washed three times with PBS containing 1% BSA (w/v) followed by 60 min incubation with 1:1000 anti-mouse Alexa Fluor 488 conjugated secondary antibody (diluted with PBS containing 5% BSA, Invitrogen, Cat# A-21202) at RT in the dark. After washing three times, cells were resuspended in 200 μL PBS containing 1% BSA for detection by Guava® easyCyte™ (Luminex, Austin, TX, USA) utilizing laser excitation and emission wavelengths of 488 nm and 530 nm, respectively. For each sample, 20,000 cellular events were collected, and the total fluorescence intensity of the positive expression cell population was calculated. Data were normalized to the WT receptor and parental HEK293T cells (Supplementary Fig. 7, Supplementary Table 6).
Statistical and reproducibility
All functional study data were analyzed using GraphPad Prism 8.3 (GraphPad Software, San Diego, CA, USA) and presented as means ± S.E.M. from at least three independent experiments. Dose-response curves were generated with a three-parameter logistic equation. The significance was determined with either a two-tailed Student’s t-test or one-way ANOVA with Dunnett’s multiple comparison test, and P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant data are available from the corresponding authors upon reasonable request. The raw data underlying Figs. 1b, c, 2d, e, 3c, 3h, 4g, h, and Supplementary Figs. 1c–e, 5a, b, and Supplementary Table 6 are provided as a Supplementary Data file. The atomic coordinates and electron microscopy maps have been deposited in the Protein Data Bank (PDB) under accession codes: 9KFI (the relaxin-3–RXFP3–Gi complex), 9KFK (the relaxin-3–RXFP4–Gi complex) and 9KFJ (the compound 4–RXFP3–Gi complex), as well as Electron Microscopy Data Bank (EMDB) under accession codes: EMD-62297 (the relaxin-3–RXFP3–Gi complex), EMD-62299 (the relaxin-3–RXFP4–Gi complex) and EMD-62298 (the compound 4–RXFP3–Gi), respectively. All relevant data are available from the authors and/or included in the manuscript or supplemental data.
References
Matsumoto, M. et al. The novel G-protein coupled receptor SALPR shares sequence similarity with somatostatin and angiotensin receptors. Gene 248, 183–189 (2000).
Liu, C. et al. Identification of relaxin-3/INSL7 as an endogenous ligand for the orphan G-protein-coupled receptor GPCR135. J. Biol. Chem. 278, 50754–50764 (2003).
Bathgate, R. A. et al. International union of pharmacology LVII: recommendations for the nomenclature of receptors for relaxin family peptides. Pharmacol. Rev. 58, 7–31 (2006).
Smith, C. M. et al. Distribution of relaxin-3 and RXFP3 within arousal, stress, affective, and cognitive circuits of mouse brain. J. Comp. Neurol. 518, 4016–4045 (2010).
Smith, C. M. et al. Central injection of relaxin-3 receptor (RXFP3) antagonist peptides reduces motivated food seeking and consumption in C57BL/6J mice. Behav. Brain Res. 268, 117–126 (2014).
Calvez, J. et al. Role of relaxin-3/RXFP3 system in stress-induced binge-like eating in female rats. Neuropharmacology 102, 207–215 (2016).
Banerjee, A. et al. Swim stress excitation of nucleus incertus and rapid induction of relaxin-3 expression via CRF1 activation. Neuropharmacology 58, 145–155 (2010).
Zhang, C. et al. Central relaxin-3 receptor (RXFP3) activation reduces elevated, but not basal, anxiety-like behaviour in C57BL/6J mice. Behav. Brain Res. 292, 125–132 (2015).
Ryan, P. J. et al. Central relaxin-3 receptor (RXFP3) activation decreases anxiety- and depressive-like behaviours in the rat. Behav. Brain Res. 244, 142–151 (2013).
Hosken, I. T. et al. Relaxin-3 receptor (Rxfp3) gene knockout mice display reduced running wheel activity: implications for role of relaxin-3/RXFP3 signalling in sustained arousal. Behav. Brain Res. 278, 167–175 (2015).
Smith, C. M. et al. Viral-mediated delivery of an RXFP3 agonist into brain promotes arousal in mice. Ital. J. Anat. Embryol. 118, 42–46 (2013).
Ma et al. Modulation of hippocampal theta oscillations and spatial memory by relaxin-3 neurons of the nucleus incertus. Learn Mem. 16, 730–742 (2009).
Albert-Gasco, H. et al. Central relaxin-3 receptor (RXFP3) activation increases ERK phosphorylation in septal cholinergic neurons and impairs spatial working memory. Brain Struct. Funct. 222, 449–463 (2017).
Ryan, P. J. et al. Relaxin-3/RXFP3 system regulates alcohol-seeking. Proc. Natl. Acad. Sci. USA 110, 20789–20794 (2013).
Ryan, P. J. et al. Relaxin-3 mRNA levels in nucleus incertus correlate with alcohol and sucrose intake in rats. Drug Alcohol Depend. 140, 8–16 (2014).
Shirahase, T. et al. Increased alcohol consumption in relaxin-3 deficient male mice. Neurosci. Lett. 612, 155–160 (2016).
Guan, D. et al. Indole-containing amidinohydrazones as nonpeptide, dual RXFP3/4 agonists: synthesis, structure-activity relationship, and molecular modeling studies. J. Med. Chem. 64, 17866–17886 (2021).
Lin, G. et al. High-throughput screening campaign identified a potential small molecule RXFP3/4 agonist. Molecules 26, 7511 (2021).
DeChristopher, B. et al. Discovery of a small molecule RXFP3/4 agonist that increases food intake in rats upon acute central administration. Bioorg. Med. Chem. Lett. 29, 991–994 (2019).
Chen, Y. et al. Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4). Nat. Commun. 14, 492 (2023).
Patil, N. A. et al. Relaxin family peptides: structure-activity relationship studies. Br. J. Pharmacol. 174, 950–961 (2017).
Cherezov, V. et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007).
Rosengren, K. J. et al. Solution structure and novel insights into the determinants of the receptor specificity of human relaxin-3. J. Biol. Chem. 281, 5845–5851 (2006).
Ballesteros, J. uanA. & Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995).
Wang, X. Y. et al. The electrostatic interactions of relaxin-3 with receptor RXFP4 and the influence of its B-chain C-terminal conformation. FEBS J. 281, 2927–2936 (2014).
Feng, W. et al. Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors. Cell Discov. 9, 81 (2023).
Wang, Y. et al. Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3. Proc. Natl. Acad. Sci. USA 121, e2400298121 (2024).
Hu, M. J. et al. Mechanism for insulin-like peptide 5 distinguishing the homologous relaxin family peptide receptor 3 and 4. Sci. Rep. 6, 29648 (2016).
Belgi, A. et al. Minimum active structure of insulin-like peptide 5. J. Med. Chem. 56, 9509–9516 (2013).
Liu, C. et al. Relaxin-3/insulin-like peptide 5 chimeric peptide, a selective ligand for G protein-coupled receptor (GPCR)135 and GPCR142 over leucine-rich repeat-containing G protein-coupled receptor 7. Mol. Pharm. 67, 231–240 (2005).
Koehl, A. et al. Structure of the micro-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A. et al. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D. Biol. Crystallogr. 65, 1074–1080 (2009).
Terwilliger, T. C. et al. Automated ligand fitting by core-fragment fitting and extension into density. Acta Crystallogr. D. Biol. Crystallogr. 62, 915–922 (2006).
Adams, P. D. et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Sobolev, O. V. et al. A global ramachandran score identifies protein structures with unlikely stereochemistry. Structure 28, 1249–1258.e1242 (2020).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We are grateful to Yun Wang, Qi Zhu and Chun Shen for technical assistance. This work was partially supported by grants from the National Natural Science Foundation of China 82073904 (M.-W.W.), 82121005 (D.Y.), and 21704064 (Q.T.Z.); STI2030-Major Project 2021ZD0203400 (Q.T.Z.); and Hainan Provincial Major Science and Technology Project ZDKJ2021028 (D.Y. and Q.T.Z.). The cryo-EM data were collected at the Cryo-Electron Microscopy Research Center and the Advanced Center for Electron Microscopy, Shanghai Institute of Materia Medica, Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
Y.C. designed expression constructs, purified the receptor complexes, screened the specimens, prepared the final samples for cryo-EM data collection, conducted map calculation, built the models of the complexes, performed signaling experiments, and participated in manuscript preparation; Q.T.Z. assisted model building, performed structural analysis, and participated in manuscript writing; S.Y.Y. and J.H.Y. took part in molecular cloning, plasmid construction and functional experiments; J.C. attempted ligand labeling experiment; D.H.Y. assisted in data collection and project management; M.-W.W. initiated the project, supervised the studies, analyzed the data, and wrote the manuscript with inputs from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Tiffany Myint and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Laura Rodríguez Pérez.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, Y., Zhou, Q., Yan, S. et al. Molecular mechanism underlying non-discriminatory recognition of relaxin-3 by RXFP3 and RXFP4. Commun Biol 8, 794 (2025). https://doi.org/10.1038/s42003-025-08220-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08220-7